Abstract
Background
Galectin-3 is an animal lectin that has been implicated in wound healing and is decreased in inflammatory bowel disease (IBD). Matrix metalloproteinase-7 (MMP7) also known as matrilysin-1, a protease shown to cleave extracellular matrix proteins, is highly expressed in IBD tissues, especially at the leading edge of gastrointestinal ulcers. The ability of MMP7 to cleave galectin-3 and influence wound healing has not been reported previously.
Aim
To determine whether MMP7 cleaves galectin-3 and modulates wound healing in intestinal epithelial cells.
Methods
The cleaved fragments of galectin-3 were identified by N-terminal sequencing and mass spectrometry. Western blotting was used to detect the cleaved galectin-3 products in a colonic epithelial cell line (T84 cells). Cell migration was studied by in vitro scratch method.
Results
We demonstrate for the first time that MMP7 cleaves galectin-3 in vitro, resulting in three cleaved fragments (20.2 kDa, 18.9 kDa and 15.5 kDa). Exogenous treatment of T84 cells with recombinant MMP7 resulted in the appearance of secreted galectin-3 cleavage fragments in the supernatant. MMP7 inhibited cell migration and resulted in wound retraction and the addition of MMP7 to galectin-3 abrogated the wound healing and cell migration induced by galectin-3.
Conclusions
We have demonstrated that galectin-3 is a substrate for MMP7. Cleavage of galectin-3 may be one mechanism by which MMP7 inhibits wound healing. This study has significance in understanding delayed wound healing in chronic intestinal diseases like intestinal ulcers and IBD where MMP7 protein expression is elevated with a decreased galectin-3 protein expression.
Keywords: MMP7, galectin-3, inflammatory bowel disease, wound healing, cell migration
INTRODUCTION
Galectins are a family of β-galactoside-binding animal lectins that are expressed in a wide range of tissues and regulate many biological functions.1–3 In the gastrointestinal tract, galectin-3 is highly expressed in enterocytes and subepithelial macrophages.4, 5 In confluent enterocytes, galectin-3 is predominantly expressed in giant inclusions at the apical membrane6 and secretion of galectin-3 from a variety of cell types via a non-classical secretory pathway has been reported.7
Among the reported functions of galectin-3 is the promotion of cell migration and wound re-epithelialization.8–11 Galectin-3 may enhance these interactions between cell-cell and cell-matrix molecules by cross-linking a number of molecules, including integrins,12 laminins11 and collagens.13 In the gastrointestinal tract, this influence on wound healing and cell migration may be reflected in the significant down-regulation of galectin-3 in intestinal epithelial cells of patients with Crohn's disease and in inflamed biopsies from inflammatory bowel disease (IBD) patients.14–16 Further correlative evidence is that increased galectin-3 expression is associated with both colon cancer neoplastic progression and metastasis.17–19
Matrix metalloproteinases (MMPs), a group of zinc-dependent endopeptidases, have demonstrable roles in extracellular matrix (ECM) degradation and remodeling.20 In addition to remodeling the ECM, MMPs have been shown to modulate various aspects of wound healing, including scar resorption and re-epithelialization in multiple systems.21
While this effect of MMPs may be via ECM proteolysis, MMPs have also been shown to have proteolytic activity against a number of galectins. For example, MMP2, MMP9, MMP13 and membrane type 1-matrix metalloproteinase (MT1-MMP) are known to cleave the N-terminal domain of galectin-3.9, 22–24
Matrix metalloproteinase-7 (MMP7) also known as matrilysin-1, is the smallest of the MMPs and has broad proteolytic activity against a variety of extracellular matrix substrates, including collagens, insoluble elastins, proteoglycans, laminin, fibronectin, and casein.25, 26 However, MMP7 has not previously been shown to cleave galectin-3. In the gastrointestinal tract, MMP7 expression is increased in epithelial cells of IBD tissues14, 27 and specifically localized to epithelial cells at the leading edge of ulcers.28 In this study, we demonstrate that MMP7 cleaves galectin-3 and impairs galectin-3-mediated wound healing in intestinal epithelial cells. Our results provide a mechanistic explanation for elevated MMP7 expression and decreased galectin-3 protein levels in IBD patients.14, 29
MATERIALS AND METHODS
Cell culture
T84 cells were be maintained in Dulbecco's modified Eagle's medium/Ham's F-12 (1:1) supplemented with 10% fetal calf serum, penicillin G sodium (100 U/ml) and streptomycin sulfate (100 μg/ml) at 37°C in an atmosphere of 5% CO2.
Reagents and antibodies
Recombinant human MMP7 was obtained from Calbiochem (San Diego, CA, USA) and recombinant human galectin-3 from Sigma (St. Louis, MO). Rabbit anti-galectin-3 polyclonal antibody specific for a synthetic peptide conjugated to KLH derived from within 200 residues to the C-terminus was obtained from Abcam Inc., (Cambridge, MA) and a mouse monoclonal anti-galectin-3 antibody (clone B2C10) specific for an epitope within the first 45 amino acids of galectin-3 was obtained from BD Biosciences (San Jose, CA). The mouse anti-MMP7 monoclonal antibody was obtained from R&D Systems (Minneapolis, MN, USA) and goat anti-GAPDH polyclonal antibody was obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). The donkey anti-mouse Alexa Fluor 488, donkey anti-rabbit Alexa Fluor 568 and Alexa Fluor 680- labeled secondary antibodies were obtained from Invitrogen Corporation (Carlsbad, CA). The nuclear stain, DAPI was from Roche Applied Science (Mannheim, Germany).
Analysis of galectin-3 proteolysis
Recombinant human galectin-3 was incubated with recombinant active MMP-7 at a molar ratio of 1:10 at 37°C in 50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl and 10 mM CaCl2. The incubation was stopped at different time intervals by adding with a volume of 6× denaturing Laemmli buffer, and the sample was loaded on a 4–12% SDS-PAGE. The resolved proteins were transferred to PVDF membranes using a Bio-Rad mini-TransBlot apparatus and stained with Coomassie Brilliant Blue R-250 (Bio-Rad, Hercules, CA).
Mass spectrometry and N-terminal sequencing of cleaved galectin-3 fragments
Recombinant galectin-3 was digested by MMP7 in 50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl and 10 mM CaCl2. The digested samples were analyzed by mass spectrometry analysis at Johns Hopkins University Mass Spectrometry/Proteomics Facility (Baltimore, MD, USA). The incubated samples were run by electrospray LC/MS on Applied Biosystems QSTAR mass spectrometer. Fragments were separated by 4–12% SDS-PAGE, and transferred onto a PVDF membrane using a 10 mM CAPS buffer (pH 11) containing 10% methanol. The PVDF membranes were stained with Coomassie Brilliant BlueR-250, dried, and individual bands excised and subjected to N-terminal sequencing. The N-terminus sequencing of the cleaved fragments was performed at the Synthesis & Sequencing Facility at Johns Hopkins University.
Confocal microscopy
Cells grown on filters were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were permeabilized with 0.3% Triton X-100 in PBS and nonspecific binding was blocked by incubating for 2 hours at room temperature in PBS containing 5% donkey serum and 0.1% Triton X-100. Cells were incubated overnight at 4°C with primary antibodies (rabbit anti-galectin-3 polyclonal and mouse anti-MMP7 monoclonal) and with secondary antibodies (donkey anti-mouse Alexa Fluor 488 and donkey anti-rabbit Alexa Fluor 568) for 1 h at room temperature in the dark. The cell nuclei were stained using DAPI. The filters were mounted between a slide and coverslips in fluorescence mounting medium from DAKO (Carpinteria, CA) and viewed in Zeiss LSM 510 META confocal microscope at Ross Confocal Facilty at Johns Hopkins University.
SDS-PAGE, western blot analysis and densitometric analysis
Cells were washed in PBS and scraped into lysis buffer (75mM NaCl, 25 mM Tris/HCl pH 7.5, 0.25% (w/v) sodium deoxycholate, 1.5% Triton X-100, 4mM CaCl2) containing protease inhibitors. The cell culture medium was concentrated using Amicon ultra-10K filters while cell lysates were sonicated on ice and microcentrifuged at 14000 × g for 10 min at 4°C to remove cellular debris. Protein quantification of cell lysates and culture medium was performed using a BCA protein assay kit (Pierce, Rockford, IL, USA). The proteins were resolved using SDS-PAGE and transferred to PVDF membranes using a Bio-Rad mini-TransBlot apparatus. Membranes were probed with specific primary antibodies and Alexa Fluor 568 conjugated secondary antibodies. Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA) was used for visualization and quantitation of immunoreactive bands.
Wound healing assay
The in vitro scratch assay was performed as previously described.30–32 Briefly, T84 cells were grown to confluency on 24-well plate and cells were serum-starved for 24 h before the experiment. The plates were marked with a reference line drawn along the diameter of each well by using a marker. The wounds were made at angle of ~ 45° with a sterile micro-tip and the cells were incubated in the presence of recombinant active MMP7 (100 nM) or recombinant galectin-3 (5 μg/ml). The cells were immediately imaged using a 20× objective on a Zeiss LSM 510 META automated inverted microscope at 37°C in the presence of 5% CO2. The wound width at 0 and 24 h were measured using ImageJ. Cell migration was calculated as % wound width covered=(width at 0h − width at 24h)/width at 0h ×100.
Statistical analysis
Wound healing assay was the composite of six separate experiments and the groups were compared using ANOVA followed by Tukey's post hoc analysis. A p < 0.05 was considered statistically significant. SAS JMP IN software was used for statistical analysis.
RESULTS
MMP7 cleaves recombinant galectin-3
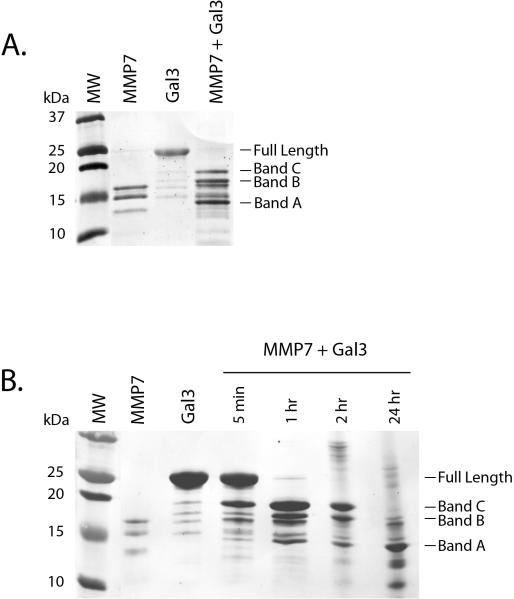
It has been previously shown that the N-terminal domain of galectin-3 is susceptible to proteolysis by MMP2, MMP9, MMP13 and MT1-MMP.9, 22, 23 We tested whether galectin-3 could be cleaved by MMP7 in vitro (Figure 1). Recombinant active MMP7 was incubated with recombinant galectin-3 and samples were collected at various time intervals, run on an SDS-PAGE and detected by staining PVDF membranes with Coomassie Brilliant Blue R-250. Four hours after MMP7 treatment, the full length galectin-3 band (~26 kDa) was undetectable but three galectin-3 cleavage products appeared (Figure 1A: bands A, B and C). Kinetic studies show that the galectin-3 cleavage products were appearing as early as 5 min after incubation (Figure 1B). At 2 hours, the full length galectin-3 band was undetectable and by 24 hours, the predominant cleavage products were mostly smaller than ~15kDa. These results suggest that MMP7 can directly cleave galectin-3 in vitro in a time dependent manner.
Figure 1. MMP7 cleaves recombinant galectin-3.
Recombinant MMP7 and galectin-3 were incubated together for various time points. Reaction products were separated on SDS-PAGE, transferred to PVDF membranes and proteins detected by Coommassie Brilliant Blue staining. A. A 4 hr reaction mixture revealed three possible galectin-3 cleavage products (Bands A, B and C). B. A time course revealed galectin-3 cleavage within 5 minutes with the early appearance of Bands B and C. Band A became the predominant MMP7-induced galectin-3 cleavage product at the 24 hr incubation time point.
Identification of MMP7-induced galectin-3 N-terminus cleavage sites
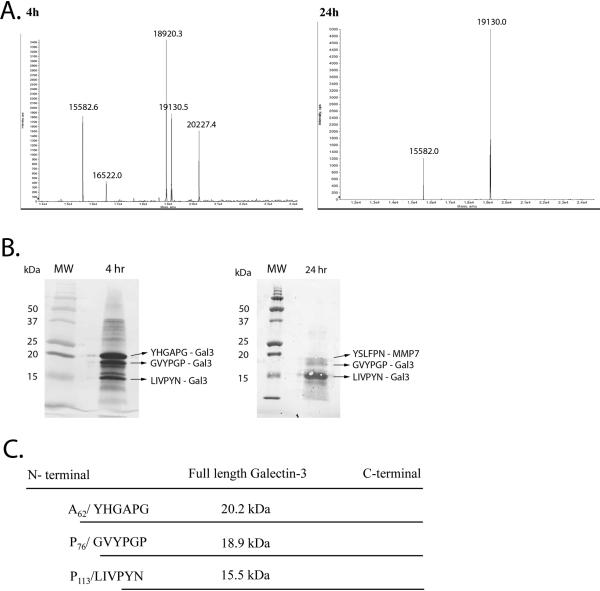
MMP7 induced cleavage of galectin-3 at multiple sites. To determine the exact cleavage sites mass spectrometry analysis was conducted on the 4 and 24 hour MMP7/galectin-3 cleavage reaction mixtures (Figure 2A). In the 4 hour reaction, 4 major proteins were identified with molecular masses of 15582.6, 18920.3, 19130.5 and 20227.4 Da. In the 24 hour reaction, two major bands were identified with molecular masses of 15582.0 and 19130.0 Da.
Figure 2. Identification of MMP7 induced galectin-3 cleavage site.
A. Mass spectra of recombinant galectin-3 and MMP7 reaction products. Recombinant MMP7 and galectin-3 were incubated together for 4 and 24 hours and mass spectrometry performed. B. Coomassie Brilliant Blue staining of the 4 and 24 hr reaction mixtures utilized for subsequent N-terminal sequencing. C. Schematic representation of MMP7-directed galectin-3 cleavage sites for the 20.2, 18.9 and 15.5 kDa galectin-3 fragments. (figure not to scale.)
To determine the specific cleavage sites of the reaction products of recombinant MMP7 with recombinant galectin-3, the 4 and 24 hour cleavage reaction products were run on SDS-PAGE, transferred to PVDF membranes and stained for protein visualization (Figure 2B). Bands were excised and N-terminal sequencing of the visualized protein bands was conducted. Sequence analysis revealed that the 19130 Da protein was recombinant MMP7, with an N-terminal sequence of YSLFPNSPK. The sequences of the remaining cleavage reaction proteins all corresponded to galectin-3. The galectin-3 cleavage site of band C (20.2 kDa) was Ala62 / Tyr63; band B (18.9 kDa) was Pro76 / Gly77 and band A (15.5 kDa) was Pro113/Leu114 (Figure 2C). This result indicates that all identified galectin-3 cleavage products result from MMP7 cleavage of amino acids in the N-terminal region.
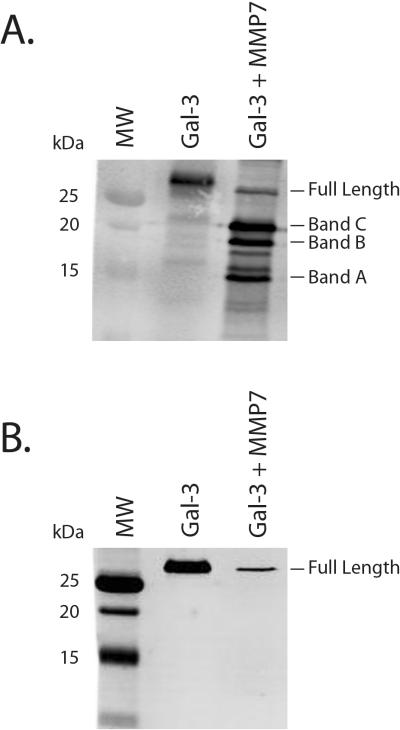
To confirm whether MMP7 cleaves galectin-3 at the N-terminus or the C-terminus, a 1 hour MMP7/galectin-3 cleavage reaction mixture was analyzed by western blotting using anti-galectin-3 antibodies specific for either the N-terminus or C-terminus. The three cleavage products as well as the full length galectin-3 were visible when probing with the C-terminus-specific galectin-3 antibody (Figure 3A). In contrast, the N-terminus-specific anti-galectin-3 antibody detected the full length galectin-3 band but none of the MMP7-directed galectin-3 cleavage products (Figure 3B). This result further suggests that MMP7 cleaves galectin-3 at three N-terminus sites and not the C-terminus region.
Figure 3. Western blot confirmation of N-terminal cleavage of galectin-3 by MMP7.
Recombinant MMP7 and galectin-3 were incubated together for 1 hr and cleavage reaction products were detected with an anti-galectin-3 C-terminus antibody (A.) and an anti-galectin-3 N-terminus antibody (B.) MMP7-induced galectin-3 cleavage products were only detected with the anti-galectin-3 C-terminus antibody.
MMP7 cleaves endogenous galectin-3 of T84 intestinal epithelial cells
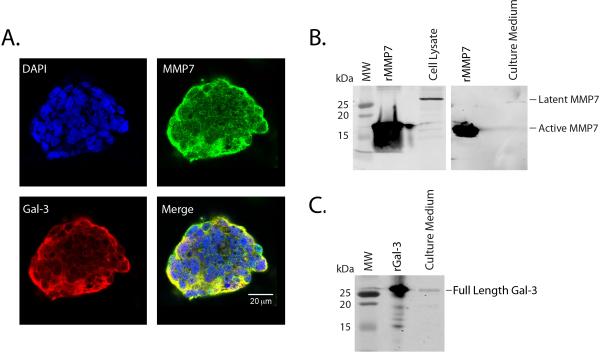
Galectin-3 and MMP7 were previously found to be expressed in intestinal epithelial cells.4, 6, 18, 19, 28, 33–35 We next analyzed the distribution of MMP7 and galectin-3 in T84 cells, a colonic epithelial cell line. Dual immunofluorescence confocal microscopy revealed that galectin-3 and MMP7 were expressed within T84 cell colonies, with superposition of galectin-3 and MMP7 immunostaining demonstrating predominant co-localization at the edge of the colonies (Figure 4A). Further Western blot analysis determined that, in unstimulated T84 cells, the MMP7 is expressed only as the latent, pro-form (28 kDa) in both cellular lysates and culture supernatants (Figure 4B) and not as active MMP7 (19kDa). The active secretion of full length galectin-3 protein (26 kDa) into the T84 cell culture supernatant was also confirmed by Western blot analysis (Figure 4C).
Figure 4. Galectin-3 and MMP7 are expressed in T84 cells.
A. T84 cell colonies were stained for anti-galectin-3 C-terminus (red), MMP7 (green) and DAPI (blue) and visualized by confocal microscopy. Predominant MMP7 and galectin-3 staining were seen in the periphery with co-localization (yellow) seen in the merged image. B. Western blot analysis detected the 28 kDa pro-MMP7 in T84 cell lysates and culture medium supernatants. C. Western blot analysis detected the full length galectin-3 protein in the T84 culture medium supernatants.
To determine if active MMP7 can cleave galectin-3 expressed by intestinal epithelial cells, T84 cells were treated with active recombinant MMP7 (19kDa) for 4h and the culture medium examined for galectin-3 cleavage by Western blot analysis. The anti-galectin-3 C-terminus-specific antibody detected the 20.2 kDa (Band C) and 15.5 kDa (Band A) cleavage products (Figure 5). This result demonstrates that MMP7 can cleave endogenous galectin-3 expressed by intestinal epithelial cells.
Figure 5. MMP7 cleaves galectin-3 secreted by T84 cells.
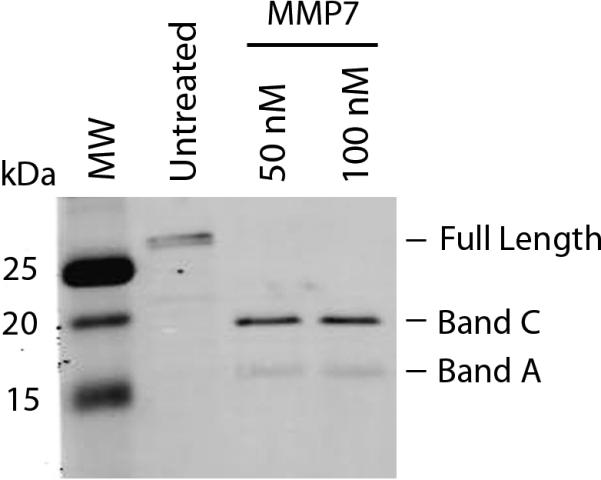
T84 cells were incubated with (50 and 100 nM) MMP7 for 4 hours and presence of galectin-3 in culture supernatants detected by Western blot analysis. The 15.5 and 20.2 kDa galectin-3 cleavage products were detected in MMP7-treated cells while only the full length galectin-3 was detected in untreated T84 cell culture supernatants.
MMP7 inhibits galectin-3-mediated wound healing in T84 cells
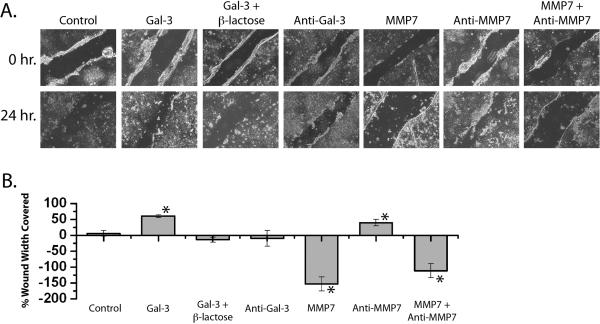
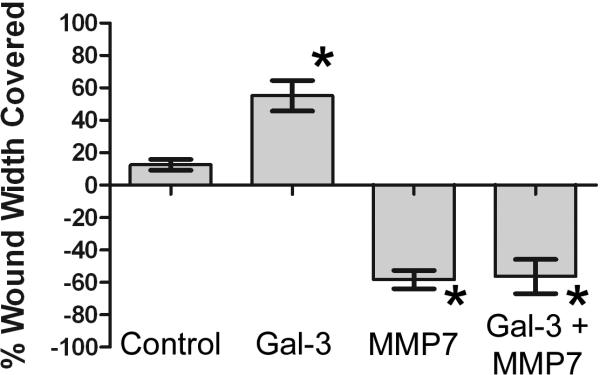
Galectin-3 has been previously shown to promote wound healing in epithelial cells.8 To assess whether MMP7 can influence galectin-3-mediated wound healing, we performed an in vitro scratch wound-healing assay. Treatment of wounded T84 cell monolayers with galectin-3 for 24 hours promoted healing with a 60.4% ± 4.4 reduction in wound width (Figure 6). The galectin-3-induced reduction in wound width was inhibited by both β-lactose (−14.1% ± 8.1), a non-specific inhibitor of galectin-3, and an anti-galectin-3 neutralizing antibody (−9.8% ± 24.8). Conversely, a 24 hour treatment of wounded T84 cells with MMP7 resulted in a significant widening of the initial wound (−153.2% ± 22.1) which was partially reversed by pretreatment with an anti-MMP7 antibody and MMP7 (−111.6 ± 21.8). The addition of the anti-MMP7 antibody alone appeared to promote wound healing (39.5 ± 10.2). Efficient knockdown of galectin-3 and MMP7 expression alone or in combination using siRNA resulted in wound width coverage that was not statistically different from untransfected cells or cells transfected with a negative control siRNA (data not shown). In a separate experiment, wounds treated with both galectin-3 and MMP7 for 24 hours demonstrated significant widening (−56.3% ± 26.0) comparable to MMP7 alone (−58.2 ± 13.8) (Fig 7), indicating that MMP7 treatment abrogated the wound healing induced by galectin-3.
Figure 6. Galectin-3 promotes and MMP7 inhibits wound healing in T84 cells.
Confluent T84 cell monolayers were wounded and immediately treated with: galectin-3, galectin-3 + β-lactose, anti-galectin-3 neutralizing antibody, active MMP7, anti-MMP7 neutralizing antibody, and active MMP7 + anti-MMP7 neutralizing antibody. A. Photomicrographs were obtained at 0 and 24 hours. B. Quantitative analysis of wound width at 0 and 24 hrs was performed and percent wound width covered calculated (n = 6 per condition; * p < 0.01).
Figure 7. MMP7 treatment negates the galectin-3-induced wound healing in T84 cells.
Confluent T84 cell monolayers were wounded in the presence of galectin-3, MMP7 and galectin-3 plus MMP7. Quantitative analysis of wound width at 0 and 24 hrs was performed and percent wound width covered calculated (n = 6 per condition; * p < 0.01).
DISCUSSION
In this study, we demonstrated that MMP7 cleaves the N-terminal domain of galectin-3 at three specific sites. This finding that MMP7 cleaves galectin-3 supports previous studies demonstrating that the galectins are, indeed, targets for MMP proteolytic cleavage. Our findings add to the list of other proteases that specifically cleave galectin-3. These other known proteases include MMP2, MMP9, MMP13 and MT1-MMP.22–24, 28, 36, 37
Previous studies have identified proteolytic cleavage sites within galectin-3.23, 24, 36, 37 MMP2, MMP9 and MMP13 all cleave galectin-3 at the Ala62 -Tyr63 peptide bond.23, 24, 36, 37 The likelihood that the resultant 20.2 kDa galectin-3 fragment is a main cleavage product of MMPs is consistent with our demonstration that MMP7 also generates this same peptide fragment. The additional MMP2-induced galectin-3 peptide bond cleavage site, Gly32-Ala33, was not proteolytically cleaved by MMP7. Similarly, the two additional MMP13-induced galectin-3 peptide bond cleavage sites, Pro92-Ser93 and Pro102-Ala103, were not proteolytically cleaved by MMP7. However, MMP7 appeared to cleave galectin-3 at two previously unidentified peptide bonds, Pro76-Gly77 and Pro113-Leu114.
The co-localization of MMP7 and galectin-3 at the peripheral edge of T84 cell colonies suggests that the proteolytic cleavage of galectin-3 by MMP7 may serve a physiologic role. The galectin-3 localization is consistent with previous findings identifying galectin-3 at the peripheral edge of HT29 cell colonies38 and at the apical surface of confluent T84 cells.6 Similarly, the MMP7 localization is consistent with previous findings demonstrating predominant MMP7 expression in migrating enterocytes in intestinal ulcers33 and ulcer borders.28 Enhanced MMP7 expression is also seen in 90% of colorectal cancers34 with enhanced expression in colorectal cancers exhibiting nodal or distal metastases.35
Our studies demonstrated that galectin-3 induced wound healing and MMP7 inhibited wound healing in confluent T84 cells. Furthermore, we demonstrated that concurrent treatment of wounded T84 cells with MMP7 and galectin-3 resulted in the overall abrogation of wound healing. The galectin-3 induced wound healing in our model is consistent with findings that galectin-3 promotes wound healing in corneal abrasions.8, 10, 12 These results indicate that galectin-3 may contribute to re-epithelialization in intestinal wounds and ulcerations and that this expression may be modified by relative MMP7 expression levels. It may be further hypothesized that the decreased expression of galectin-315, 16 and increased expression of MMP714, 28, 39 in active IBD tissues may represent a pathophysiologic state of reduced re-epithelialization ability.
Interestingly, our finding that MMP7 inhibits wound healing is in contrast to previous studies of airway epithelial cells. Dunsmore et. al.40 demonstrated that MMP7 is required for re-epithelialization in wounded tracheal explants. In lung epithelial cells, MMP7-mediated re-epithelialization appeared to be regulated, in part, by cleavage of E-cadherin.41 However, in our confluent colonic epithelial T84 cell culture system, MMP7 had no effect on E-cadherin expression or localization (data not shown). It should be noted that the exogenous administration of MMP7 to confluent wounded T84 cells does not exclude the possibility that the proteolytic cleavage of other cell-cell and cell-matrix proteins may have contributed to the inhibited wound healing in our model. There is sufficient evidence that MMP7 cleaves a number of proteins that may participate in re-epithelialization and wound healing.26, 42 However, determining the exact contributions of MMP7 cleavage of galectin-3 in comparison to other molecules was beyond the scope of this study.
Of note, in contrast to conditions seen in IBD, the increased expression of both galectin-3 and MMP7 were found in advanced colorectal cancer and positively correlated with metastatic potential. However, the increased expression of galectin-3 in advanced colorectal cancer may be related to an increased expression of galectin-3 cleavage products rather than the full-length galectin-3. When breast (infiltrating ductal carcinoma) cancer tissues were assessed using antibodies recognizing cleaved versus full-length galectin-3, full-length galectin-3 was seen in ductules with normal morphology while the cleaved galectin-3 was localized to both normal and invasive cells.36, 37 Further evidence indicates that the expression of full-length galectin-3 in tumors does not result in increased tumorigenicity, rather, galectin-3 cleavage is a required process for tumor progression.
Overall, these results support previous studies that galectins are proteolytic cleavage targets of MMPs. Furthermore, this study supports the hypothesis that the proteolytic cleavage of galectins by MMP7 may be a mechanism by which MMPs influence re-epithelialization in wound healing and inflammation as well as cell migration in cancer progression and metastasis. This interaction between galectin-3 and MMP7 may be physiologically relevant in IBD, where an increased MMP7 and a decreased galectin-3 expression in inflamed tissues has been reported. Further studies will be necessary to elucidate the complex interactions between MMPs and their targets in the mechanisms of wound healing and cell migration in intestinal processes associated with gastrointestinal inflammation and cancer.
ACKNOWLEDGEMENTS
We thank John Gibas of the Ross Confocal Facility, Johns Hopkins University. We are grateful to Dr. Mark Donowitz for helpful suggestions. We also thank the Mass Spectrometry/Proteomics Facility and Synthesis & Sequencing Facility at Johns Hopkins University, Baltimore, MD, USA. This research was supported by the K08 DK0708046 grant and the NIH core grant R24 DK064388 (to JHK) and the CCFA Research Fellowship Award (to KJR).
REFERENCES
- 1.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 2.Malemud CJ. Matrix metalloproteinases: role in skeletal development and growth plate disorders. Front Biosci. 2006;11:1702–1715. doi: 10.2741/1916. [DOI] [PubMed] [Google Scholar]
- 3.Malemud CJ. Matrix metalloproteinases (MMPs) in health and disease: an overview. Front Biosci. 2006;11:1696–1701. doi: 10.2741/1915. [DOI] [PubMed] [Google Scholar]
- 4.Lotz MM, Andrews CW, Jr., Korzelius CA, et al. Decreased expression of Mac-2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc Natl Acad Sci U S A. 1993;90:3466–3470. doi: 10.1073/pnas.90.8.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazowski E, Dotan I, Tulchinsky H, et al. Galectin-3 expression in pouchitis in patients with ulcerative colitis who underwent ileal pouch-anal anastomosis (IPAA) Pathol Res Pract. 2009;205:551–558. doi: 10.1016/j.prp.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Huflejt ME, Jordan ET, Gitt MA, et al. Strikingly different localization of galectin-3 and galectin-4 in human colon adenocarcinoma T84 cells. Galectin-4 is localized at sites of cell adhesion. J Biol Chem. 1997;272:14294–14303. doi: 10.1074/jbc.272.22.14294. [DOI] [PubMed] [Google Scholar]
- 7.Cooper DN, Barondes SH. Evidence for export of a muscle lectin from cytosol to extracellular matrix and for a novel secretory mechanism. J Cell Biol. 1990;110:1681–1691. doi: 10.1083/jcb.110.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Z, Said N, Amin S, et al. Galectins-3 and -7, but not galectin-1, play a role in re-epithelialization of wounds. J Biol Chem. 2002;277:42299–42305. doi: 10.1074/jbc.M200981200. [DOI] [PubMed] [Google Scholar]
- 9.Ochieng J, Green B, Evans S, et al. Modulation of the biological functions of galectin-3 by matrix metalloproteinases. Biochim Biophys Acta. 1998;1379:97–106. doi: 10.1016/s0304-4165(97)00086-x. [DOI] [PubMed] [Google Scholar]
- 10.Saravanan C, Cao Z, Head SR, et al. Detection of differentially expressed wound-healing-related glycogenes in galectin-3-deficient mice. Invest Ophthalmol Vis Sci. 2009;50:5690–5696. doi: 10.1167/iovs.08-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paret C, Bourouba M, Beer A, et al. Ly6 family member C4.4A binds laminins 1 and 5, associates with galectin-3 and supports cell migration. Int J Cancer. 2005;115:724–733. doi: 10.1002/ijc.20977. [DOI] [PubMed] [Google Scholar]
- 12.Saravanan C, Liu FT, Gipson IK, et al. Galectin-3 promotes lamellipodia formation in epithelial cells by interacting with complex N-glycans on alpha3beta1 integrin. J Cell Sci. 2009;122:3684–3693. doi: 10.1242/jcs.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrichs J, Manninen A, Muller DJ, et al. Galectin-3 regulates integrin alpha2beta1-mediated adhesion to collagen-I and -IV. J Biol Chem. 2008;283:32264–32272. doi: 10.1074/jbc.M803634200. [DOI] [PubMed] [Google Scholar]
- 14.Matsuno K, Adachi Y, Yamamoto H, et al. The expression of matrix metalloproteinase matrilysin indicates the degree of inflammation in ulcerative colitis. J Gastroenterol. 2003;38:348–354. doi: 10.1007/s005350300062. [DOI] [PubMed] [Google Scholar]
- 15.Jensen-Jarolim E, Gscheidlinger R, Oberhuber G, et al. The constitutive expression of galectin-3 is downregulated in the intestinal epithelia of Crohn's disease patients, and tumour necrosis factor alpha decreases the level of galectin-3-specific mRNA in HCT-8 cells. Eur J Gastroenterol Hepatol. 2002;14:145–152. doi: 10.1097/00042737-200202000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Muller S, Schaffer T, Flogerzi B, et al. Galectin-3 modulates T cell activity and is reduced in the inflamed intestinal epithelium in IBD. Inflamm Bowel Dis. 2006;12:588–597. doi: 10.1097/01.MIB.0000225341.37226.7c. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M, Inufusa H, Adachi T, et al. Involvement of galectin-3 expression in colorectal cancer progression and metastasis. Int J Oncol. 1999;15:143–148. doi: 10.3892/ijo.15.1.143. [DOI] [PubMed] [Google Scholar]
- 18.Sanjuan X, Fernandez PL, Castells A, et al. Differential expression of galectin 3 and galectin 1 in colorectal cancer progression. Gastroenterology. 1997;113:1906–1915. doi: 10.1016/s0016-5085(97)70010-6. [DOI] [PubMed] [Google Scholar]
- 19.Schoeppner HL, Raz A, Ho SB, et al. Expression of an endogenous galactose-binding lectin correlates with neoplastic progression in the colon. Cancer. 1995;75:2818–2826. doi: 10.1002/1097-0142(19950615)75:12<2818::aid-cncr2820751206>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 20.Matrisian LM. The matrix-degrading metalloproteinases. Bioessays. 1992;14:455–463. doi: 10.1002/bies.950140705. [DOI] [PubMed] [Google Scholar]
- 21.Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. Int J Biochem Cell Biol. 2008;40:1334–1347. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth M, Osenkowski P, Hesek D, et al. Cleavage at the stem region releases an active ectodomain of the membrane type 1 matrix metalloproteinase. Biochem J. 2005;387:497–506. doi: 10.1042/BJ20041324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochieng J, Fridman R, Nangia-Makker P, et al. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry. 1994;33:14109–14114. doi: 10.1021/bi00251a020. [DOI] [PubMed] [Google Scholar]
- 24.Guevremont M, Martel-Pelletier J, Boileau C, et al. Galectin-3 surface expression on human adult chondrocytes: a potential substrate for collagenase-3. Ann Rheum Dis. 2004;63:636–643. doi: 10.1136/ard.2003.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki K, Hattori Y, Umenishi F, et al. Purification and characterization of extracellular matrix-degrading metalloproteinase, matrin (pump-1), secreted from human rectal carcinoma cell line. Cancer Res. 1990;50:7758–7764. [PubMed] [Google Scholar]
- 26.Wilson CL, Matrisian LM. Matrilysin: an epithelial matrix metalloproteinase with potentially novel functions. Int J Biochem Cell Biol. 1996;28:123–136. doi: 10.1016/1357-2725(95)00121-2. [DOI] [PubMed] [Google Scholar]
- 27.Makitalo L, Kolho KL, Karikoski R, et al. Expression profiles of matrix metalloproteinases and their inhibitors in colonic inflammation related to pediatric inflammatory bowel disease. Scand J Gastroenterol. 2010 doi: 10.3109/00365520903583863. [DOI] [PubMed] [Google Scholar]
- 28.Saarialho-Kere UK, Vaalamo M, Puolakkainen P, et al. Enhanced expression of matrilysin, collagenase, and stromelysin-1 in gastrointestinal ulcers. Am J Pathol. 1996;148:519–526. [PMC free article] [PubMed] [Google Scholar]
- 29.Shkoda A, Werner T, Daniel H, et al. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–1125. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 30.Kalabis J, Rosenberg I, Podolsky DK. Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J Biol Chem. 2006;281:6434–6441. doi: 10.1074/jbc.M512905200. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg IM, Goke M, Kanai M, et al. Epithelial cell kinase-B61: an autocrine loop modulating intestinal epithelial migration and barrier function. Am J Physiol. 1997;273:G824–832. doi: 10.1152/ajpgi.1997.273.4.G824. [DOI] [PubMed] [Google Scholar]
- 32.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 33.Salmela MT, Pender SL, Karjalainen-Lindsberg ML, et al. Collagenase-1 (MMP-1), matrilysin-1 (MMP-7), and stromelysin-2 (MMP-10) are expressed by migrating enterocytes during intestinal wound healing. Scand J Gastroenterol. 2004;39:1095–1104. doi: 10.1080/00365520410003470. [DOI] [PubMed] [Google Scholar]
- 34.Newell KJ, Witty JP, Rodgers WH, et al. Expression and localization of matrix-degrading metalloproteinases during colorectal tumorigenesis. Mol Carcinog. 1994;10:199–206. doi: 10.1002/mc.2940100404. [DOI] [PubMed] [Google Scholar]
- 35.Adachi Y, Yamamoto H, Itoh F, et al. Contribution of matrilysin (MMP-7) to the metastatic pathway of human colorectal cancers. Gut. 1999;45:252–258. doi: 10.1136/gut.45.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nangia-Makker P, Raz T, Tait L, et al. Galectin-3 cleavage: a novel surrogate marker for matrix metalloproteinase activity in growing breast cancers. Cancer Res. 2007;67:11760–11768. doi: 10.1158/0008-5472.CAN-07-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nangia-Makker P, Balan V, Raz A. Regulation of tumor progression by extracellular galectin-3. Cancer Microenviron. 2008;1:43–51. doi: 10.1007/s12307-008-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remy L, Trespeuch C, Bachy S, et al. Matrilysin 1 influences colon carcinoma cell migration by cleavage of the laminin-5 beta3 chain. Cancer Res. 2006;66:11228–11237. doi: 10.1158/0008-5472.CAN-06-1187. [DOI] [PubMed] [Google Scholar]
- 39.Vaalamo M, Karjalainen-Lindsberg ML, Puolakkainen P, et al. Distinct expression profiles of stromelysin-2 (MMP-10), collagenase-3 (MMP-13), macrophage metalloelastase (MMP-12), and tissue inhibitor of metalloproteinases-3 (TIMP-3) in intestinal ulcerations. Am J Pathol. 1998;152:1005–1014. [PMC free article] [PubMed] [Google Scholar]
- 40.Dunsmore SE, Saarialho-Kere UK, Roby JD, et al. Matrilysin expression and function in airway epithelium. J Clin Invest. 1998;102:1321–1331. doi: 10.1172/JCI1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGuire JK, Li Q, Parks WC. Matrilysin (matrix metalloproteinase-7) mediates E-cadherin ectodomain shedding in injured lung epithelium. Am J Pathol. 2003;162:1831–1843. doi: 10.1016/S0002-9440(10)64318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulmer TA, Keeler V, Andre S, et al. The tumor-associated antigen 90K/Mac-2-binding protein secreted by human colon carcinoma cells enhances extracellular levels of promatrilysin and is a novel substrate of matrix metalloproteinases-2, -7 (matrilysin) and -9: Implications of proteolytic cleavage. Biochim Biophys Acta. 2010;1800:336–343. doi: 10.1016/j.bbagen.2009.07.030. [DOI] [PubMed] [Google Scholar]