Abstract
The presence of ubiquitinated protein inclusions is a hallmark of most adult onset neurodegenerative disorders. Although the toxicity of these structures remains controversial, their prolonged presence in neurons is indicative of some failure in fundamental cellular processes. It therefore may be possible that driving the elimination of inclusions can help re-establish normal cellular function. There is growing evidence that macroautophagy has two roles; first, as a non-selective degradative response to cellular stress such as starvation, and the other as a highly selective quality control mechanism whose basal levels are important to maintain cellular health. One particular form of macroautophagy, aggrephagy, may have particular relevance in neurodegeneration, as it is responsible for the selective elimination of accumulated and aggregated ubiquitinated proteins. In this review, we will discuss the molecular mechanisms and role of protein aggregation in neurodegeneration, as well as the molecular mechanism of aggrephagy and how it may impact disease.
Keywords: autophagy, protein aggregates, neurodegeneration, ubiquitination, p62, ALFY, aggresome, neurons
1. Introduction
Intracellular protein aggregation is a hallmark of a wide array of neurological disorders, including tauopathies, synucleinopathies, TDP-43 proteinopathies and polyglutamine disorders. Although the relationship between protein aggregation with disease pathogenesis is unclear (Caughey and Lansbury, 2003; Jellinger, 2009; Ross and Poirier, 2005; Spires-Jones et al., 2009; Williams and Paulson, 2008), several studies using mouse genetics, lentiviral technologies and RNA interference have shown that elimination of the accumulation-prone proteins permits symptomatic reversal in different neurodegenerative models (Harper, 1996; Lin et al., 2009; Regulier et al., 2003; Xia et al., 2004; Yamamoto et al., 2000; Zu et al., 2004). Often correlating with the regression of symptoms is the disappearance of aggregated protein, indicating that neurons can eliminate these proteins, despite their heterogeneous structure and cellular distribution. Moreover, while these studies do not establish a causative link between protein aggregation and neurodegeneration, they clearly suggest that elimination of accumulated proteins may alleviate underlying cellular dysfunction, and potentiate recovery across different disorders.
So, how are aggregation-prone proteins disposed of by neurons? Unlike many cells used to study protein aggregation, neurons are post-mitotic and cannot dilute their cytosol by cell division or asymmetrically distribute inclusions (Rujano et al., 2006). Instead, neurons must rely upon two distinct cellular mechanisms of protein degradation: the ubiquitin-proteasome system (UPS) and autophagy-mediated lysosomal degradation. The UPS is a highly conserved system by which proteins are targeted to the proteasome upon covalent modification by ubiquitin (Glickman and Ciechanover, 2002). Studies examining the role of the UPS in neurodegeneration are extensive, and are covered elsewhere in this issue. In this review, we will discuss the emerging importance of lysosome-mediated degradation of aggregated cytosolic proteins by macroautophagy. Predominantly known as a nonselective degradative response to starvation, macroautophagy also is required for the selective elimination of organelles, infective agents, and more recently, protein aggregates. In this review, we will examine how aggrephagy, the selective elimination of aggregates by macroautophagy, can play a role in eliminating protein aggregates that have been implicated in neurodegenerative disease.
2. Protein aggregation in neurodegeneration
The ability of proteins to aggregate or come together as a larger complex is a fundamental process through which proteins exert their normal function. In the context of neurodegeneration, protein aggregates generally refer to oligomeric complexes of misfolded or unfolded proteins that can be structured or amorphous, which are insoluble and metabolically stable under physiologic conditions (Kopito, 2000). Together with the pattern of neuronal dysfunction and degeneration, the presence of these structures, termed histologically as intracellular inclusions, bodies, tangles or threads, is often part of the diagnostic repertoire of a pathogenic process (Table 1). Their abnormal presence and prevalence across numerous disorders have led to models implicating their role in pathogenesis, and have begun to influence how disorders are classified. For example, the discovery of TDP-43 as the major component of the ubiquitinated inclusions in amyotrophic lateral sclerosis (ALS) (Arai et al., 2006; Tan et al., 2007; Zhang et al., 2008) and forms of frontotemporal lobar degeneration (FTLD-TDP) (Liscic et al., 2008; Mackenzie et al., 2010) has strengthened the link between these diseases.
Table 1.
Neurological protein conformation disorders.
| Disease name |
Affected gene | Intracellular Ubiquitinated Lesion |
Major component |
Cell Type | Distribution of Inclusions |
Citations |
|---|---|---|---|---|---|---|
| HD | HD/IT-15 | DN, NII, GCI, DNS | polyQ Htt | Neurons, Glia | Ctx, hc, str, bs | (DiFiglia et al., 1997; Gutekunst et al., 1999; Maat-Schieman et al., 1999) |
| DRPLA | ATN1 | NCI, NII, DNS, GII | polyQ Atn1 | Neurons, Glia | str,bs, Ctx, dn, gp, lgn, sn, hc, cb |
(Hayashi et al., 1998a; Hayashi et al., 1998b; Yazawa, 2000) |
| SBMA | AR | NII, granules | polyQ AR | Motoneurons | bs,hyp, sn | (Adachi et al., 2005; Li et al., 1998a; Li et al., 1998b) |
| SCA1 | SCA1 | NCI, NII, DNS | polyQ Atxn1 | Neurons | Ctx, str, sn, bs, sc | (Durr et al., 1996; Duyckaerts et al., 1999; Servadio et al., 1995) |
| SCA2 | SCA2 | NCI, GCI, NII | polyQ Atxn2 | Neurons, Glia | Ctx, str, dn, gp, thal, bs, Pkj |
(Huynh et al., 1999; Huynh et al., 2000; Koyano et al., 1999; Pang et al., 2002) |
| MJD/SCA3 | SCA3 | NCI, NII, DNS | polyQ Atxn3 | Neurons | sc, dn, gp, thal, Ctx | (Durr et al., 1996; Paulson et al., 1997; Rub et al., 2006; Schmidt et al., 1998) |
| SCA6 | CACNA1A | NCI, NII | polyQ CACNA1A | Neurons | Ctx, hc, str, bd, cb, sc | (Ikeuchi et al., 1997; Ishikawa et al., 2001; Zhuchenko et al., 1997) |
| SCA7 | SCA7 | NCI, NII | polyQ Atxn7 | Neurons | lgn, bs, sn, hc, str, cb, thal, ctx |
(David et al., 1997; Einum et al., 2001; Garden et al., 2002a; Holmberg et al., 1998) |
| SCA17 | TBP | DNS, NII | polyQ TBP | Neurons | Thal, cb, bs | (Friedman et al., 2007; Nakamura et al., 2001; Rolfs et al., 2003) |
| PD DLB |
See (Moore et al., 2005) |
LB, LN** | α-synuclein | Neurons | sn>ctx, amyg amyg, ctx, hc, sn |
(Burke et al., 2008; Duda et al., 2000; Lippa et al., 2007; Markopoulou et al., 2008) |
| MSA | GCI, NCI, GII, NFT | α-synuclein, tubulin |
Glia, Neurons | Widespread; bg, cb, bs |
(Papp and Lantos, 1994; Probst-Cousin et al., 1996; Wakabayashi and Takahashi, 2006) |
|
| AD | See (Spires-Jones et al., 2009) | NFT, Threads, DN | Tau | Neurons | Ctx, hc, str, thal | (Onorato et al., 1989; Shukla and Bridges, 1999; Taraszewska et al., 1996) |
| PSP | Threads, NFTs | Tau | Neurons | str, gp, thal, cb, bs, ctx |
(Cervos-Navarro and Schumacher, 1994; Verny et al., 1996) |
|
| ALS familial | See (Wood et al., 2003) |
Hyaline inclusions | SOD1 | Neurons, Glia, Motoneurons |
sc, ctx, bs | (Mackenzie et al., 2007; Shibata et al., 1996; Wijesekera and Leigh, 2009) |
| ALS sporadic | n/a | Skeins, LB-like Bunina bodies |
TDP-43 Cystatin-C, Tf |
Neurons, Glia, Motoneurons |
bs, ctx | (Arai et al., 2006; Leigh et al., 1991; Mackenzie et al., 2007; Okamoto et al., 2008; Tan et al., 2007; Wood et al., 2003) |
| CMT2E | NEFL | NF whorls, NF islets*** | NF | Peripheral nerves |
(Fabrizi et al., 2004; Fabrizi et al., 2007; Jordanova et al., 2003) |
|
| FTLD | MAPT* | NFT, DN | Tau | Neurons, Glia | Ctx, hc, amyg, str, gp, thal, bs, cb |
(Cairns et al., 2007) |
| FTLD –U (CHMP2B) |
CHMP2B | NCI, GCI, DN, NII | Unknown (p62-& Ub-pos) |
Neurons, Glia | Ctx, hc, str, sc, bs | (Liscic et al., 2008; Schumacher et al., 2009; van der Zee et al., 2007) |
| FTLD –U types 1–3 type3 PGRN type4 |
Unknown PGRN VCP |
NCI, GCI, DN, NII | TDP-43 (p62-& Ub-pos) |
Neurons, Glia | Ctx, hc, str, sc, bs | (Davidson et al., 2007; Forman et al., 2006a; Forman et al., 2006b; Liscic et al., 2008; van der Zee et al., 2007; Watts et al., 2004) |
| FTLD with Pick bodies |
Unknown | Pick bodies | Tau, NF | Neurons, Glia | Ctx, hc, str, sc, bs | (Dickson, 2001; Uchihara et al., 2003) |
| Alexander’s disease |
GFAP | Rosenthal fibers | α-crystallin, GFAP |
Astrocytes | Widespread, ctx, ic, ec |
(Borrett and Becker, 1985; Brenner et al., 2001; Iwaki et al., 1989) |
Notes:
Other yet identified mutations may still exist (Cairns et al., 2007);
Mutations in LRRK2 may not present with LB pathology (Marti-Masso et al., 2009; Santpere and Ferrer, 2009; Zimprich et al., 2004);
not examined for ubiquitin in human samples. Ubiquitinated inclusions are observed in studies using cultured neurons(Lin and Schlaepfer, 2006).
Abbreviations: AD, Alzheimer’s disease; ALS, Amyotrophic lateral sclerosis; amyg, amygdala; ATN1, atrophin 1; ATXN, ataxin; AR, androgen receptor; bs, brain stem; CACNA1A, calcium channel, voltage-dependent, P/Q type, alpha 1A subunit ; cb, cerebellum; CMT2E, Charcot-Marie Tooth type 2E; CHMP2B, chromatin modifying protein 2B; Ctx, cortex; DLB, Dementia with Lewy Bodies; DN, Dystrophic neurites; DNS, Diffuse neuronal staining; DRPLA, dentatorubropallidoluysian atrophy; ec, external capsule; FTLD-U, frontotemporal lobar degeneration with ubiquitinated inclusions; GCI, Glial intracytoplasmic inclusions; GFAP, Glial fibrillary acid protein, GII, Glial intranuclear inclusions; gp, globus pallidus; hc, hippocampus; HD, Huntington’s disease; Htt, Huntingtin; ic, internal capsule; LB, Lewy body; lgn, lateral geniculate nucleus; LN, Lewy neurites; MAPT, microtubule-associated protein tau; MJD, Machado-Joseph’s disease; MSA, Multiple systems atrophy; NCI, Neuronal cytoplasmic inclusions; NEFL, neurofilament light chain; NF, neurofilament; NFT, Neurofibrillary tangles; NII, Neuronal intranuclear inclusions; PD, Parkinson’s disease; Pkj, Purkinje cells; PGRN, progranulin; PSP, Progressive supranuclear palsy; sc, spinal cord; SCA, spinocerebellear ataxia; sn, substantia nigra; str, striatum; TBP, TATA binding protein; TDP-43, TAR DNA binding protein-43Tf, Transferrin; thal, thalamus; Ub, ubiquitin; VCP, valosin-containing protein.
Most of our fundamental understanding of protein aggregation has been derived from animal models, cell lines and in vitro studies. From these studies, protein aggregation has emerged as a complex multistep process reflected in the variability of protein aggregate structure, size and intracellular localization (Woulfe, 2008). Since all of these variables can be observed within a single pathologic sample (DiFiglia et al., 1997; Galvin et al., 2001; Gray et al., 1987; Jellinger, 2009; Katsuse et al., 2003; Liscic et al., 2008), it is critical to keep in mind which structure we are examining when studying pathogenesis.
2.1 The formation of protein inclusions in neurons
The maturation of misfolded or unfolded protein into protein aggregates can vary across different disorders, but generally protein aggregation results from proteins that fold into an abnormal conformation, leading to the formation of oligomeric intermediates (Merlini et al., 2001). These intermediates can aggregate and further mature into small protein aggregates. These small protein aggregates can form into a wide variety of structures (Dobson, 2003). Due to their structural stability, amyloid fibrils are the most commonly studied structure, although they may not be the most prevelant (Dobson, 2003). These smaller aggregates, both structured and unstructured, continue to grow and multimerize into larger aggregates or inclusions (Grune et al., 2004; Kopito, 2000). Different kinds of aggregates can be found in the neuropil, soma and nuclei, depending upon the protein and disorder examined (DiFiglia et al., 1997; Geschwind, 2003; Gray et al., 1987; Gutekunst et al., 1995; Jellinger, 2009; van der Zee et al., 2007; Wakabayashi and Takahashi, 2006).
Larger cytoplasmic inclusions can evolve further and coalesce into an aggresome, a pericentriolar, membrane-free cytoplasmic inclusion formed specifically at the microtubule organizing center (MTOC) containing misfolded, ubiquitinated proteins caged within intermediate filaments such as vimentin or keratin (Johnston et al., 1998; Kopito, 2000). It has been proposed that the aggresome is a protective structure, formed to sequester proteins that cannot be degraded by the proteasome and packaged for degradation by autophagy (Johnston et al., 1998; Kopito, 2000). Although studies in heterologous systems clearly demonstrate that disease-causing proteins become packaged in this manner (Iwata et al., 2005; Johnston and Madura, 2004; Waelter et al., 2001; Wong et al., 2008), this may not be the case for neurons. For instance, neuronal cytoplasmic inclusions (NCIs) are ubiquitinated but rarely vimentin-positive, possibly because vimentin is expressed predominantly in immature neurons and become replaced by neurofilaments (NFs) as neurons mature (Bennett et al., 1981; Cochard and Paulin, 1984). Nonetheless, in Multiple Systems Atrophy, glial cytoplasmic inclusions are also vimentin-negative (Wakabayashi and Takahashi, 2006) (Table 1). Further, abnormal accumulation of NFs is found across several neurodegenerative diseases and mutations within the NF light chains have been implicated in their accumulation in Charot Marie Tooth Type 2 (CMT2) neuropathies; however, NFs are the major constituent of the inclusion and do not form a ‘cage’ as described for an aggresome (Perrot and Eyer, 2009; Roy et al., 2005). Moreover, although the readily dividing, stable cell lines used to study protein aggregation possess MTOCs, studies indicate that after neurogenesis (Doxsey et al., 2005; Wang et al., 2009), neuronal centrosomes no longer function as MTOCs, and no longer extend microtubules or recruit γ-tubulin (Stiess et al., 2010). Despite the lack of MTOCs, however, microtubule nucleation is still required for proper neuronal function and maintenance (Ahmad et al., 1994; Yu et al., 1994), but rather than at a central perinuclear point, microtubules nucleate across multiple sites throughout the cell (Stiess et al., 2010). Larger inclusions therefore still may form in a microtubule-dependent manner (McNaught et al., 2002), but may not fulfill the strict definition of an aggresome. Moreover, the lack of a single MTOC also may explain why aggregates are not limited to the soma.
One indication that neurons create aggresome-like structures is found in studies with histone deacetylase 6 (HDAC6). HDAC6 is a ubiquitin-binding microtubule deacetylase, that is required to recruit ubiquitinated, misfolded proteins to the aggresome (Iwata et al., 2005; Kawaguchi et al., 2003). HDAC6 may not be a structural element, however, since depletion of HDAC6 did not disrupt preformed inclusions (Iwata et al., 2005). Unfortunately, reports on HDAC6 immunoreactivity of NCIs in disease have been limited- although HDAC6-positive Lewy Bodies from neocortical and brain stem samples from Parkinson’s disease (PD) and Dementia with Lewy Bodies (DLB) brains, respectively, were reported (Kawaguchi et al., 2003), and HDAC6 was shown to be slightly elevated in Alzheimer’s Disease (AD) cortex and hippocampus possibly through an interaction with tau (Ding et al., 2008; Perez et al., 2009), its presence in other disorders has not been explored. Overall, despite structural differences, the function of inclusion body formation in neurons may still share a functional commonality with aggresomes. In a study by Lucas and colleagues (Ortega et al., 2010), they found that global impairment of the UPS in an HD mouse model diminished upon the appearance of neuronal inclusions. Interestingly, the inhibition of inclusion formation inhibited the amelioration of the UPS.
2.2 The toxicity of protein aggregates
Although protein aggregation is common across many disorders, the role it may or may not play in toxicity across different diseases is a complex discussion that cannot be fully addressed in this review. In general, however, there are three alternative hypotheses as to the possible role of protein aggregation: Protein inclusions are directly or indirectly toxic; protein inclusions are protective, and abrogate the toxicity due to a toxic monomeric or oligomeric species through their sequestration; and protein inclusions are benign by-standers that may correlate with the disease but have little influence on the disease process. In light of the duration of adult onset neurodegenerative disorders and the complexity that each disease presents, it is likely that a combination of the three hypotheses may account for the overall course of each unique disease.
Despite the features of protein aggregation that may be unique to each disease, protein aggregation also shares several common traits. The major component of the inclusions is often ubiquitously expressed, such as huntingtin (Htt), α-synuclein, tau and TDP-43 (Aronin et al., 1995; Dauer et al., 2002; Georgieff et al., 1993; Jakes et al., 1994; Ou et al., 1995; Perrot and Eyer, 2009; Trottier et al., 1995). Moreover the inclusions are found throughout the brain, often in neurons and glia, with little direct correlation to the pattern of neurodegeneration that occurs. For example, in the polyQ expansion disorder Huntington’s disease (HD), both the neuronal intranuclear inclusions (NII) and dystrophic neurites are found throughout the cortex, while the degeneration is predominantly in the striatum (DiFiglia et al., 1997; Gutekunst et al., 1995; Vonsattel and DiFiglia, 1998). Indeed, the medium spiny neurons which are most vulnerable in HD, demonstrate little to no NIIs (Gutekunst et al., 1995). In an inducible model of hereditary tauopathy, neuronal loss occurred in areas where neurofibrillary tangles were absent, not where they were present (Santacruz et al., 2005). These and other similar observations lend support to two hypotheses: Protein inclusions are not toxic; or, rather than a cell autonomous toxicity, aggregates may promote toxicity to neurons within a circuit or through glia (Di Giorgio et al., 2007; Garden et al., 2002; Gu et al., 2005; Nagai et al., 2007; Perrot and Eyer, 2009), possibly promoting ‘murder rather than suicide’ (Ross, 2004; Wexler et al., 1991).
Although the toxicity of aggregation is under debate, aggregation of proteins not implicated in any diseases become cytotoxic (Bucciantini et al., 2002; Ordway et al., 1997), indicating that some abnormal conformation is disadvantageous. There is growing evidence that the toxic conformer may be the smaller aggregate species (Caughey and Lansbury, 2003). Studies with one subset of aggregated species, amyloid oligomers, find that rather than the actual protein that has oligomerized, the structure of the aggregates may dictate their toxicity (Kayed et al., 2003). Differing hypotheses have been proposed to account for the toxicity. Both the relatively disorganized prefibrillary aggregate, and the more ordered amyloid structures, can lead to altered or aberrant interactions with other proteins resulting in sequestration and dysregulation or loss of function (Chalmers and Love, 2007; Ihara et al., 2007; Iqbal et al., 2008; Lippens et al., 2007; Suhr et al., 2001). An alternative hypothesis is the ‘amyloid pore’ hypothesis which states that ordered aggregates form pores in the cellular membranes thereby causing ionic imbalance in cells (Lashuel et al., 2002; Volles and Lansbury, 2002). Lastly, protein aggregates have been implicated in interfering with general protein homeostasis by directly or indirectly inhibiting protein degradation or evoking ER stress.
In contrast, the larger, visible inclusions that are an integral part of distinguishing different neurodegenerative diseases may be a structure that sequesters toxic species from causing greater harm (Arrasate et al., 2004; Ortega et al., 2010) or inert ‘bystanders’(Tran and Miller, 1999). As originally proposed for the aggresome (Kopito, 2000), these large inclusions may be a ‘first step’ towards targeting the accumulating proteins for degradation (see below). If this is the case, attempts to prevent aggregation or promote disaggregation without driving elimination of the aggregating protein may demonstrate little to no benefit for neurons, since not only is the amount of accumulated protein unchanged, but the inhibition of inclusion formation may impede further the degradative capacity that remains.
2.3 The accumulation of ubiquitinated proteins
The most common feature observed in proteinopathies is that the accumulating proteins are invariably positive for ubiquitin (Alves-Rodrigues et al., 1998; Liscic et al., 2008; Tan et al., 2007). Indeed for some disorders, such as sALS, prior to identification of the aggregating protein, ubiquitin immunoreactivity was the only means by which abnormal protein deposits could be identified (Liscic et al., 2008).
In healthy cells, protein misfolding readily occurs; one estimate revealed that over 30% of the newly synthesized proteins are misfolded and aggregated (Schubert et al., 2000). Those proteins that are not refolded become ubiquitinated and targeted to the proteasome (Barral et al., 2004; Bukau et al., 2006; Schubert et al., 2000). Despite the large volume of protein turnover required, proteins do not accumulate since ubiquitinated proteins are rapidly targeted to the proteasome and eliminated, with the rate-limiting event being the conjugation of ubiquitin itself (Chau et al., 1989; Glotzer et al., 1991; Hershko, 1991; Hershko and Ciechanover, 1998).
Why under neurodegenerative conditions ubiquitinated substrates accumulate remains unclear. A direct or indirect inhibition of proteasome function has been proposed but remains controversial (Bedford et al., 2008b; Bence et al., 2001; Bowman et al., 2005; Ding et al., 2002; Grune et al., 2004; Kabashi et al., 2008; Kabashi et al., 2004; Maynard et al., 2009; Ortega et al., 2010). Although conditional gene knockout studies have shown that inhibition of the UPS in mice leads to accumulation of ubiquitinated proteins, inclusions and neurodegeneration (Bedford et al., 2008), elimination of core components of macroautophagy, in flies (Lindmo et al., 2006; Nezis et al., 2008; Simonsen et al., 2008) and in mice lead to similar results (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010), indicating that disruption of protein degradation is generally detrimental to neurons. Using a form of mass spectroscopy analyses (Kirkpatrick et al., 2005; Kirkpatrick et al., 2006), a recent study by Kopito and colleagues found that different ubiquitin chains of K48, K63 and K11 accumulated in patient samples and mouse models of HD (Bennett et al., 2007). Although these findings do not explain why these chains accumulate, this global change in ubiquitin indicates that neurons are unable to process proteins properly. Taken together, although the role of protein aggregation in neurodegeneration continues to be a debate, these data are suggestive that driving elimination of the accumulated and aggregated ubiquitinated structures will help to re-establish homeostasis to the cell. There is growing evidence that a cellular pathway that can degrade such complex aggregated structures is a selective form of macroautophagy, known as aggrephagy.
3. Mammalian macroautophagy
Autophagy is a general term for the degradation of cytosolic components by the lysosome, and can be separated into three types: microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy. In microautophagy, cytosolic material is brought into the lysosome through direct invagination of the lysosomal membrane and subsequent budding of vesicles into the lysosomal lumen (Xie and Klionsky, 2007). Microautophagy has been studied most extensively in yeast (Uttenweiler and Mayer, 2008) and there is little mechanistic understanding of microautophagy in mammalian systems (Xie and Klionsky, 2007). CMA involves a direct import through the lysosomal membrane of substrates containing a KFERQ motif, which are recognized by a group of molecular chaperones (Cuervo et al., 1997). CMA has not been described in yeast, and was first described in hepatocytes by Dice in the late 1980’s (Chiang et al., 1989). Recently a growing role of CMA and most notably its dysfunction in neurodegeneration has emerged, and is also further reviewed in this issue.
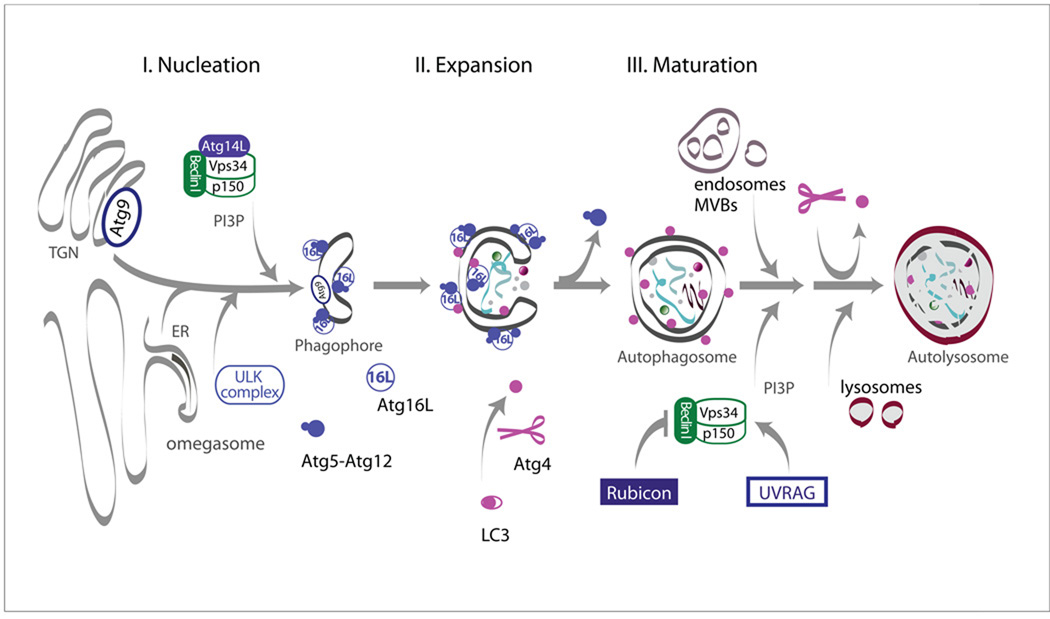
Macroautophagy is the process whereby cytoplasmic constituents become sequestered into a de novo synthesized double membrane structure called the isolation membrane or phagophore, which closes to become an autophagosome (Seglen et al., 1990). To achieve degradation, the autophagosome then fuses with the lysosome directly, or first forms an amphisome by fusing with vesicles of the endocytic pathway (Figure 1). The core players involved in macroautophagy, first identified in yeast, is conserved throughout evolution and named the Autophagy-related (Atg) proteins (Klionsky et al., 2003). Despite the numerous advances made over the last decade in understanding the molecular mechanisms of autophagy, key questions remain unanswered (Hamasaki and Yoshimori, 2010; Reggiori, 2006). Macroautophagy, however, generally can be divided into three main steps: 1) Nucleation; 2) Expansion; and 3) Maturation (Figure 1). The molecular machineries required for these steps have been extensively reviewed elsewhere (He and Klionsky, 2009; He and Levine, 2010; Longatti and Tooze, 2009; Simonsen and Tooze, 2009) and only the main players involved will be described here.
Figure 1. Schematic summary of macroautophagy.
Although the origin of the autophagosomal membrane is still a matter of discussion, the early steps of autophagosome formation (nucleation and expansion) are known to require the following proteins: i) the Atg1/unc-51-like kinase (ULK) complex; ii) the Vps34/class III phosphatidylinositol 3-kinase (PI3K) complex I; iii) the transmembrane protein Atg9 and its cycling from the TGN and iv) the two ubiquitin-like proteins Atg12 and LC3 (the mammalian homologue of Atg8) and their conjugation systems (see text for details). Autophagosome maturation involves fusion with different endocytic compartments (early endosomes, multivesicular bodies and late endosomes) to form amphisomes containing both autophagosomal and endosomal content or autophagosomes might fuse directly with lysosomes. Finally the sequestered cytoplasmic material becomes degraded and the resulting macromolecules recycled back to the cytosol to be reused by the cell.
3.1 Nucleation and Expansion
Several key proteins are required for the early steps of autophagosome formation: i) the Atg1/Unc-51-like kinase (ULK) complex; ii) the Vps34/class III phosphatidylinositol 3-kinase (PI3K) complex I; iii) the transmembrane protein Atg9 and its associated cycling machinery; and iv) the two ubiquitin-like proteins Atg12 and the mammalian homologs of Atg8 and their conjugation systems.
One of the main questions in macroautophagy has focused on the origin of the autophagosome membrane. The endoplasmic reticulum (ER), Golgi network and mitochondria have all been implicated (Reggiori, 2006). Recently live cell imaging and EM-based studies have centered upon the ER: Upon nutrient deprivation, a structure rich in phosphatidylinositol-3-phosphate (PI3P) called the omegasome forms in the ER preceding the formation of autophagosomes (Axe et al., 2008; Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009) (Figure 1). Nonetheless, Atg9, a transmembrane protein that resides in the trans golgi network (TGN) is also essential for phagophore formation (Young et al., 2006), but exactly how Atg9 traffics to the phagophore is not yet clear. Finally an alternative model, suggesting that the outer membrane of ER-associated mitochondria participates in autophagosome biogenesis also was proposed recently (Hailey et al., 2010). Continued studies using high resolution microscopy and genetics will make much needed progress in this field.
The mechanism and regulation by which the autophagosomes nucleate is also not completely understood. Inhibition of the mammalian Target of Rapamycin (mTOR) is a critical trigger to evoke autophagy upon nutrient deprivation (Jung et al., 2010; Neufeld, 2010). mTOR is a serine/threonine kinase that, aside from autophagy, has been implicated in maintenance of basic cellular functions including cell growth, cell proliferation, protein translation and protein synthesis. Under nutrient rich conditions, mTOR phosphorylates ULK1, the closely related ULK2 and the associated Atg13, thereby inhibiting ULK function and preventing autophagosome formation. When mTOR activity is inhibited by starvation or treatment with rapamycin, the ULK complex becomes dephosphorylated and activated to stimulate formation of autophagic membranes (Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009; Jung et al., 2010; Neufeld, 2010). However, the Atg1/ULK complex is also essential for mTOR-independent autophagy (Hara et al., 2008) as it plays an important role in recruting other Atg proteins to the sites of autophagosome formation (Mizushima, 2010), as well as in regulation of starvation-induced cycling of mAtg9, the only integral membrane protein in the core autophagy machinery (Young et al., 2006).
Autophagosome nucleation also requires the Vps34/class III PI3K complex I which generates PI3P. This complex contains four core components; the Vps34/class III PI3K, the Vps15-like serine/threonine kinase p150, the mammalian Atg6 protein Beclin1 and Atg14L (also known as Barkor) (Simonsen and Tooze, 2009) (Figure 1). Inhibition of Vps34 activity by 3-methyladenine or wortmannin (Hendil et al., 1990; Seglen and Gordon, 1982; Wu et al., 2010) can be used to inhibit macroautophagy since it suppresses autophagosome nucleation. In addition, several Beclin-associated proteins like Ambra-1, Bif-1 and Bcl-2 have been found to regulate the activity of this complex (He and Levine, 2010; Simonsen and Tooze, 2009).
Once nucleated, the phagophore elongates around the cargo and closes to form an autophagosome. Studies in yeast and mammalian cells have shown that elongation requires the ubiquitin-like proteins Atg12 and Atg8 and their respective conjugation machineries (Ohsumi and Mizushima, 2004) (Figure 1). Six mammalian homologs to Atg8 have been described: Microtubule associated protein 1 light chain 3 (MAP1LC3) A, B and C; the GABA receptor associated protein (GABARAP); and the GABARAP-like proteins 1 (GABARAPL1, also known as GATE-16) and GABARAPL2. There is also a possible seventh homolog, GABARAPL3, however thus far its expression has not been observed. Although all Atg8 homologs can conjugate to lipid, MAP1LC3B is the most extensively studied and will be referred to simply as LC3 (Tanida et al., 2004). Atg12 and LC3 become activated by the E1-like enzyme Atg7 and then conjugated by E2-like enzymes (Atg10 and Atg3, respectively) to Atg5 or phosphatidylethanolamine (PE), respectively. The Atg5-12 conjugate then associates with Atg16L in the phagophore membrane, to act as a possible E3-like ligase for the recruitment and lipidation of LC3 (Hanada et al., 2007). This results in two measurable forms of LC3: The cytosolic form (called LC3-I), which is first cleaved at its C-terminus by the cysteine protease Atg4, and the PE-conjugated form (called LC3-II) (Tanida et al., 2004). In contrast to the other proteins required for nucleation and expansion of autophagic membranes, LC3-II remains membrane bound throughout the process and is therefore a widely used marker for autophagosomes. It is worth noting that examples of Beclin1-, as well as Atg5- and Atg7-independent autophagy have been reported in cells subjected to certain stressors (Chu et al., 2007; Nishida et al., 2009; Scarlatti et al., 2008), but nothing is known about the role of these non-canonical autophagy pathways in neurons. The molecular mechanisms involved in the final closure of the phagophore to form an autophagosome are also not known, although a role for LC3 has been suggested (Nakatogawa et al., 2007).
While mTOR inhibition is required for general macroautophagy, the potentiation of macroautophagic degradation of aggregated protein can occur in an mTOR independent manner (Filimonenko et al., 2010; Sarkar et al., 2007; Yamamoto et al., 2006). The mechanisms underlying mTOR-independent autophagosome formation is not fully understood. It is possible that rather than enhancing the nucleation of autophagosomes which is mTOR-dependent, the efficiency of membrane building may be enhanced by increasing the production of PI3P (Yamamoto et al., 2006). This is further substantiated by findings that co-stimulation of mTOR-independent and mTOR-dependent autophagy can enhance the degradation of mutant Huntingtin and aggregate-prone α-Synuclein (Sarkar et al., 2007).
3.2 Maturation
Maturation of autophagosomes into fully degradative autolysosomes requires the fusion of autophagosomes to other vesicle compartments. To prepare the autophagosome for fusion, LC3-II is removed from the outer leaflet of the outer membrane by the LC3 cleavage enzyme, Atg4 (Figure 1) (Tanida et al., 2004). Further maturation may occur by a single or multi-step fusion process. In yeast, autophagosomes fuse directly with the vacuole, whereas in mammalian cells autophagosome maturation may require sequential fusion steps with different endocytic populations, to first create an amphisome before fusion with lysosomes (Longatti and Tooze, 2009; Simonsen and Tooze, 2009). A stepwise model of autophagosomes maturation is supported by recent evidence showing that depletion of the early endocytic protein coat protein complex I (COPI) and subunits of the endosomal sorting complex required for transport (ESCRT), impedes autophagic degradation (Filimonenko et al., 2007; Lee et al., 2007; Razi et al., 2009). In both cases, defective autophagosome maturation was accompanied by an accumulation of autophagosomes and p62- and ubiquitin-positive protein aggregates. Interestingly, overexpression of an ESCRT-III subunit mutant (Vps2B/CHMP2BIntron5) corresponding to a mutation found in patients with frontotemporal lobar degeneration with ubiquitinated inclusions (FTDL-U) caused a similar phenotype in murine cortical neurons (Lee et al., 2007) or in HeLa cells (Filimonenko et al., 2007), suggesting that ESCRT-mediated control of autophagy is physiologically important.
Further highlighting the intimacy between the autophagic and endocytic pathways is a second complex that engages Vps34, p150 and Beclin1 known as the class III PI3K complex II. This complex can contain the positive regulator UVRAG (ultraviolet irradiation resistant-associated gene) or the negative regulator Rubicon (RUN domain and cysteine-rich domain containing, Beclin1-interacting protein), and both are involved in regulation of endocytic trafficking and autophagosome maturation (Itakura et al., 2008; Matsunaga et al., 2009; Sun et al., 2008; Zhong et al., 2009). In addition, numerous Rab proteins (Fader et al., 2008; Gutierrez et al., 2004; Hirota and Tanaka, 2009; Itoh et al., 2008; Jager et al., 2004; Munafo and Colombo, 2002; Ravikumar et al., 2008), as well as the Vps-C complexes HOPS (homotypic vacuole fusion and protein sorting) and CORVET (class C core vacuole/endosome tethering) (Lindmo et al., 2006; Nickerson et al., 2009) also have been implicated in autophagosome maturation. Finally, the recent identification of the PI3P-binding protein FYCO1 (FYVE and coiled-coil domain-containing protein 1) as an LC3- and Rab7-binding protein mediating microtubule plus end-directed transport of autophagosomes (Pankiv et al., 2010) may provide a link between the autophagic and endocytic machineries involved in regulation of autophagosome maturation.
The transport of autophagosomes for fusion with lysosomes has been found to depend on microtubules and dynein (Fass et al., 2006; Kimura et al., 2008). Such transport is especially important in neurons, where autophagosomes might form distant from lysosomes (Wang et al., 2006). The accumulation of autophagosomes within axons is often associated with various neurodegenerative disorders (Boland and Nixon, 2006; Yue et al., 2009). Mutations of the dynein machinery, albeit toxic themselves, also can increase accumulation and enhance the toxicity of polyQ proteins in flies and mice (Batlevi et al., 2010; Ravikumar et al., 2005).
Other players that may be selectively involved in maturation of ubiquitin-containing autophagosomes, including the AAA-ATPase valosin containing protein (VCP/p97) and HDAC6, have recently been identified (Lee et al., 2010; Tresse et al., 2010). Interestingly, mutations in VCP linked to Inclusion body myopathy associated with Paget's disease of the bone and fronto-temporal dementia (IBMPFD) also was found to impair autophagy, suggesting that impaired autophagosome maturation contributes to the pathology of this disease (Ju et al., 2009; Tresse et al., 2010). HDAC6, initially identified to mediate transport of misfolded proteins to the aggresome (Kawaguchi et al., 2003), was recently implicated in maturation of ubiquitin-positive autophagosomes (Lee et al., 2010). HDAC6 overexpression in fly eyes exposed expressing expanded polyQ proteins also was found to be neuroprotective (Pandey et al., 2007).
4. Aggrephagy - the mammalian counterpart to Cvt?
The mechanisms responsible for cargo selection and selective autophagy is currently an area of intense study and cargo-specific names like aggrephagy (selective degradation of protein aggregates), pexophagy (selective degradation of peroxisomes) and mitophagy (selective degradation of mitochondria) have been given (Klionsky et al., 2007; Knaevelsrud and Simonsen, 2010). Selectivity is achieved by a cargo receptor and a specificity adaptor that together connect the cargo to the core autophagic machinery. Typically, these autophagy receptors are not required for starvation-induced autophagy, but more research is needed to understand the mechanisms involved in selective macroautophagy and how these processes are regulated.
4.1 The yeast Cvt pathway
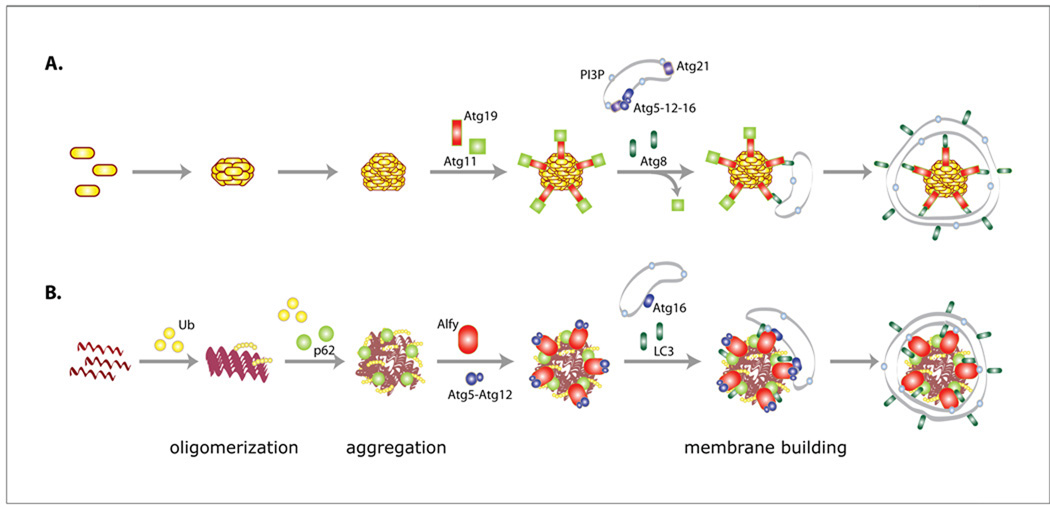
Although relatively novel in mammalian systems, selective macroautophagy has long-been described in yeast. Known as cytosol to vacuole targeting (Cvt), Cvt is a metabolic process that rely on receptor and adaptor proteins to traffic two vacuolar hydrolases, aminopeptidase 1 (ApeI) and the less studied α-mannosidase (Ams1) to the vacuole (Figure 2) (Lynch-Day and Klionsky, 2010). Ape1 is synthesized as a precursor (prApe1), which in a step-wise manner multimerizes into the higher order Ape1 complex prior to being sequestered into small, autophagosome-like, Cvt vesicles. Unlike autophagosomes, these vesicles form tightly around the ApeI complex, devoid of bulk cytosol. Association of the Ape1 complex with the Cvt vesicle membrane requires interaction of the Ape1 complex with the cargo receptor Atg19. Atg19 binds specifically to the ApeI complex, and possesses an Atg8-interacting W/Y/FxxL/I/V (WxxL) motif (Noda et al., 2008). WxxL motifs, found in several autophagy receptors, interact with Atg8/LC3 family members and are commonly referred to as LC3-interacting regions (LIR) (Pankiv et al., 2007) or LC3 recognition sequences (LRS) (Ichimura et al., 2008).
Figure 2. Schematic comparison between models of Cvt and aggrephagy.
A. Cvt. The precursor Ape1 is oligomerized into a 12-mer then further aggregated into the Ape1 complex. The autophagy receptor protein Atg19 then binds to the ApeI complex and attracts Atg11 which brings the complex to the phagophore. Atg11 is displaced by PE-bound Atg8 and Cvt vesicle forms around the complex. The PI3P-binding proteins Atg20, Atg21 and Atg24 are also involved in the Cvt pathway. B. Aggrephagy. The aggregate-prone protein becomes ubiquitinated and oligomerize. p62 binds to the ubiquitinated proteins and drives the formation of aggregates. Aggresome-like larger aggregates may continue to form but are not shown. The inclusion attracts Alfy, which is important for recruitment of Atg5-Atg12 to the inclusion. The presence of Alfy also stabilizes the interaction of LC3 with the inclusion. Alfy binds to PI3P in the autophagic membrane and may facilitate binding of Atg5-Atg12 to the membrane associated Atg16, creating the Atg5-Atg12-Atg16 complex, which might work as an E3-like ligase to permit LC3 conjugation to PE in the membrane, and formation of the autophagomes around the inclusion.
Prior to binding to Atg8, however, the prApe1-Atg19 complex binds directly to the adaptor Atg11, which transports the complex to the pre-autophagosomal structure (PAS) through an interaction between Atg11 and the actin cytoskeleton (Monastyrska and Klionsky, 2006). Atg11 also interacts with the core Atg proteins Atg9 and the Atg1-Atg13 kinase complex (He and Klionsky, 2006; Yorimitsu and Klionsky, 2005), suggesting that Atg11 acts as an important scaffolding protein. Atg11 is not required for non-selective autophagy, but seems to be involved in other forms of selective autophagy, like mitophagy and pexophagy. Interestingly, Atg11 over-expression increases recruitment of Atg8 and Atg9 to the PAS, resulting in more Cvt vesicles, suggesting that Atg11 levels regulate the rate of selective autophagy (Lynch-Day and Klionsky, 2010). Once at the PAS, Atg11 is displaced by Atg8 (Shintani et al., 2002). In addition to Atg11 and Atg19, the PI3P-binding proteins Atg20, Atg21 and Atg24 are also selectively involved in Cvt (Nice et al., 2002; Stromhaug et al., 2004). Further studies in yeast have revealed that selective autophagy is not limited to Cvt. Mitophagy requires the receptor protein Atg32, which is located in the outer mitochondrial membrane, and like Atg19, has an Atg8-binding LIR and interacts with Atg11 (Kanki et al., 2009; Okamoto et al., 2009). Similar regulation has also has been shown with Atg30 and pexophagy (Manjithaya et al., 2010).
4.2 Aggrephagy
Over the past several years, it has become increasingly clear that macroautophagy also plays a critical role in the selective elimination of aggregated proteins in mammalian cells. Studies indicate that similar to Cvt, the process of aggrephagy requires proteins that are exclusive for substrate selection and targeting (Bjorkoy et al., 2005; Filimonenko et al., 2010; Kirkin et al., 2009; Pankiv et al., 2007), as well as the core autophagic machinery (Filimonenko et al., 2010; Filimonenko et al., 2007; Yamamoto et al., 2006) (Figure 2). Moreover, similar to Cvt vacuoles, aggregate-containing autophagosomes appear largely devoid of other material, suggesting that the vesicle membrane expands tightly around its cargo (Filimonenko et al., 2010).
The nonspecific nature of macroautophagy in mammalian systems was challenged with the identification of the autophagy cargo receptors p62/SQSTM1 (Bjorkoy et al., 2005; Pankiv et al., 2007) and NBR1 (neighbor of BRCA1 gene 1) (Kirkin et al., 2009). p62 and NBR1 link ubiquitinated proteins via their ubiquitin-associated (UBA) domain to the core autophagic proteins LC3/GABARAP through their LIR domain (Kirkin et al., 2009; Pankiv et al., 2007). A PB1 domain permits both proteins to interact with other binding partners. Both proteins also homo-oligomerize, p62 through its PB1 domain, and NBR1 through its coiled-coil domain (Lamark et al., 2009). In contrast to Cvt, for which prApe1 oligomerizes independently of Atg19 (Lynch-Day and Klionsky, 2010), p62 and NBR1 are required for the formation of ubiquitin-positive inclusions (Bjorkoy et al., 2005; Clausen et al., 2010; Kirkin et al., 2009; Nezis et al., 2008), and p62 have been associated with inclusions in several human neurological disorders, including Lewy bodies, neurofibrillary tangles and Huntingtin aggregates (HD) (Kuusisto et al., 2001; Zatloukal et al., 2002). Interestingly, while nestin-specific deletion of Atg7 led to intraneuronal ubiquitinated inclusions, their appearance was suppressed when the mice were crossed with p62 knockout mice (Komatsu et al., 2007). Thus, p62 and NBR1 may structurally maintain larger aggregates of accumulating ubiquitinated proteins (Korolchuk et al., 2009; Lamark et al., 2009). Interestingly, the loss of larger inclusions did not impede neurotoxicity, indicating that smaller aggregated species may not require p62 to form. Consistent with this observation, the accumulation of ubiquitinated aggregated proteins have been reported in a subset of brain regions in p62 knockout mice starting from 6 months of age (Wooten et al., 2008).
Both p62 and NBR1 are continuously degraded as autophagic substrates, and thus often are used as indirect indicators of general macroautophagic activity (Bjorkoy et al., 2005; Kirkin et al., 2009; Pankiv et al., 2007). p62 also is a cargo receptor for other ubiquitinated substrates, like midbody remnants after mitosis (Pohl and Jentsch, 2009), peroxisomes (Kim et al., 2008; Platta and Erdmann, 2007), mitochondria (Geisler et al., 2010), intracellular bacteria (Deretic, 2010; Zheng et al., 2009) and ribosomal proteins (Ponpuak et al., 2010).
Recently, the 400kDa, PI3P-binding Autophagy-linked FYVE domain protein (Alfy) was identified to be required for aggrephagy, but not for starvation-induced autophagy (Filimonenko et al., 2010). Alfy interacts directly with p62 through its PH-BEACH domain (Clausen et al., 2010) and with Atg5 through its five WD40 repeats (Filimonenko et al., 2010). In cells, Alfy is required to scaffold the Atg5-Atg12-Atg16 E3-ubiquitin ligase-like complex and LC3 to the ubiquitin- and p62-positive polyQ inclusion (Filimonenko et al., 2010). These data suggest that Alfy functions as a scaffolding adaptor protein, similar to Atg11 in the Cvt pathway: Alfy connects the receptor-bound cargo to the core autophagic machinery and permits the autophagic membrane to form closely to the cargo, excluding bulk cytoplasm. As mentioned above, overexpression of Atg11 increases the formation of Cvt vesicles and increases turnover of prApe1. Interestingly, overexpression of Alfy or its C-terminal p62- and Atg5-binding region was found to decrease the number of protein inclusions in primary neurons expressing polyQ Htt and be neuroprotective in a Drosophila eye model of polyQ disease, suggesting that increased Alfy levels can stimulate aggrephagy (Filimonenko et al., 2010). The requirement of Alfy for aggrephagy is consistent with earlier observations in Drosophila deficient in the Alfy homologue Blue Cheese (bchs). Bchs mutant flies had a reduced lifespan, accelerated accumulation of ubiquitin-positive protein aggregates and neuronal degeneration (Finley et al., 2003). It will be interesting to learn whether Alfy, as p62, is involved in selective autophagy of other cargo such as the midbody ring structure or mitochondria.
4.3 Regulation of aggrephagy
Very little is understood about the regulation of aggrephagy. Since acute inhibition of macroautophagy during starvation can lead to cell death (Scarlatti et al., 2009), and since aggrephagy shares the core autophagic machinery, it would be important that cells can regulate an aggrephagic response. Interestingly, protein levels of Alfy are highest in brain and lowest in liver, thus inversely correlating with responsiveness to starvation (Simonsen et al., 2004). Alfy also undergoes stress-induced nucleocytoplasmic shuttling; although Alfy is usually found in the nucleus and decorating the nuclear membrane, it becomes recruited to cytoplasmic inclusions upon cellular stress like starvation, UPS inhibition and expression of aggregate-prone proteins (Filimonenko et al., 2010; Simonsen et al., 2004). Although it is uncertain what signals to Alfy to exit the nucleus, it is tempting to speculate that nuclear localization of Alfy might be a means by which the cell limits readily available levels of Alfy to impede aggrephagy in healthy cells. This hypothesis is supported by experiments showing that overexpression of Alfy increases clearance of polyQ inclusions in neurons (Filimonenko et al., 2010).
The aggrephagy cargo receptor p62 is also known to be a stress response protein, and both its mRNA and protein levels are induced upon exposure to various stressors (Ishii et al., 1997; Nagaoka et al., 2004). p62 also undergoes nucleocytoplasmic shuttling and may recruit Alfy from the nucleus to cytoplasmic inclusions (Clausen et al., 2010; Filimonenko et al., 2010). It was recently found that p62 expression is upregulated by the transcription factor NF-E2-related factor 2 (Nrf2) in response to oxidative stress (Jain et al., 2010) and that increased p62 levels again contribute to the activation of Nrf2, creating a positive loop resulting in expression of anti-oxidant genes (Fan et al., 2010; Jain et al., 2010; Komatsu et al., 2010; Lau et al., 2010). The p62-mediated Nrf2 activation is due to p62 blocking the interaction between Nrf2 and the Kelch-like ECH-associated protein 1 (Keap1), which under basal conditions leads to ubiquitination and UPS-mediated degradation of Nrf2 (Kobayashi et al., 2004). Interestingly, the drug deprenyl, a candidate for neuroprotection in PD, can mediate nuclear accumulation of Nrf2 and Nrf2-induced activation of oxidative stress-related proteins (Nakaso et al., 2006). Thus, activation of the p62-Nrf2 system may be a useful therapeutic strategy for PD, as well as other neurodegenerative disorders.
A complex network of post-translational modifications of aggregated proteins seems to regulate their clearance by autophagy. As discussed above, K63-linked ubiquitination may be critical to permit p62 and NBR1 to recognize cargo (Kirkin et al., 2009; Long et al., 2008; Wooten et al., 2008) and inclusions labelled with K63-linked ubiquitin chains have been associated with autophagic degradation (Tan et al., 2008). Furthermore, HDAC6 also preferentially binds K63-linked ubiquitin chains in vivo (Olzmann et al., 2007). The specific ubiquitin ligases involved in aggrephagy are at present unknown.
Aside from ubiquitination, other post-translational modifications, like acetylation and phosphorylation, also have been shown to enhance the degradation of aggregate-prone proteins by autophagy. When the normally occuring acetylation of the mutant Htt protein was prevented, its oligomerization and autophagy-dependent degradation was inhibited. A modest dependence on p62 was observed, however the mechanism by which trafficking to the autophagosomes is promoted remains unclear (Jeong et al., 2009). Phosphorylation of Htt by the inflammatory kinase IKK was found to enhance its degradation, but also to regulate additional post-translational modifications of Htt, like ubiquitination, SUMOylation, and acetylation (Thompson et al., 2009). Thus, a complex network of post-translational modifications is likely involved in regulation of protein aggregate clearance. How these changes modify autophagic degradation in neurons will give us further insight into how proteins are selected for degradation by aggrephagy or other macroautophagic pathways.
5. Concluding remarks
Although the toxicity of protein inclusions may be debatable, a strong argument exists that the elimination of the aggregating proteins and inclusions may be an effective means to ameliorate pathogenesis in different neurodegenerative models. Recently, macroautophagy has emerged as a highly selective quality control mechanism whose basal levels are important to prevent accumulation of protein. The mechanisms responsible for selective autophagy of various cargos is currently an area of intense study, but the cargo receptor p62, together with the adaptor protein Alfy, seem to be critical for selective autophagic degradation of protein aggregates and inclusions. Thus, stimulation of aggrephagy by modifying these proteins might be a beneficial approach in neurodegeneration. Nonetheless, care must be taken to degrade only those inclusions that represent ‘cellular garbage’ and not cellular protein bodies that represent functional cellular structures. Furthermore, up-regulation of any form of autophagy is unlikely to be effective if dysfunctional macroautophagy is a root cause. It is thus critical that we continue further study in macroautophagy, especially in neurons, if we are to use it to combat effectively neurodegenerative disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmad FJ, et al. Inhibition of microtubule nucleation at the neuronal centrosome compromises axon growth. Neuron. 1994;12:271–280. doi: 10.1016/0896-6273(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Alves-Rodrigues A, et al. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 1998;21:516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Aronin N, et al. CAG expansion affects the expression of mutant Huntingtin in the Huntington's disease brain. Neuron. 1995;15:1193–1201. doi: 10.1016/0896-6273(95)90106-x. [DOI] [PubMed] [Google Scholar]
- Arrasate M, et al. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, et al. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Batlevi Y, et al. Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila. Proc.Natl.Acad.Sci.U.S.A. 2010;107:742–747. doi: 10.1073/pnas.0907967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bennett GS, et al. Different proteins associated with 10-nanometer filaments in cultured chick neurons and nonneuronal cells. Science. 1981;212:567–569. doi: 10.1126/science.6163217. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–519. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- Bukau B, et al. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Chalmers KA, Love S. Phosphorylated Smad 2/3 colocalizes with phospho-tau inclusions in Pick disease, progressive supranuclear palsy, and corticobasal degeneration but not with alpha-synuclein inclusions in multiple system atrophy or dementia with Lewy bodies. J Neuropathol Exp Neurol. 2007;66:1019–1026. doi: 10.1097/nen.0b013e31815885ad. [DOI] [PubMed] [Google Scholar]
- Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chiang HL, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Chu CT, et al. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3:663–666. doi: 10.4161/auto.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TH, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- Cochard P, Paulin D. Initial expression of neurofilaments and vimentin in the central and peripheral nervous system of the mouse embryo in vivo. J Neurosci. 1984;4:2080–2094. doi: 10.1523/JNEUROSCI.04-08-02080.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, et al. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- Dauer W, et al. Resistance of alpha -synuclein null mice to the parkinsonian neurotoxin MPTP. Proc Natl Acad Sci U S A. 2002;99:14524–14529. doi: 10.1073/pnas.172514599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, et al. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Ding H, et al. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J Neurochem. 2008;106:2119–2130. doi: 10.1111/j.1471-4159.2008.05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Doxsey S, et al. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Fader CM, et al. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Fan W, et al. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, et al. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. Journal of Biological Chemistry. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, et al. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin JE, et al. Synucleinopathies: clinical and pathological implications. Arch Neurol. 2001;58:186–190. doi: 10.1001/archneur.58.2.186. [DOI] [PubMed] [Google Scholar]
- Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, et al. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22:4897–4905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Georgieff IS, et al. Expression of high molecular weight tau in the central and peripheral nervous systems. J Cell Sci. 1993;105(Pt 3):729–737. doi: 10.1242/jcs.105.3.729. [DOI] [PubMed] [Google Scholar]
- Geschwind DH. Tau phosphorylation, tangles, and neurodegeneration: the chicken or the egg? Neuron. 2003;40:457–460. doi: 10.1016/s0896-6273(03)00681-0. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glotzer M, et al. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gray EG, et al. Alzheimer's disease: paired helical filaments and cytomembranes. Neuropathol Appl Neurobiol. 1987;13:91–110. doi: 10.1111/j.1365-2990.1987.tb00174.x. [DOI] [PubMed] [Google Scholar]
- Grune T, et al. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and 'aggresomes' during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Gu X, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, et al. Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc Natl Acad Sci U S A. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, et al. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. Journal of Cell Science. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Yoshimori T. Where do they come from? Insights into autophagosome formation. FEBS Letters. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Hanada T, et al. The ATG12-ATG5 conjugate has a novel e3-like activity for protein lipidation in autophagy. J Biol Chem. 2007 doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper P, editor. Huntington's Disease. London, U.K.: W.B. Saunders Company Ltd.; 1996. [Google Scholar]
- Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Autophagy and neurodegeneration. ACS Chem Biol. 2006;1:211–213. doi: 10.1021/cb600182h. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu.Rev.Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Current Opinion in Cell Biology. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendil KB, et al. Both endocytic and endogenous protein degradation in fibroblasts is stimulated by serum/amino acid deprivation and inhibited by 3-methyladenine. Biochemical Journal. 1990;272:577–581. doi: 10.1042/bj2720577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. The ubiquitin pathway for protein degradation. Trends Biochem Sci. 1991;16:265–268. doi: 10.1016/0968-0004(91)90101-z. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci. 2009;66:2913–2932. doi: 10.1007/s00018-009-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Ihara M, et al. Sept4, a component of presynaptic scaffold and Lewy bodies, is required for the suppression of alpha-synuclein neurotoxicity. Neuron. 2007;53:519–533. doi: 10.1016/j.neuron.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Iqbal K, et al. Cytosolic abnormally hyperphosphorylated tau but not paired helical filaments sequester normal MAPs and inhibit microtubule assembly. J Alzheimers Dis. 2008;14:365–370. doi: 10.3233/jad-2008-14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, et al. Low micromolar levels of hydrogen peroxide and proteasome inhibitors induce the 60-kDa A170 stress protein in murine peritoneal macrophages. Biochemical and Biophysical Research Communications. 1997;232:33–37. doi: 10.1006/bbrc.1997.6221. [DOI] [PubMed] [Google Scholar]
- Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Molecular Biology of the Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, et al. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Molecular Biology of the Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, et al. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005 doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Jager S, et al. Role for Rab7 in maturation of late autophagic vacuoles. Journal of Cell Science. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Jain A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. Journal of Biological Chemistry. 2010 doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakes R, et al. Identification of two distinct synucleins from human brain. FEBS Lett. 1994;345:27–32. doi: 10.1016/0014-5793(94)00395-5. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Formation and development of Lewy pathology: a critical update. J Neurol. 2009;256 Suppl 3:270–279. doi: 10.1007/s00415-009-5243-y. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Madura K. Rings, chains and ladders: ubiquitin goes to work in the neuron. Prog Neurobiol. 2004;73:227–257. doi: 10.1016/j.pneurobio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Johnston JA, et al. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, et al. mTOR regulation of autophagy. FEBS Letters. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuse O, et al. Developmental stages of cortical Lewy bodies and their relation to axonal transport blockage in brains of patients with dementia with Lewy bodies. J Neurol Sci. 2003;211:29–35. doi: 10.1016/s0022-510x(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kim PK, et al. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, et al. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct.Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, et al. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, et al. How shall I eat thee? Autophagy. 2007;3:413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Letters. 2010 doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat.Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, et al. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy. 2009;5:862–863. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- Kuusisto E, et al. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- Lamark T, et al. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, et al. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- Lau A, et al. A non-canonical mechanism of Nrf2 activation by autophagy deficiency: a direct interaction between Keap1 and p62. Mol.Cell Biol. 2010 doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, et al. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Current Biology. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, et al. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K, et al. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp Cell Res. 2006;312:2018–2027. doi: 10.1016/j.yexcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Lippens G, et al. Tau aggregation in Alzheimer's disease: what role for phosphorylation? Prion. 2007;1:21–25. doi: 10.4161/pri.1.1.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscic RM, et al. ALS and FTLD: two faces of TDP-43 proteinopathy. Eur J Neurol. 2008;15:772–780. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, et al. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death.Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Letters. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, et al. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–1373. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat.Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McNaught KS, et al. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci. 2002;16:2136–2148. doi: 10.1046/j.1460-9568.2002.02301.x. [DOI] [PubMed] [Google Scholar]
- Merlini G, et al. Protein aggregation. Clin Chem Lab Med. 2001;39:1065–1075. doi: 10.1515/CCLM.2001.172. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Monastyrska I, Klionsky DJ. Autophagy in organelle homeostasis: peroxisome turnover. Mol Aspects Med. 2006;27:483–494. doi: 10.1016/j.mam.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka U, et al. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. Journal of Neurochemistry. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- Nakaso K, et al. Novel cytoprotective mechanism of anti-parkinsonian drug deprenyl: PI3K and Nrf2-derived induction of antioxidative proteins. Biochemical and Biophysical Research Communications. 2006;339:915–922. doi: 10.1016/j.bbrc.2005.11.095. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, et al. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Current Opinion in Cell Biology. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice DC, et al. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, et al. Vps-C complexes: gatekeepers of endolysosomal traffic. Current Opinion in Cell Biology. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Okamoto K, et al. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway JM, et al. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- Ortega Z, et al. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J Neurosci. 2010;30:3675–3688. doi: 10.1523/JNEUROSCI.5673-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SH, et al. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pankiv S, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. Journal of Cell Biology. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Perez M, et al. Tau--an inhibitor of deacetylase HDAC6 function. J Neurochem. 2009;109:1756–1766. doi: 10.1111/j.1471-4159.2009.06102.x. [DOI] [PubMed] [Google Scholar]
- Perrot R, Eyer J. Neuronal intermediate filaments and neurodegenerative disorders. Brain Res Bull. 2009;80:282–295. doi: 10.1016/j.brainresbull.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat.Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- Ponpuak M, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat.Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, et al. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, et al. Early endosomes and endosomal coatomer are required for autophagy. Journal of Cell Biology. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F. 1. Membrane origin for autophagy. Curr.Top.Dev.Biol. 2006;74:1–30. doi: 10.1016/S0070-2153(06)74001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulier E, et al. Early and reversible neuropathology induced by tetracycline-regulated lentiviral overexpression of mutant huntingtin in rat striatum. Hum Mol Genet. 2003;12:2827–2836. doi: 10.1093/hmg/ddg305. [DOI] [PubMed] [Google Scholar]
- Ross CA. Huntington's disease: new paths to pathogenesis. Cell. 2004;118:4–7. doi: 10.1016/j.cell.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol. 2005;6:891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- Roy S, et al. Axonal transport defects: a common theme in neurodegenerative diseases. Acta Neuropathol. 2005 109;:5–13. doi: 10.1007/s00401-004-0952-x. [DOI] [PubMed] [Google Scholar]
- Rujano MA, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. Journal of Biological Chemistry. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, et al. Does autophagy have a license to kill mammalian cells? Cell Death.Differ. 2009;16:12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, et al. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, et al. Non-selective autophagy. Semin Cell Biol. 1990;1:441–448. [PubMed] [Google Scholar]
- Shintani T, et al. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. Journal of Cell Biology. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires-Jones TL, et al. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci. 2009;32:150–159. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Stiess M, et al. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 2010;327:704–707. doi: 10.1126/science.1182179. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, et al. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr ST, et al. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J Cell Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc.Natl.Acad.Sci.U.S.A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CF, et al. TDP-43 immunoreactivity in neuronal inclusions in familial amyotrophic lateral sclerosis with or without SOD1 gene mutation. Acta Neuropathol. 2007;113:535–542. doi: 10.1007/s00401-007-0206-9. [DOI] [PubMed] [Google Scholar]
- Tan JM, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- Tanida I, et al. LC3 conjugation system in mammalian autophagy. Int.J.Biochem.Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. Journal of Cell Biology. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PB, Miller RJ. Aggregates in neurodegenerative disease: crowds and power? Trends Neurosci. 1999;22:194–197. doi: 10.1016/s0166-2236(99)01409-5. [DOI] [PubMed] [Google Scholar]
- Tresse E, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y, et al. Cellular localization of the Huntington's disease protein and discrimination of the normal and mutated form. Nat Genet. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- Uttenweiler A, Mayer A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol.Biol. 2008;445:245–259. doi: 10.1007/978-1-59745-157-4_16. [DOI] [PubMed] [Google Scholar]
- van der Zee J, et al. Frontotemporal lobar degeneration with ubiquitin-positive inclusions: a molecular genetic update. Neurodegener Dis. 2007;4:227–235. doi: 10.1159/000101847. [DOI] [PubMed] [Google Scholar]
- Volles MJ, Lansbury PT., Jr Vesicle permeabilization by protofibrillar alpha-synuclein is sensitive to Parkinson's disease-linked mutations and occurs by a pore-like mechanism. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Waelter S, et al. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Takahashi H. Cellular pathology in multiple system atrophy. Neuropathology. 2006;26:338–345. doi: 10.1111/j.1440-1789.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- Wang QJ, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler NS, et al. Molecular approaches to hereditary diseases of the nervous system: Huntington's disease as a paradigm. Annual Review of Neuroscience. 1991;14:503–529. doi: 10.1146/annurev.ne.14.030191.002443. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, et al. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten MW, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- Woulfe J. Nuclear bodies in neurodegenerative disease. Biochim Biophys Acta. 2008;1783:2195–2206. doi: 10.1016/j.bbamcr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Wu YT, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. Journal of Biological Chemistry. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, et al. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, et al. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, et al. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 Suppl 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. Journal of Cell Science. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Yu W, et al. Microtubule fragmentation and partitioning in the axon during collateral branch formation. J Neurosci. 1994;14:5872–5884. doi: 10.1523/JNEUROSCI.14-10-05872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, et al. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta. 2009;1793:1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am.J.Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. TDP-43-immunoreactive neuronal and glial inclusions in the neostriatum in amyotrophic lateral sclerosis with and without dementia. Acta Neuropathol. 2008;115:115–122. doi: 10.1007/s00401-007-0285-7. [DOI] [PubMed] [Google Scholar]
- Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat.Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, et al. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]