Abstract
Genomic-level analyses of DNA from non-invasive sources would facilitate powerful conservation and evolutionary studies in natural populations of endangered and otherwise elusive species. However, the typical low quantity and poor quality of DNA that is extracted from non-invasive samples have generally precluded such work. Here we apply a modified DNA capture protocol that, when used in combination with massively-parallel sequencing technology, facilitates efficient and highly-accurate resequencing of megabases of specified nuclear genomic regions from fecal DNA samples. We validated our approach by comparing genetic variants identified from corresponding fecal and blood DNA samples of six western chimpanzees (Pan troglodytes verus) across more than 1.5 megabases of chromosome 21, chromosome X, and the complete mitochondrial genome. Our results suggest that it is now feasible to conduct genomic studies in natural populations for which constraints on invasive sampling have otherwise long been a barrier. The data we collected also provided an opportunity to examine western chimpanzee genetic diversity at unprecedented scale. Despite high mitochondrial genome diversity (π = 0.585%), western chimpanzees have a low ratio (0.42) of X chromosomal (π = 0.034%) to autosomal (chromosome 21 π = 0.081%) sequence diversity, a pattern that may reflect an unusual demographic history of this subspecies.
Keywords: molecular ecology, population genetics, non-invasive sampling, conservation genomics
INTRODUCTION
Genetic research related to the conservation, evolution, and behavior of non-human, non-model organisms, especially research on natural populations of endangered mammals, has yet to benefit extensively from the recent availability of massively-parallel sequencing technology. One common impediment to large-scale genetic studies of endangered species is a lack of high-quality DNA. Often, it is undesirable or impossible to trap or dart animals to collect invasive blood or tissue samples that would yield high-quality DNA, as trapping or darting risks harming the animal and disrupting behavioral data collection. The international transport of invasive samples is also regulated by the Convention on International Trade in Endangered Species (CITES), sometimes adding administrative complexity to genetic research with these organisms.
In principle, DNA for genetic analyses can also be isolated from non-invasive samples such as feces and shed hair. Such samples can be collected readily without harm (sometimes even without direct observation of the animal) and are thus ideal in many respects for genetic studies of natural populations. However, DNA isolated from non-invasive samples is often fragmented and low in quantity, and therefore extremely challenging to use in genetic analyses (Taberlet et al. 1999). Additionally, representation of the nuclear genome may be incomplete in DNA isolated from shed hair, which can restrict analyses of such samples to the mitochondrial genome (Jeffery et al. 2007). In turn, while fecal samples can be excellent sources of nuclear genome DNA (originating from intestinal wall epithelial cells), the fecal extract may contain chemicals that inhibit PCR (Kohn & Wayne 1997; Nechvatal et al. 2008). PCR is necessary for nearly all types of traditional genetic analyses. Moreover, endogenous (from the study animal) DNA extracted from fecal samples is thought to be overwhelmed by DNA from exogenous sources, especially gut bacteria, since bacteria account for the majority of fecal dry weight, at least in humans (Stephen & Cummings 1980).
Due to these limitations, genetic analyses of DNA from non-invasive samples using traditional techniques have largely been restricted to mitochondrial DNA sequencing and the genotyping of small numbers of microsatellite loci (even these efforts often had to address allelic dropout-related challenges; Arandjelovic et al. 2009; Buchan et al. 2005; McKelvey & Schwartz 2004). While such work has provided important insights into taxonomy, population structure, and the relationship between relatedness and behavior in natural populations in a number of species (e.g., DeSalle & Amato 2004; Kohn & Wayne 1997; Piggott & Taylor 2003; Vigilant & Guschanski 2009), a new approach is required for genomic-level analyses of non-invasive DNA, which will facilitate large-scale genetic studies in natural populations.
Recently-developed massively-parallel sequencing technologies require only small input quantities of short DNA fragments, thereby obviating two traditional limitations of fecal DNA genetic analyses. Theoretically, one could perform massively-parallel shotgun sequencing of fecal DNA to obtain endogenous sequence data. However, because the vast majority of the DNA sequenced would be from exogenous sources, in practice, the fecal DNA shotgun sequencing approach is limited to microbiome diversity analyses (Qin et al. 2010) rather than population genetic analyses of the endogenous species. Thus, the primary remaining challenge to performing genomic-scale studies with DNA from fecal samples is to enrich the endogenous DNA against the overwhelming preponderance of exogenous DNA. The ability to carry out such studies would represent a powerful new tool for conservation and evolutionary ecology studies (Kohn et al. 2006; Ouborg et al. 2010).
Here we describe a method for capturing targeted genomic regions from fecal DNA samples that facilitates efficient resequencing of megabases of the nuclear genome with massively-parallel sequencing technology. To demonstrate the accuracy of this approach, we generated and compared fecal and blood DNA resequencing data for more than 1.5 Mb of the genome for each of six chimpanzees (Pan troglodytes), and also compared these data to ~35 kb of resequencing data collected from blood DNA using traditional PCR and sequencing methods.
MATERIALS AND METHODS
Samples
Fecal and blood samples from adult chimpanzee individuals were collected during routine veterinary examinations at the New Iberia Research Center, Lafayette, LA. These individuals are captive born but primarily or exclusively of western chimpanzee ancestry (based on pedigree analysis and comparisons of mitochondrial DNA and Y chromosome sequences to those from individuals of known capture locations; Stone et al. 2002). The chimpanzee diet was manufactured by PMI Nutrition International, LLC (“New Iberia Primate Diet”; minimum crude protein = 20%, minimum crude fat = 5%, and maximum crude fiber 10%). For fecal samples, 2g of stool was collected within one hour of defecation in a 15 mL tube with 10 mL RNALater (Ambion) and shaken vigorously. Once in the laboratory, the samples were stored at −80°C prior to DNA extraction.
The QIAamp DNA Stool Mini Kit (Qiagen) was used for DNA isolation. Compared to other approaches, this protocol provides superior DNA yields with relatively limited chemical inhibition of PCR (Nechvatal et al. 2008). For each extraction, 1.4 mL of the RNALater-feces mixture was centrifuged for 1 min at 1000g. Following removal of the supernatant, the remaining fecal sample was extracted according to the manufacturer instructions for maximizing the final proportion of non-bacterial DNA. In turn, whole blood was collected in EDTA Vacutainers (BD Biosciences) and stored at −80°C prior to DNA extraction with the Gentra Puregene Blood Kit (Qiagen).
We used quantitative PCR to estimate the proportion of endogenous DNA (starting concentrations estimated with a Nanodrop ND-1000 spectrophotometer) in the fecal DNA extracts, using primers unique in the chimpanzee nuclear genome (forward 5′-3′ CAATCAAGACGTCCAGCTCA and reverse 5′-3′ TAGAACTGCTGCCCCACTTT), and evaluated against a standard curve constructed from the blood DNA of one individual (Flint, 93A009). The samples were run in 25 μL reactions using iQ SYBR Green Supermix (Bio-Rad) on a Bio-Rad iCycler Thermal Cycler with an initial denaturation of 95°C for 7 min, followed by 40 cycles of 95°C for 30 sec and 60°C for 45 sec. Test samples were run in triplicate and standards run in duplicate. As expected, the proportion of endogenous DNA extracted from the six fecal samples was low: average 0.018, range 0.005 to 0.052 (Table S1).
Targeted DNA regions
We used the Galaxy browser (Giardine et al. 2005) to download the panTro2 assembly of the chimpanzee reference genome sequence (The chimpanzee sequencing and analysis consortium 2005) and the associated RepeatMasker (Jurka 2000) and SimpleRepeats (Benson 1999) tracks from the UCSC Genome Bioinformatics Site (Rhead et al. 2010). Using the R statistical environment (R Development Core Team 2010), chromosome 21 and X sequences were masked for (i) repeats, (ii) chimpanzee whole genome assembly comparison (WGAC) and whole genome shotgun sequence detection (WSSD) segmental duplications (Cheng et al. 2005), (iii) human genome (hg18; converted to panTro2 with LiftOver in Galaxy) segmental duplications (Bailey et al. 2002), (iv) chimpanzee copy number variants (Perry et al. 2008), (v) the first 10 Mb of the X chromosome to avoid the pseudoautosomal region, (vi) gaps, and (vii) “N”s.
Agilent SureSelect (Gnirke et al. 2009) “baits” of length 120 bp each were designed from remaining contiguous sequences of ≥ 2 kb for chromosomes 21 and X and for the complete mitochondrial genome at 4x tiling coverage (i.e., starting positions of baits every 30 bp), with every other probe designed as the reverse complement of the reference sequence. 55,000 baits corresponding to the first 394 filtered regions of chromosome 21 (1,052,310 bp), the first 209 filtered regions of chromosome X (550,471 bp), and the complete mitochondrial genome (16,554 bp) were selected for the SureSelect Capture Library (ELID 0254881).
DNA capture
For blood DNA samples, DNA captures were performed following manufacturer instructions (Agilent SureSelect Target Enrichment System, Illumina Single-End Sequencing Platform Library Prep Protocol, Version 1.2 April 2009). For fecal DNA samples, the endogenous DNA represents only a small proportion of the total DNA (1.8%, on average, in the chimpanzee samples in this study). Therefore, a series of protocol adjustments were required: 1. To avoid “allelic dropout” issues, by ensuring sufficient representation (i.e., copy number) of the targeted endogenous regions in the input DNA. 2. To obtain sufficient sequence read coverage for accurate SNP identification, by achieving an exceptional level of enrichment for the targeted regions. These protocol adjustments are detailed in the succeeding paragraphs.
To ensure sufficient initial representation of targeted endogenous regions without performing multiple ligation reactions, we performed a size selection prior to adapter ligation. From each sample, DNA was extracted multiple times; usually 4 but up to 8 extractions were performed, as necessary to obtain ≥ 15 μg of total DNA (~150 ng endogenous chimpanzee DNA). The DNA was sheared to median fragment size ~200 bp (total fragment size range ~100–500 bp) using the Covaris Model S2 system (following operating conditions in the Agilent SureSelect Protocol), in 3 μg total DNA/100 μL Buffer AE (Qiagen) aliquots. The aliquots were combined, concentrated to 25 μL, and electrophoresed on a 2% low-melt agarose gel (Bio-Rad). Three excisions were performed per sample, corresponding roughly to 125–175 bp, 175–225 bp, and 225–275 bp, and purified using the Qiaquick Gel Extraction Kit (Qiagen) followed by the Qiaquick PCR Purification Kit (Qiagen) to remove any impurities remaining following gel extraction, eluted with 30 μl Buffer EB. Following visualization using the Agilent 2100 Bioanalyzer (DNA 1000 kit), 1.5 μg of the purified DNA excision closest to 200 bp in size was prepared for adapter ligation as recommended in the SureSelect Protocol. Excess fragmented, size-selected product can be stored at −80°C for future experiments. Adapter ligation was performed as recommended, except with 3 μL rather than 6 μL Illumina adapter oligo mix. A second round of gel purification was then used to remove unligated adapters. Here, all visible product was excised from the gel and purified as above.
Much larger quantities of adapter-ligated fecal DNA, compared to typical reactions with pure endogenous DNA, are needed for SureSelect biotinylated RNA bait hybridization. For each sample we performed 16 PCR amplifications of 50 μL volume using primers specific to the universal adapter sequences (Illumina Primers 1.1 and 2.1) with 1 μL of the gel-purified eluate in each reaction, using the Agilent recommended PCR conditions (14 total cycles). Unused eluate can be stored at −80 °C for future experiments. Products were purified with the Qiaquick PCR Purification Kit (4 columns with 4 PCR amplified products in each), visualized with the Bioanalyzer, and quantified with a Nanodrop ND-1000 spectrophotometer. 20 μg of purified product (5 μg per column) was concentrated to 15 μL.
To accommodate the increased quantity of DNA (20 μg rather than 389 ng), we used a larger than suggested final volume (71 μL rather than 27 μL) for the SureSelect hybridization. Hybridization buffer was prepared as recommended (49 μL total volume). The 15 μL DNA sample (20 μg) was mixed with 5 μL SureSelect Block #1, 5 μL SureSelect Block #2, and 1.2 μL SureSelect Block #3 and heat-denatured as recommended in the SureSelect protocol. The SureSelect Oligo Capture Library was prepared as recommended except with 1 μL of undiluted RNase Block. The full volumes of the hybridization buffer and DNA sample/blockers were then mixed with the Capture Library/RNAse block, and hybridized for 26 hours at 65°C.
Based on a pilot experiment, we found that with one round of capture and following the Agilent-recommended washing protocol, there was insufficient enrichment of targeted regions for confident SNP identification (chromosome 21 enrichment = 646, effective enrichment = 5.8; chromosome X enrichment = 1844, effective enrichment = 16.6; compare to values in Table S1; enrichment calculations described below in “Sequencing and analysis”). Therefore, we increased the washing stringency and performed a second round of DNA capture before sequencing. To do so, after the first round of hybridization (using the process described above), binding to the streptavidin-coated magnetic beads was performed as recommended. We then washed the beads twice with SureSelect Wash Buffer #1 for 7 min each at room temperature, followed by 6 washes with SureSelect Wash Buffer #2 for 10 min each at 65°C. Captured DNA was then eluted from the beads and desalted per instructions. For each sample, we then performed 4 PCR amplifications of 25 μL volume each using 0.5 μL of the SureSelect GA Primer Mix and 1 μL of the eluted library. PCR conditions were as suggested in the Agilent protocol except using 13, rather than 18, cycles. Following PCR, the 4 reactions were combined and purified using a MinElute spin column, eluted with 10 μL Buffer EB, and visualized and quantified with the Bioanalyzer.
For the second round of capture, 10 ng of the post-PCR eluted first round library (much less input DNA is needed for the second round of capture because there has already been one round of enrichment for the targeted regions, and by minimizing this quantity we limit the number of PCR cycles necessary in the above step) was hybridized to another SureSelect Oligo Capture Library aliquot using volumes recommended in the original protocol, at 65°C for 22 hours. Following binding to beads, the extended wash protocol and PCR protocol (4 reactions, 13 cycles) were again performed as described above. These 4 PCR amplifications were combined and purified using the Qiaquick PCR Purification kit, eluted with 30 μL Buffer EB, and visualized and quantified on the Agilent Bioanalyzer prior to sequencing.
Sequencing and enrichment estimation
Each prepared library was sequenced for 76 cycles on one flowcell lane using an Illumina Genome Analyzer II, (GAII) at a concentration of 9 pM and with the Single Read Cluster Generation Kit V4, Sequencing Kit V4, and software SCS 2.6. Sequence data are available at the NCBI Short Read Archive, accession number SRA012374.
Sequence reads were aligned to a subset of the chimpanzee genome (panTro2), comprising chromosomes 21, X and the full mitochondrial genome, using the Burrows-Wheeler Alignment tool (BWA) with default alignment parameters (Li & Durbin 2009). The alignment data were processed using the SAMtools (Sequence Alignment/Map) software package (Li et al. 2009). To assess the efficiency of the DNA capture, we first calculated the enrichment of targeted regions in each sample, by comparing the number of sequencing reads per base mapped to targeted regions with the number of reads per base mapped to regions of the same chromosomes that were not targeted – but that met the same filtering criteria for target selection (i.e., regions that satisfied all the criteria of the captured sequences, but were not targeted only because we were limited to 55,000 SureSelect baits). For the fecal samples, to account for the fact that endogenous DNA is overwhelmed by DNA from exogenous sources, we also calculated the effective enrichment as the estimated proportion of endogenous DNA (previously estimated by quantitative PCR) times the enrichment of targeted regions in the sequence data. The efficiency values for each sample are reported in Table S1.
Sequence data analysis
Mutation rates and genetic diversity in the mitochondrial genome are usually substantially greater than those in the nuclear genome. Western chimpanzee mitochondrial diversity in the hypervariable region exceeded the limits of the default BWA parameters for number of allowed alignment mismatches, at least when reads from the six individuals in our study were aligned to the chimpanzee reference mitochondrial genome. This resulted in the exclusion of good reads and the loss of data. Rather than ad hoc adjustment of these parameters, which could be problematic in future studies where mitochondrial genome diversity is unknown, we performed de novo assembly of the mitochondrial genome using the sequence read data from each sample, using the ABySS (Assembly By Short Sequences) software package (Simpson et al. 2009). Contigs from the assembly were filtered for coverage and aligned against the mtDNA reference sequence. The final mtDNA sequence was assembled from contigs with high read coverage and identified as mitochondrial in origin based on the alignment analysis. These assembled mitochondrial genome consensus sequences were used in all subsequent analyses. We note that nuclear copies of mitochondrial DNA sequences (Numts; Bensasson et al. 2001; Lopez et al. 1994) may also be captured by the biotinylated RNA baits and sequenced using our approach. However, because the mitochondrial copy number is many times greater than the nuclear copy number (and the rates of capture and sequencing are proportional), true mitochondrial genome reads will grossly outnumber those of Numts and the consensus sequence will therefore be unaffected.
For chromosomes 21 and X, we removed reads that mapped to multiple locations or that mapped poorly by excluding all reads with mapping quality score < 10. Before calling genotypes, we also simulated all 76bp reads that could arise from our targeted regions, and aligned them to the entire chimpanzee genome using BWA. To limit the possibility of erroneous genotype calls from cross-hybridization to non-targeted regions with small-scale sequence homology, we removed all targeted regions where at least one simulated read could be mapped (with up to eight mismatches) to an alternative genomic location (19 of the 394 chromosome 21 regions removed; 21 of the 209 chromosome X regions removed).
To consider only unique original fragments in the SNP identification analyses (i.e., to avoid any post-ligation and post-capture amplification biases), we used a Perl script to select one read per strand for each start position at random (for a theoretical maximum filtered coverage 152 bp [76 bp sequence reads, two strands]). The unselected reads were excluded from further analysis. Per-targeted site summary data for all samples are provided in Dataset S1, available at http://giladlab.uchicago.edu/data/datasetS1/. For all sites in the targeted regions, we used R to count the number of times each of the four nucleotides was present in the mapped reads. This analysis was performed separately for reads mapping to the plus and minus strands. Since low coverage makes it difficult to call genotypes confidently, we filtered out all sites with less than 10 mapped reads (with any of the four nucleotides at that position) on each strand. Of the remaining sites for each sample, heterozygous sites were identified as those for which the proportions of the most common nucleotide from the reads on both strands were≤ 0.8; otherwise, sites were considered homozygous for the most common nucleotide. We found that these criteria (especially the requirement for evidence of heterozygosity among the reads mapped to both strands) resulted in high quality SNP calls (Fig. S1). Genotype calls at all SNP sites for each sample are provided in Dataset S2 (also available at http://giladlab.uchicago.edu/data/datasetS1/). When calculating allele frequency distributions and genotype distance matrices, only sites where genotypes could be called across all individuals in the analysis were considered. Ancestral and derived alleles were identified based on comparison to the human and rhesus macaque (Macaca mulatta) reference genome sequences.
All programming scripts used for data processing and analysis are available at http://giladlab.uchicago.edu/data/fecalcode/.
Validation
We randomly selected 20 targeted regions on chromosome 21 for PCR and Sanger sequencing analysis. Amplification primers (contained within the targeted regions) were designed with Primer3 (Rozen & Skaletsky 2000) to amplify ~2 kb fragments. Amplified products were purified with Shrimp Alkaline Phosphatase and Exonuclease I (USB Corp.), and then cycle sequenced with internal primers and analyzed on an Applied Biosystems 3730XL capillary sequencer at the University of Chicago Cancer Research Center DNA Sequencing Facility. Primer sequences and PCR conditions are presented in Table S2.
RESULTS
To facilitate genomic-scale nucleotide sequencing of DNA isolated from feces, we optimized a sequence capture approach based on Agilent’s SureSelect technology (Gnirke et al. 2009). To develop and test our approach, we collected fecal and blood samples from each of six unrelated individuals of the western chimpanzee subspecies (P. troglodytes verus) and extracted DNA from each sample separately. As expected, the proportion of endogenous DNA extracted from the six fecal samples was low (average 0.018; range 0.005 to 0.052; estimated by quantitative PCR; Table S1). To capture, enrich, and ultimately obtain endogenous genomic sequence from the fecal DNA, we designed SureSelect probes that targeted ~1 Mb of sequence from chimpanzee chromosome 21 (comprised of 394 distinct genomic regions), ~550 kb of chromosome X (209 regions), and the complete mitochondrial genome.
Fecal DNA capture and sequencing
To ensure sufficient representation of the targeted genomic regions, in our optimized capture protocol for fecal DNA we hybridized a larger-than-typical quantity of total input DNA to the SureSelect probes (20 μg in our protocol in contrast to only ~400 ng required for the standard SureSelect protocol). We also used a more stringent washing protocol following hybridization, and performed two successive rounds of capture. Fecal DNA libraries were prepared for all six chimpanzees, and each library was sequenced for 76 cycles on one lane of an Illumina Genome Analyzer II (GAII) flow cell. Corresponding blood DNA libraries were prepared using the standard SureSelect single-round capture protocol and sequenced similarly.
For all samples, the majority of the sequenced reads were aligned successfully to the targeted genomic regions (minimum 82%; Table S1). Enrichment levels, calculated as the per-base number of reads mapped to targeted regions on chromosomes 21 and X divided by the per-base number of reads mapped to non-targeted genomic regions meeting the same filtering criteria, ranged from 347,908 to 2,238,909 for the fecal DNA samples. Even when we corrected for the low ratios of endogenous to exogenous DNA in the fecal samples, the effective enrichments were still excellent (range 2,375 to 35,409), and on average 3.1 times greater than the enrichment levels for the blood DNA samples (range 2,753 to 6,048; Table S1), probably reflecting the two rounds of fecal DNA capture.
To avoid potential biases arising from post-ligation and post-capture amplification steps in the SureSelect protocol, when multiple aligned reads had the same starting position and originated from the same strand we sub-sampled one read at random. The resulting mean filtered sequence coverage per targeted base ranged among the samples from 54–114 on chromosome 21 and from 42–115 on chromosome X (Table S1; note that following our sub-sampling approach, the theoretical maximum coverage per base is 152, see Materials and Methods). Filtered sequence coverage was minimal or zero for only a small percentage of targeted base positions in each sample (range 2.8 to 8.4%), making impossible the accurate identification of genetic variants at those positions. However, per-base filtered coverage was strongly correlated across samples (Fig. S2), meaning that the minority of sites without genotype data tended to be the same across samples rather than randomly distributed. Variation in DNA to biotinylated RNA bait hybridization efficiency is likely responsible for this phenomenon.
Identification, accuracy and validation of single nucleotide polymorphisms (SNPs)
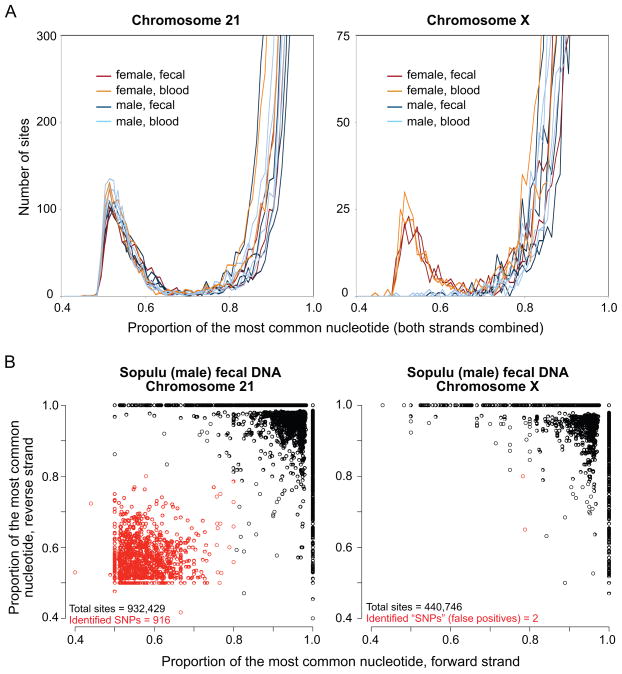
To assess the quality of the data and facilitate effective identification of genetic variants, we first considered frequency distributions of the proportion of the most common nucleotide at each targeted site. For chromosome 21, these distributions were similar across all samples with distinct peaks for intermediate-proportion nucleotides, whereas such peaks in the chromosome X distributions were observed only in females (Fig. 1A), reflecting the copy number difference between sexes.
Fig. 1. Data quality and SNP identification.
(A) Frequency distributions of the proportion of the most common nucleotide at each targeted site that has filtered read coverage sufficient for SNP identification (≥ 20 total reads with at least 10 from each strand; proportion bins = 0.01) for each sample, separately by chromosome. For the overwhelming majority of sites, the most common nucleotide proportion equals 1 (the Y axis is cut off). There is a dearth of sites with intermediate-proportion nucleotides on the X chromosome in male samples. (B) Plots of the most common nucleotide proportion by mapped strand, for each site with filtered read coverage ≥ 10 on each strand for one selected sample (Sopulu, fecal DNA), separately by chromosome. Heterozygous sites were identified as those with most common nucleotide proportion ≤ 0.8 on both strands (red circles). For the overwhelming majority of sites, the most common nucleotide proportions equal 1 on both strands: 868,450 of 934,229 sites (93%) on chromosome 21, and 413,224 of 440,746 sites (94%) on chromosome X.
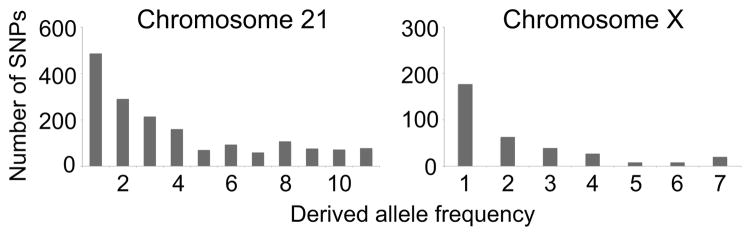
We proceeded by studying the effect of different sequence coverage cutoffs on the identification of SNPs (Fig. S1), and chose to classify as heterozygous those sites for which the proportion of the most common nucleotide was ≤ 0.8 on both strands, conditional on a minimum coverage of 10 reads per strand (Materials and Methods; Fig. 1B). By examining the percentage of spuriously-identified “heterozygous” X chromosome sites in males, we estimated the average false-positive rates to be 0.0007% for fecal DNA and 0.0010% for blood DNA. Overall, we identified an average of 838 (±84) heterozygous sites per sample on chromosome 21 and an average of 168 (±14) heterozygous sites on chromosome X in females (Table 1). Thus, the estimated proportions of incorrectly-identified heterozygous sites in our study are 0.8%, 2.0%, 1.1%, and 2.7% for fecal DNA chromosome 21, fecal DNA chromosome X in females, blood DNA chromosome 21, and blood DNA chromosome X in females, respectively (note that this estimate varies depending on the underlying genetic diversity of each chromosome, in contrast to the overall false-positive rate, which is expected to be relatively independent of the genetic background). The level of genetic diversity suggested by the average number of heterozygous sites is consistent with results from previous studies of western chimpanzee nucleotide diversity, as are the derived SNP allele frequency distributions (Fig. 2; (Gilad et al. 2003; Kaessmann et al. 1999; Verrelli et al. 2006; Yu et al. 2003).
Table 1.
Sample-level heterozygosity.
| Individual | Source | Chromosome 21 | Chromosome X | ||||
|---|---|---|---|---|---|---|---|
| Sites (bp)1 | Hets.2 | π, %3 | Sites (bp)1 | Hets.2 | π, %3 | ||
| 93A009 Flint (male) | Blood | 953,738 | 837 | 0.088 | 446,949 | 5 | 0.001 |
| Fecal | 926,914 | 779 | 0.084 | 437,025 | 5 | 0.001 | |
| A2A009 Sopulu (male) | Blood | 970,322 | 968 | 0.100 | 461,782 | 6 | 0.001 |
| Fecal | 932,429 | 916 | 0.098 | 440,746 | 2 | 0.001 | |
| X161 Judd (male) | Blood | 969,471 | 800 | 0.083 | 460,384 | 2 | 0.000 |
| Fecal | 951,672 | 778 | 0.082 | 460,706 | 4 | 0.001 | |
| 91A016 Coty (male) | Blood | 969,566 | 848 | 0.087 | 462,098 | 5 | 0.001 |
| Fecal | 941,109 | 791 | 0.084 | 457,146 | 1 | 0.000 | |
| 91A010 Peanut (female) | Blood | 964,745 | 764 | 0.079 | 467,563 | 178 | 0.038 |
| Fecal | 904,775 | 712 | 0.079 | 462,743 | 173 | 0.037 | |
| A1A005 Kierra (female) | Blood | 967,457 | 979 | 0.101 | 468,931 | 172 | 0.037 |
| Fecal | 904,089 | 886 | 0.098 | 447,698 | 147 | 0.033 | |
Number of sites with filtered read coverage sufficient for SNP identification.
Number of heterozygous sites in each sample identified using the criteria described in Materials and Methods (the X chromosome heterozygous sites in males are false positives).
Pairwise nucleotide diversity, the percentage of heterozygous sites (π for the X chromosome in males is an approximation of the false-positive error rate).
Fig. 2. Derived allele frequency distributions.
For all segregating sites with filtered read coverage sufficient for SNP identification in the results from the fecal DNA samples of all individuals, we determined how many chromosomes (n = 12 total for chromosome 21; n = 8 total for chromosome X) carried the derived allele (derived allele frequency). Plots depict the number of SNPs in each derived allele frequency bin. Ancestral/derived allele states were estimated by comparisons to the human and rhesus macaque reference genome sequences.
We observed slight but consistent differences between the fecal and blood DNA results (Table 1). First, there were generally more chromosome 21 and X sites with filtered sequence coverage sufficient for SNP identification in the blood DNA samples. In part, we can attribute this difference to the considerable proportions of sequence reads that align to the mitochondrial genome, especially in the fecal DNA results (Table S1). This is a property of the design of the capturing assay and can be altered easily – for example, by reducing the density of RNA baits targeting the mitochondrial genome (i.e., rather than 4x tiling coverage as for all targeted regions in our study, 0.5x coverage for the mitochondrial genome would likely be sufficient given its relatively large input copy number). Second, we observed slightly lower heterozygosity in the fecal DNA samples (Table 1). This result does not seem to reflect significant differences in the false-positive or false-negative rates, because the effect is all but eliminated when we consider only those sites with sequence coverage sufficient for SNP identification in both the feces and blood samples for each individual (Table S3). Instead, genetic diversity is likely marginally greater at sites with insufficient sequence coverage in the fecal samples, probably because some regions that include polymorphisms may hybridize less well to the capturing probes than regions whose sequence is identical to the designed probes; this effect may be amplified by the two rounds of capture.
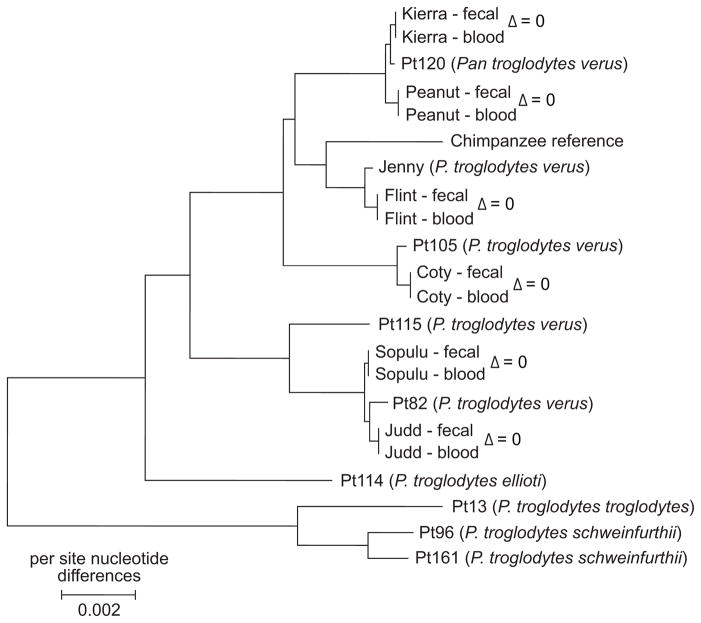
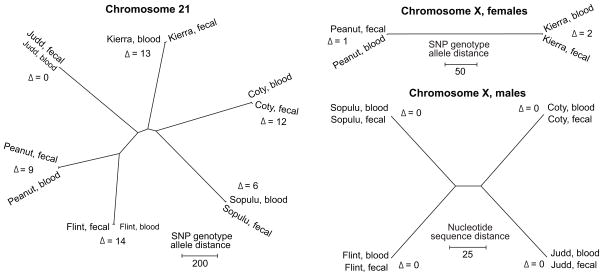
Overall, however, the fecal and blood DNA results from the same individual are strongly concordant. The matched fecal and blood DNA consensus mitochondrial genomes have zero nucleotide sequence differences across all 16,554 sites for each individual. Phylogenetic analysis of these sequences indicates that all chimpanzees in our study have western chimpanzee matrilineal ancestry, as expected (Fig. 3). Focusing on the nuclear genome sequences, we compared genotype allele distances across pairs of fecal and blood DNA samples, and found very few genotype discrepancies between sample pairs (Fig. 4). For example, at 850,702 sites (1,701,404 genotype allele calls) on chromosome 21 with coverage sufficient for SNP identification across all samples, there were an average of 9 (0.0005%) spurious genotype allele differences between fecal and blood DNA samples from the same individual, compared to an average of 1,095 differences between individuals. This is a conservative estimate of the false-negative rate associated with SNP identification in the fecal DNA samples, as it assumes that there are no falsely-identified SNPs in the blood DNA samples. Moreover, we are certain that the excellent agreement between fecal and blood DNA sequences does not reflect contamination because for five of the six individuals (except for Flint, used in protocol optimization), fecal DNA was isolated, captured, and sequenced before the corresponding blood DNA was extracted.
Fig. 3. Neighbor-joining phylogeny of chimpanzee complete mitochondrial genome sequences.
Blood and fecal DNA results from this study (underlined) are compared with previously-published complete mitochondrial genome sequences from chimpanzees of known subspecies (Stone et al. in press) and that of the chimpanzee reference sequence (panTro2). Based on the estimated phylogeny, the chimpanzees in this study have western chimpanzee (P. t. verus) matrilineal ancestry, as expected. Nucleotide sequence distances (Δ) are given for each same-individual pair of samples. There are no nucleotide sequence differences between the mitochondrial genomes of the matched fecal DNA and blood DNA samples of each chimpanzee. The neighbor-joining phylogeny (of evolutionary distances computed using the Maximum Composite Likelihood method; positions containing gaps were eliminated) was estimated using the MEGA4 software (Tamura et al. 2007).
Fig. 4. Neighbor-joining trees constructed from distance matrices.
Based on the chromosome 21 SNP genotype distance matrix (genotypes coded as 0, 1, and 2), female chromosome X genotype distance matrix (genotypes coded as 0, 1, and 2), and male chromosome X consensus nucleotide distance matrix (nucleotides coded as 0 and 1; false-positive “heterozygous” sites on male X chromosomes per Table 1 were excluded) for sites with coverage sufficient for SNP identification in all samples. There is high correspondence between fecal and blood DNA samples from the same individuals relative to samples from other individuals. Genotype or nucleotide sequence distances (Δ) are given for each same-individual pair of samples.
Finally, we considered whether systematic capture and sequencing biases could result in false-negative SNP identification errors shared across the fecal and blood DNA data. To examine this possibility, we used traditional PCR and Sanger sequencing methods to analyze 35,481 bp of 20 randomly selected targeted regions from the blood DNA of one individual (Flint). Initially, we observed several apparent false-positives in the GAII-based genotype calls in one targeted region. However, the GAII data also indicated a SNP in the same region as one PCR primer, suggesting possible allele-specific amplification. After designing a new primer for this region and resequencing (Fig. S3), for this and the other examined regions we observed excellent consistency between the Sanger sequencing results and both the fecal and blood DNA GAII genotype calls (Fig. 5; 50 total SNPs identified from the Sanger sequencing data, with 1 false-negative SNP in each of the blood and fecal DNA results from the corresponding GAII sequencing data and 0 false-positive SNPs).
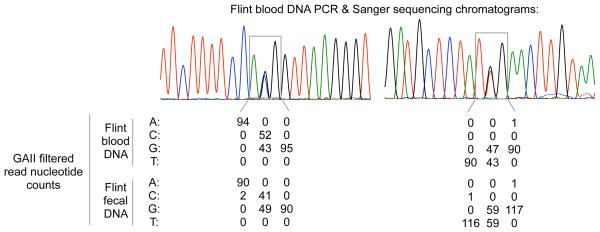
Fig. 5. PCR and Sanger sequencing assessment of SNP identification accuracy.
Two selected sequence chromatograms from PCR and Sanger sequencing validation of SNPs using blood DNA from Flint, with a summary of DNA capture/GAII sequence data for the variable and adjacent sites from the chromatograms. In total, 50 SNPs were identified from 35,481 bp of chromosome 21 Sanger sequencing data. In the corresponding GAII sequencing data, there were 0 false positive SNPs, and 1 false-negative SNP in each of the blood and fecal DNA results (for different SNPs).
DISCUSSION
We developed and tested a method for sequencing megabases of DNA from fecal samples that combines an optimized DNA capture approach with massively-parallel sequencing. We sequenced more than 1.5 Mb of chromosomes 21 and X, and the complete mitochondrial genome of six chimpanzees using one Illumina GAII flow cell lane per sample. As four of these individuals were male, our results demonstrate that single-copy nuclear loci (i.e., chromosome X in males) can be sequenced readily. Therefore, the Y chromosome could also be targeted and sequenced in future applications of this method.
The ability to generate genomic-scale nucleotide sequence data from non-invasive samples is expected to facilitate powerful new studies related to the conservation, behavior, demography, and evolutionary ecology of endangered species For example, while the current study was designed for the purpose of methodological development, not to address a specific biological question, the collected data do also provide an opportunity to examine western chimpanzee genetic diversity at unprecedented scale. Western chimpanzees and humans are thought to have similar levels of autosomal genetic diversity (Fischer et al. 2004; The chimpanzee sequencing and analysis consortium 2005; Yu et al. 2003), a notion supported by our chromosome 21 data (Table 2). In contrast, estimated X chromosomal genetic diversity is relatively lower in western chimpanzees, despite much higher mitochondrial diversity. Such a pattern may reflect differences in the demographic histories of humans and western chimpanzees (Bustamante & Ramachandran 2009; Ellegren 2009). If so, then to help generate explanatory hypotheses it might be valuable to consider mating behavior and dispersal pattern differences between chimpanzee subspecies, which might themselves have markedly different demographic histories. A comparison of our observations in western chimpanzees to the limited data available for central chimpanzees (Pan troglodytes troglodytes) suggests that central chimpanzees have considerably higher levels of autosomal and X chromosomal genetic diversities yet a lower level of mitochondrial diversity. Moreover, there may be much less of a disparity between autosomal and X chromosome genetic diversity in central chimpanzees (Fischer et al. 2004; Kaessmann et al. 1999; Stone et al. 2002; Verrelli et al. 2008; Verrelli et al. 2006).
Table 2.
Comparison of western chimpanzee and human nucleotide diversity.
| Species – Population1 | Autosomes | Chromosome X | Mitochondrial genome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n2 | Sites (bp) | S3 | π, %4 | n2 | Sites (bp) | S3 | π, %4 | n2 | Sites (bp) | S3 | π, %4 | |
| Chimpanzee – P. t. verus (western) | 12 | 861,142 | 2,062 | 0.081 | 8 | 420,610 | 401 | 0.034 | 6 | 15,564 | 191 | 0.585 |
| Human – Biaka (Africa) | 28 | 112,399 | 574 | 0.121 | 14 | 97,728 | 280 | 0.095 | 10 | 15,573 | 104 | 0.208 |
| Human – San (Africa) | 19.5 | 112,399 | 501 | 0.126 | 9 | 97,728 | 220 | 0.085 | 6 | 15,582 | 102 | 0.298 |
| Human – Basque (Europe) | 32 | 112,399 | 338 | 0.087 | 16 | 97,728 | 200 | 0.071 | 5 | 15,585 | 15 | 0.040 |
| Human – Han (Asia) | 32 | 112,399 | 354 | 0.081 | 16 | 97,728 | 174 | 0.058 | 4 | 15,583 | 61 | 0.196 |
Chimpanzee data are from this study; only the fecal DNA data were considered. Only sites with read coverage sufficient for SNP identification across all samples were analyzed. Human nuclear data are from Wall et al. (2008). Human mitochondrial data (for corresponding populations with n ≥ 4) are from Kivisild et al. (2006), for which population samples were chosen without prior knowledge of mitochondrial haplotype. The chimpanzee complete mitochondrial genome sequences were trimmed to the orthologous region for which sequences were available for the human samples.
Number of chromosomes.
Number of segregating sites.
Average pairwise nucleotide diversity, percent.
Our approach requires a priori availability of genome sequence, from which DNA capture probes are designed. The rapidly increasing capacities and reduced cost of newer sequencing technologies (Bentley et al. 2008; Drmanac et al. 2010; Eid et al. 2009; Harris et al. 2008; Margulies et al. 2005; Shendure et al. 2005) suggest that very soon it will be feasible for an individual research group to sequence the genome of their study organism from just one high-quality DNA sample. Yet for many species, even this step may not be necessary as there are existing plans by at least one consortium to sequence rapidly the genomes of 10,000 vertebrate species (Genome 10k Community of Scientists 2009).
Eventually, it is likely that sequencing capacity will reach the point at which a research group could sequence and assemble the entire genome of a host animal from a total fecal DNA extract without a capture step. However, since the vast majority of sequence data would be from exogenous sources, such a study design would likely remain computationally and economically inefficient. For example, the current capacity of the Illumina GAII is ~30 million sequence reads per lane. For each sample in our study, we collected one lane of sequence data using 76 bp, single-end reads. With shotgun sequencing, if 1% of the fecal DNA is endogenous and the size of the genome is 3 billion bp, then it would be necessary to use >2,600 lanes (with 76 bp reads) to achieve 20-fold coverage of the host genome (the minimum coverage level we required for calling SNPs). Even with dramatic improvement in sequencing capacity, the volume of data produced and the computational requirements for analysis would be enormous for one sample, and larger still for a population study. Therefore, at least for the foreseeable future, to take advantage of increases in sequencing capacity it seems a better solution to combine a DNA capture approach – such as the one described in this paper – with multiplex sequencing (Craig et al. 2008; Cronn et al. 2008).
Looking forward to continued methodological and technical improvements, we note that several of the challenges associated with the genomic analysis of fecal DNA are similar in scope (if not necessarily in magnitude) to those associated with the genomic analysis of ancient DNA, especially the fragmented nature of the DNA, the limited quantity of endogenous DNA, and the preponderance of DNA from exogenous sources such as soil microbes (Green et al. 2010; Paabo et al. 2004; Prufer et al. 2010). Indeed, Burbano et al. (2010) recently applied a DNA capture approach to sequence a set of exons and the complete mitochondrial genome from the DNA of one Neandertal individual. They also used two successive rounds of capture in their protocol, and ultimately achieved a mean sequence coverage of ~4.8 per targeted nuclear base (Burbano et al. 2010). Future developments in either area of research – fecal DNA or ancient DNA – may benefit the other.
In this study, we circumvented the traditional limitations of non-invasive DNA analysis by developing and demonstrating the feasibility of a genomic-scale fecal DNA sequencing method. We hope that applications of this method contribute to the conservation and scientific understanding of endangered species.
Supplementary Material
Fig. S1. Heterozygous site single nucleotide polymorphism identification criteria.
Fig. S2. Sequence coverage correspondence.
Fig. S3. Validation of identified single nucleotide polymorphisms by PCR and Sanger sequencing.
Table S1. Sequencing statistics summary.
Table S2. PCR amplification and sequencing primers.
Table S3. Sample-level heterozygosity, for sites with coverage sufficient for SNP identification from both fecal and blood DNA results for each individual.
Acknowledgments
We thank Babette Fontenot, Stephanie Ruiz and the New Iberia Research Center for collecting and providing the fecal and blood chimpanzee samples. The University of Louisiana at Lafayette New Iberia Research Center is funded by NIH NCRR grants RR015087, RR014491, and RR016483. We thank Katelyn Michelini for efficient operation of the GAII, John Zekos for handling the sequence data, Athma Pai for assistance with ancestral allele determination, Ran Blekhman and Jack Degner for assistance with scripts for read sub-sampling and read mapping simulation, Matthew Stephens for suggesting the use of strand information when identifying heterozygous sites, and Susan Alberts, Luis Barreiro, Fred Ernani, Emily LeProust, Edward Louis, Athma Pai, and Jenny Tung for helpful discussions and comments. This study was funded by NIGMS grant GM077959 to Y.G. G.H.P. is supported by NIH fellowship F32GM085998.
Footnotes
Data deposition: All sequence data have been deposited at the National Center for Biotechnology Information short read archive (www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) as study no. SRA012374.
References
- Arandjelovic M, Guschanski K, Schubert G, et al. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol Ecol Resour. 2009;9:28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang D, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JC, Archie EA, van Horn RC, Moss CJ, Alberts SC. Locus effects and sources of error in noninvasive genotyping. Mol Ecol Notes. 2005;5:680–683. [Google Scholar]
- Burbano HA, Hodges E, Green RE, et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–725. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Ramachandran S. Evaluating signatures of sex-specific processes in the human genome. Nat Genet. 2009;41:8–10. doi: 10.1038/ng0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R, Liston A, Parks M, et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle R, Amato G. The expansion of conservation genetics. Nat Rev Genet. 2004;5:702–712. doi: 10.1038/nrg1425. [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Ellegren H. The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 2009;25:278–284. doi: 10.1016/j.tig.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Fischer A, Wiebe V, Paabo S, Przeworski M. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- Genome 10k Community of Scientists. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Bustamante CD, Lancet D, Paabo S. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 2003;73:489–501. doi: 10.1086/378132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TD, Buzby PR, Babcock H, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- Jeffery KJ, Abernethy KA, Tutin CE, Bruford MW. Biological and environmental degradation of gorilla hair and microsatellite amplificaiton success. Biol J Linn Soc. 2007;91:281–294. [Google Scholar]
- Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Paabo S. Extensive nuclear DNA sequence diversity among chimpanzees. Science. 1999;286:1159–1162. doi: 10.1126/science.286.5442.1159. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Shen P, Wall DP, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. Genomics and conservation genetics. Trends Ecol Evol. 2006;21:629–637. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Kohn MH, Wayne RK. Facts from feces revisited. Trends Ecol Evol. 1997;12:223–227. doi: 10.1016/s0169-5347(97)01050-1. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKelvey KS, Schwartz MK. Genetic errors associated with population estimation using non-invasive molecular tagging: Problems and new solutions. J Wildl Manage. 2004;68:439–448. [Google Scholar]
- Nechvatal JM, Ram JL, Basson MD, et al. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Meth. 2008;72:124–132. doi: 10.1016/j.mimet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma RK, Hedrick PW. Conservation genetics in transition to conservation genomics. Trends Genet. 2010;26:177–187. doi: 10.1016/j.tig.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Paabo S, Poinar H, Serre D, et al. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- Perry GH, Yang F, Marques-Bonet T, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggott MP, Taylor AC. Remote collection of animal DNA and its applications in conservation management and understanding the population biology of rare and cryptic species. Wildlife Res. 2003;30:1–13. [Google Scholar]
- Prufer K, Stenzel U, Hofreiter M, et al. Computational challenges in the analysis of ancient DNA. Genome Biol. 2010;11:R47. doi: 10.1186/gb-2010-11-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2010. [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AM, Cummings JH. Mechanism of action of dietary fibre in the human colon. Nature. 1980;284:283–284. doi: 10.1038/284283a0. [DOI] [PubMed] [Google Scholar]
- Stone AC, Battistuzzi F, Kubatko LS, et al. More reliable estimates of divergence times in Pan using complete mtDNA sequences and accounting for population structure. Proc Roy Soc B. 00:1–12. doi: 10.1098/rstb.2010.0096. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone AC, Griffiths RC, Zegura SL, Hammer MF. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc Natl Acad Sci U S A. 2002;99:43–48. doi: 10.1073/pnas.012364999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Waits LP, Luikart G. Noninvasive genetic sampling: look before you leap. Trends Ecol Evol. 1999;14:323–327. doi: 10.1016/s0169-5347(99)01637-7. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- The chimpanzee sequencing and analysis consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Verrelli BC, Lewis CM, Jr, Stone AC, Perry GH. Different selective pressures shape the molecular evolution of color vision in chimpanzee and human populations. Mol Biol Evol. 2008;25:2735–2743. doi: 10.1093/molbev/msn220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, Tishkoff SA, Stone AC, Touchman JW. Contrasting histories of G6PD molecular evolution and malarial resistance in humans and chimpanzees. Mol Biol Evol. 2006;23:1592–1601. doi: 10.1093/molbev/msl024. [DOI] [PubMed] [Google Scholar]
- Vigilant L, Guschanski K. Using genetics to understand the dynamics of wild primate populations. Primates. 2009;50:105–120. doi: 10.1007/s10329-008-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Cox MP, Mendez FL, et al. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, et al. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Heterozygous site single nucleotide polymorphism identification criteria.
Fig. S2. Sequence coverage correspondence.
Fig. S3. Validation of identified single nucleotide polymorphisms by PCR and Sanger sequencing.
Table S1. Sequencing statistics summary.
Table S2. PCR amplification and sequencing primers.
Table S3. Sample-level heterozygosity, for sites with coverage sufficient for SNP identification from both fecal and blood DNA results for each individual.