Abstract
The Jamaican click beetle Pyrophorus plagiophthalamus (Coleoptera: Elateridae) is unique among all bioluminescent organisms in displaying a striking light color polymorphism [Biggley, W. H., Lloyd, J. E. & Seliger, H. H. (1967) J. Gen. Physiol. 50, 1681–1692]. Beetles on the island vary in the color of their ventral light organs from yellow–green to orange and their dorsal organs from green to yellow–green. The genetic basis for the color variation involves specific amino acid substitutions in the enzyme luciferase. Here, we show that dorsal and ventral light color in P. plagiophthalamus are under separate genetic control, we resolve the allelic basis for color variation, and, through analyses of luciferase sequence variation, we demonstrate that natural selection has produced a long-term adaptive trend for longer wavelength (more orange) ventral light on Jamaica. Our results constitute a novel example connecting the selective fixation of specific nucleotides in nature to their precisely determined phenotypic effects. We also present evidence suggesting that a recently derived ventral orange luciferase allele on the island has deterministically increased in frequency. Thus, the current luciferase polymorphism for P. plagiophthalamus appears to be mirroring the long-term anagenic trend on Jamaica, revealing a possible ongoing adaptive color transition in progress.
When we were at Bahia, an elater or beetle (Pyrophorus luminosus, Illig.) seemed the most common luminous insect. The light in this case was also rendered more brilliant by irritation. I amused myself one day by observing the springing powers of this insect, which have not, as it appears to me, been properly described.
C. Darwin (1)
Darwinian natural selection is a central axiom of evolutionary biology. Documenting the action of selection in the wild is the mantra of the adaptationist program. However, undue fervor for the adaptive process can lead to “just so” story telling (2). When one adaptive scenario is discredited, it is quickly replaced by another instead of questioning that phenotypic form may not always be due to natural selection. Justifying repeated testing for the ecological significance of a trait requires a priori rejection of the appropriate null hypothesis of neutrality (2).
Advances in molecular genetics and genomics are opening new avenues for connecting phenotype with genotype. These methods are providing an increasingly available means for addressing the pitfall of the adaptationist program of uncritical acceptance of natural selection. By elucidating the basis for a trait at the level of the gene(s), statistical tests for selection can be performed on patterns of nucleotide variation to reject neutral evolution (3–5). Given this evidence, investigation for ecological causation can proceed unfettered. Here, we demonstrate the utility of such a molecular approach for a striking visual trait, bioluminescent color, through analysis of luciferase, the responsible gene.
Bioluminescent color in the Jamaican click beetle, Pyrophorus plagiophthalamus, provides a rare opportunity to document the entire adaptive recursion from gene to phenotype to ecological performance to demographic output (fitness) to population level change (evolution). P. plagiophthalamus is unique among bioluminescent organisms in displaying a dramatic light color polymorphism (6, 7). Beetles differ in the color of light they emit from their ventral organs from yellow–green to orange, and their paired dorsal organs from green to yellow–green (Figs. 1 and 2B). The biology of P. plagiophthalamus is suggestive of intersexual selection. Beetles use their light organs during mating in a similar manner as fireflies, although male click beetles do not flash. Males fly through the forest at night, continuously luminescing from their ventral organs searching for receptive females (U.S. and J.L.F., unpublished observation). Females remain stationary in trees or bushes, occasionally responding by using their dorsal organs. Different species of Pyrophorus beetles, although not polymorphic, differ from each other in ventral color (refs. 8 and 9, see Materials and Methods). Thus, ventral color (the male signal) has changed repeatedly in the genus and the extant polymorphism on Jamaica could represent an adaptive, sexually selected color shift in progress. But nonadaptive explanations could also account for the pattern of bioluminescence. Ventral color may be neutral or could normally be constrained by stabilizing selection. The observed interspecific shifts in Pyrophorus and the polymorphism in P. plagiophthalamus may simply reflect genetic drift occurring during periods of relaxed selection or population bottlenecks.
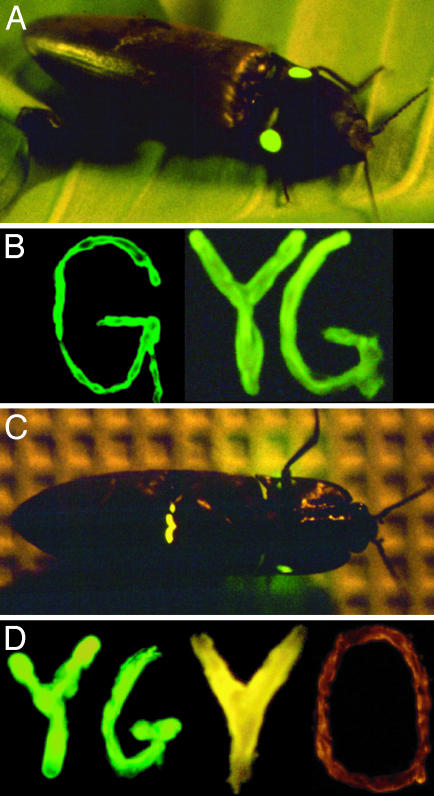
Fig. 1.
(A) Paired dorsal light organs of P. plagiophthalamus. (B) Cloned P. plagiophthalamus dorsal green (dGR) and yellow–green (dYG) luciferase alleles expressed in Escherichia coli streaked on plates. (C) Ventral light organ of a yellow bioluminescing beetle. The dorsal organ is also visible but does not bioluminesce during flight and is seen here due to the “irritated” state of the specimen. (D) Cloned ventral yellow–green (vYG), yellow (vYE), and orange (vOR) alleles.
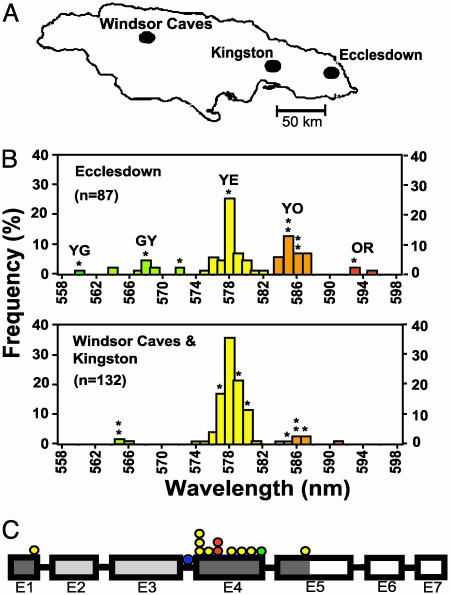
Fig. 2.
(A) Jamaican collecting sites. (B) Histogram of population frequencies for ventral organ phenotypes at Ecclesdown and pooled total for Kingston and Windsor Caves sites. Shown are peak wavelength emissions for individual beetles (in nm) as measured by a spectrophotometer. Asterisks indicate color phenotypes for 18 Jamaican beetles cloned and sequenced in this study. (C) Schematic diagram of luciferase gene for exons 1–7(E1–7). Yellow circles, nine replacement substitutions affecting light color distinguishing dGR and vYE alleles. Yellow–green circle, mutation distinguishing YG from other color alleles. Orange circles, two polymorphisms accounting for vOR allele. Blue circle, third intron difference between vYE and vOR alleles. Dark-shaded boxes, CR region; light gray boxes, NCR region.
P. plagiophthalamus possesses several features permitting a strong molecular test for natural selection on bioluminescent color to discount neutrality. First, the target gene responsible for color, luciferase, is known (10). Second, bioluminescent color can be quantified for individual beetles by using a spectrophotometer both in vivo from their intact light organs (refs. 6–12, see Materials and Methods) and in vitro from luciferase mRNA isolated and cloned from the same organs and expressed as cDNA in bacteria (refs. 12 and 13, see Materials and Methods). Thus, whole organism bioluminescence can be replicated in a Petri dish (Fig. 1). This has permitted the identification and characterization of functionally relevant nucleotides affecting color (refs. 10–12 and Table 1, which is published as supporting information on the PNAS web site). Third, light color is encoded by luciferase genes endogenous to the beetle genome. Bioluminescence is not caused by microorganisms housed in the light organs, nor is it influenced by the integument surrounding the organs or the source of luciferin (the substrate) used in the light-generating reaction (7). Amino acid changes in luciferase that affect its binding to luciferin are the sole cause for color variation. Fourth, mutations in luciferase have been found to interact in an additive fashion to determine color (10–12). Fifth, the 3D crystal structure of beetle luciferase has been determined, and the putative enzyme-substrate binding site has been identified (14). Finally, luciferase is not part of a multienzyme pathway, does not undergo posttranslational modification, and has no other known biological function in beetles besides light production (10–12). Structure–function–phenotype relationships for luciferase are therefore not complicated by pleiotropy or epistasis.
Here, we complete the causal link in P. plagiophthalamus between phenotype and genotype by (i) establishing that dorsal and ventral light color are under separate genetic control, and (ii) resolving the allelic basis for color variation. We then use the power of the P. plagiophthalamus luciferase system to test for natural selection on ventral light color at the molecular level.
Materials and Methods
Beetle Collection and Spectrophotometry. Beetles were collected at night at three different locations on Jamaica from March 7 to March 15, 1997 (Fig. 2A). In vivo spectral emission was quantified for dorsal and ventral organs of beetles (n = 219) by using a portable spectrophotometer (Spectra-Match GT, CVI Laser, Albuquerque, NM). The tip of the fiber optic input cable was held flush to a beetle's organ, and the individual was gently agitated to induce bioluminescence. Spectral emission curves were recorded on a computer using software provided by the manufacturer. A smoothing function was applied to each curve to determine peak emission wavelength (i.e., wavelength of light with highest intensity). Reported values are the means for at least three trials per organ.
Luciferase Cloning and Sequencing. Luciferase clones were generated separately for 18 P. plagiophthalamus beetles in two different ways: (i) RT-PCR of expressed mRNA isolated from each beetle's dorsal and ventral organs, and (ii) PCR of genomic DNA isolated from the heads of the same specimens. Comparisons of cDNA and genomic sequences from the same individual allowed us to deduce the genotypes of beetles and to establish the existence of separate dorsal and ventral luciferase loci. Four variable allozymes (Pgm, Mdh, Acon, and Mpi) were also scored to verify that P. plagiophthalamus represents a single species and to assess population structure on Jamaica. Messenger RNA (mRNA) was isolated from dissected light organs by using PolyAT tract System-1000 kits (Promega). RT-PCR was performed by using Access System kits (Promega) as per the manufacturer's instructions, with modified luciferase primers designed to amplify the entire coding region of the gene (≈1.6 kb). The forward (5′-GCG AGT GTT AAT TCT ACA CAT ATG ATG AAG AGA GAG-3′) and reverse primers (5′-ATT TCA TAT TGG TCT AGG TAGGAT CCC ATC ACT AGC-3′) contained NdeI and BamHI restriction sites (boldface indicates restriction enzyme recognition sequences, italics indicate modified base pairs), permitting directional cloning into pET5a expression vectors (Promega). To control for artifacts, two separate RT-PCR cloning runs were performed on the mRNA isolated from each light organ. Luciferase cDNA expression was induced with isopropyl-1-thio-ββ-d-galactopyranoside in transformed Escherichia coli BL21(DE3)pLysE competent cells (Invitrogen) and spectral emission of clones was quantified by using a grating spectrometer (10). Genomic DNA was PCR amplified 35 cycles (100-μl volume; 94°C, 30 s; 56°C, 30 s, 74°C, 2-min cycle) by using luciferase primers specific for a fragment encompassing exons 4–7. We also cloned a 656-bp segment of the third intron of the ventral luciferase gene. PCR products were cloned into the vector pCR 2.1–TOPO (Invitrogen). cDNA clones (10 per organ) and genomic DNA clones (10–12 per individual) were sequenced in both the 5′ and 3′ directions on an ALF sequencer (Amersham Biosciences). Luciferase sequences were deposited in GenBank under accession nos. AF543372, AF543378, AF543399, AF543402, AF543413, AF545853, and AF545854.
Phylogenetic Analysis. Maximum parsimony gene trees were constructed by using paup*b8 (15) to identify derived mutations unique to Jamaica (Table 1). Dorsal and ventral alleles for P. plagiophthalamus were compared with those for Pyrophorus mellifluus, the sister taxon to P. plagiophthalamus (16) from the Dominican Republic [dorsal organ peak bioluminescence (d) = 549 nm, ventral organ (v) = 554 nm], by using Pyrophorus noctilucus (Trinidad; d = 548 nm, v = 584 nm), Pyrophorus luscus (Belize; d = 547 nm, v = 578 nm) and Pyrearinus termitilluminans (17) to root trees. Sequences involved in intergenic recombination were identified by the method of Betran et al. (18). In addition, Shimodaira–Hasegawa tests (19) were conducted to test for tree topology incongruence, as implemented in paup*b8 (15).
Sequence Analysis and Tests for Selection. A variant of the McDonald–Kreitman test (20) was performed on the luciferase data by using a two-tailed Fisher's exact statistic. To test for diversifying selection on dorsal vs. ventral organ color, we used a modified QTL sign test (21). Nonsynonymous to synonymous substitution rate ratios (ω = dN/dS) were estimated by using fixed-sites, codon-substitution models for prepartitioned data (22). We discounted the bias of the extant color polymorphism in P. plagiophthalamus from our estimate of ω by using consensus sequences for the dorsal green and ventral yellow allele that were devoid of polymorphic sites and that differed only by fixed substitutions unique to Jamaican beetles. This allowed us to distinguish any past evolutionary trend for color from current variation. Pairwise nucleotide diversity values (π) were derived by using dnasp (23). F statistics for allozymes were calculated according to the method of Weir and Cockerham (24), with 95% confidence intervals estimated by bootstrapping across loci.
Coalescence Simulations. Computer simulations of the coalescence process were performed as in Hudson et al. (25) to test whether the pattern of genetic variation for the ventral orange allele departed from neutral expectation. However, instead of calculating the probability of observing a subset of i sequences with no polymorphism from a total sample of n alleles having j segregating sites, we allowed the i alleles to be variable and differ by k derived substitutions from the other n – i sequences. We also simulated two intragenic recombination events [the minimum number estimated from the ventral orange and yellow alleles by using dnasp (23)], with k = 4, j = 12, i = 10, and n = 50, representing the four fixed substitutions between ventral orange and yellow alleles in coding regions and the third intron, the 12 segregating sites for the ventral orange and yellow alleles, and the 10 ventral orange genes sequenced within a total sample size set to 50 to match the frequency of the ventral orange allele (0.201) estimated for the Ecclesdown site.
Results
Phenotypic Color Variation. Spectrophotometer measurements of in vivo light organ bioluminescence for P. plagiophthalamus revealed the existence of three dorsal color phenotypes: green (544–548 nm), lime (550–556 nm), and yellow–green (557–562 nm). Analysis of ventral organs indicated five color phenotypes: yellow–green, green–yellow, yellow, yellow–orange, and orange (Fig. 2B gives ranges for ventral phenotypes). The frequencies of ventral color phenotypes differed significantly among collecting sites (G contingency test = 46.1, P < 0.0001, 4 df, yellow–green and green–yellow and orange and yellow–orange classes were pooled for the analysis). There were more yellow/orange and green/yellow classes of beetles at Ecclesdown than elsewhere (Figs. 2 A and B).
Genetics of Dorsal and Ventral Organs. Three lines of evidence indicated that dorsal and ventral color in P. plagiophthalamus are under independent genetic control. First, spectrophotometer measurements of intact light organs indicated that dorsal and ventral color were not correlated in beetles (r = 0.01, P = 0.88, 219 df). Second, different luciferase sequences were cloned from the dorsal vs. ventral light organs of the same beetles (Table 1). Finally, comparisons of these cDNA sequences with genomic clones generated from the same beetles confirmed the existence of separate dorsal and ventral loci (data not shown), ruling out posttranscriptional modification or splicing as the cause for the observed sequence differences between light organs.
Allelic Basis for Color Variation. Spectrophotometric and sequence analysis of luciferase cDNA clones isolated from dorsal light organs revealed two dorsal color alleles: green (dGR = 548 nm peak wavelength emission) and yellow–green (dYG = 559 nm; Fig. 1B and Table 1). Analysis of ventral organ cDNA clones resolved three ventral alleles: yellow–green (vYG = 563 nm), yellow (vYE = 579 nm), and orange (vOR = 592 nm; Fig. 1D and Table 1). The results for the ventral locus generally concurred with those of Wood et al. (10). However, Wood et al. (10) reported a ventral green allele that we did not detect and that we believe represented either a cloning artifact or the rare, low-level expression of a dorsal green gene in the ventral organ.
A 1:1 relationship was found between phenotype and genotype for both dorsal and ventral light organs. For example, separate cDNA libraries generated from the ventral light organs of eight different yellow–orange colored beetles contained one vYE and one vOR allele per beetle. Similarly, all four green–yellow ventral organ cDNA libraries had just one vYG and one vYE allele each. The sole ventral orange organ cloned from a beetle generated only vOR cDNA alleles, four yellow organs generated only vYE alleles, and the single yellow–green beetle analyzed only vYG alleles. At most, two different cDNA sequences (alleles) were detected per dorsal and ventral organ for each of the 18 beetles molecularly analyzed. Genomic clones from the same specimens substantiated the cDNA findings, implying a Mendelian basis for dorsal and ventral color. Unfortunately, P. plagiophthalamus is difficult to breed in the laboratory; therefore, we cannot directly test for Mendelian inheritance through genetic crosses. Also, total genomic beetle DNA has proven recalcitrant to complete restriction enzyme digestion, precluding Southern blot analysis to verify gene copy number.
Quantitative Trait Nucleotides (QTN) Affecting Color. Wood et al. (10) identified 12 mutations affecting bioluminescent color (Table 1). These data were based on analysis of four P. plagiophthalamus luciferase color clones (GR, YG, YE, and OR) isolated from a cDNA library constructed from the pooled ventral organs of 100 beetles. As discussed above, the GR allele of Wood et al. (10) likely represented a dorsal sequence. Nevertheless, our expanded analysis of cDNA clones generated separately from the dorsal and ventral light organs of 18 beetles did not reveal any additional mutations affecting color. Phylogenetic analysis of luciferase gene trees was informative for inferring the dorsal vs. ventral locus origins for 11 of the 12 color mutations (Fig. 2C and Table 1). We could not resolve the origin of the mutation at base pair 967 distinguishing YG from the other alleles. Nine of the derived replacements originated in the ventral locus (seven fixed substitutions plus two polymorphisms at base pairs 739 and 740 accounting for the vOR allele; Table 1). All nine ventral mutations generated longer wavelength light compared with the ancestral state. In comparison, the two derived dorsal substitutions affecting color at base pairs 121 and 1,165 both resulted in shorter wavelength light. The majority of mutations affecting color reside in the fourth exon of the gene, with a couple of minor variants also present in exons 1 and 5 (Fig. 2C and Table 1). In vitro mutagenesis and modeling of the folding structure of luciferase (K.V.W., unpublished data) support these as the principal functional regions affecting color.
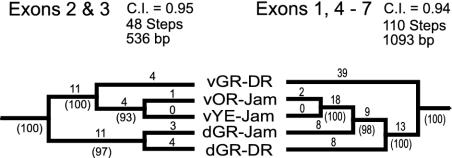
Intergenic Recombination (Gene Conversion). Comparisons of luciferase gene trees implied that P. plagiophthalamus has undergone twp episodes of dorsal/ventral intergenic recombination (Fig. 3). A basal event exchanged much of the ventral gene, except for exons 2 and 3, for dorsal sequence (Fig. 3 and Table 1). A second, more recent event helped generate the vYG and dYG alleles.
Fig. 3.
Noncongruent maximum parsimony gene trees between exons 2 and 3 vs. exons 1 and 4–7 for dorsal (d) and ventral (v) luciferase genes. Results imply a basal dorsal to ventral intergenic recombination event in P. plagiophthalamus involving exons 1 and 4–7. P level for partition–homogeneity test = 0.001; P levels for Shimodaira–Hasegawa tests for alternative tree topologies were both <0.001. Branch lengths, consistency indexes (c.i.), and bootstrap support percentages for nodes (10,000 repetitions) are given. JAM, Jamaican P. plagiophthalamus beetles; DR, P. mellifluus from Dominican Republic. Trees were rooted by using dorsal and ventral sequences generated for outgroup Pyrophorus species and a larval Pyrearinus termitilluminans allele (Materials and Methods).
McDonald–Kreitman (M–K) Test for Selection. As a first test for natural selection on luciferase, we compared ratios of polymorphism to divergence (P/D) for nonsynonymous vs. synonymous mutations (N/S) between dorsal and ventral P. plagiophthalamus genes (a variant of the M–K test for paralogous loci within a species instead of an orthologous gene among species; ref. 20). Polymorphism resulting from the recent intergenic recombination event between dYG and vYG alleles was eliminated by excluding converted YG sites from the analysis to avoid violating the M–K test assumption of nucleotide independence (see shaded base pairs in Table 1). For comparative purposes, we a priori divided the luciferase gene into two partitions designated NCR (no color, no intergenic recombination; exons 2 and 3) for regions of no apparent significance for color that were unaffected by the basal P. plagiophthalamus intergenic recombination event, and CR (color, intergenic recombination; exons 1 and 4 and exon 5 < 1,166 bp) for the areas containing functionally important sites involved in the basal dorsal-to-ventral exchange (Fig. 2C). Consistent with selection, the M–K test was significant for the phenotypically relevant CR region (P ≤ 0.011; distribution of sites: P/S = 6, P/N = 6, D/S = 1, D/N = 16), but not the NCR region (P ≥ 0.845; P/S = 4, P/N = 4, D/S = 13, D/N = 16). However, the P/D ratios for the CR region were atypical of directional selection, implying a paucity of silent fixations. Selection for preferred codons cannot explain the pattern because codon usage was not biased within or different between CR and NCR sequences [scaled χ2 for CR (G+C content 40% with Yates' correction) = 0.167; NCR (G+C 37%) = 0.108]. Also, silent substitution rates in the CR region were not unusual in other beetles (CR = 13 autapomorphies per 137 silent sites in P. mellifluus, P. noctilucus and P. luscus vs. 21 autapomorphies per 209 silent sites for rest of gene; P = 1.00, Fisher exact test). Instead, we hypothesize that the unusual P/D ratio was caused by the basal intergenic recombination event in P. plagiophthalamus purging the CR region of dorsal/ventral differences and ventral polymorphism, followed by the rapid fixation of a series of selectively driven ventral mutations.
Substitution Rate Ratios. To investigate the rapid fixation hypothesis, we compared nonsynonymous to synonymous substitution rate ratios (ω) between CR and NCR regions for derived dorsal and ventral substitutions unique to Jamaican beetles. The ω ratio of 3.49 for the CR region (dN = 0.0217 ± 0.0069, dS = 0.0062 ± 0.0062) was significantly greater than the 0.040 value (dN = 0.0023 ± 0.0023, dS = 0.058 ± 0.0241) for the NCR region (χ2 log-likelihood ratio test = 13.2, P = 0.0013, 2 df, model C vs. E of Yang and Swanson, ref. 22). Ten autapomorphic replacement substitutions were largely responsible for the high ω ratio in the CR region of luciferase (Fig. 2C and Table 1). Consistent with the rapid fixation hypothesis, 9 of the 10 replacements occurred in the ventral P. plagiophthalamus gene (P < 0.021 for two-tailed binomial test). The sole silent change at base pair 1,051 was in the dorsal gene. A release from stabilizing selection, rather than positive Darwinian selection, could conceivably explain the accelerated rate of nonsynonymous substitution in the ventral CR region. However, this interpretation is at odds with the ω ratio for the ventral CR region being >1 [χ2 = 7.62, P = 0.0058, 1 df, model M2 of Yang and Swanson (22) with ω fixed to 1 vs. estimated from data; alternatively, P = 0.049 for a binomial test that all nine derived substitutions in the ventral CR region are replacement changes based on the expected proportions of synonymous (0.284) vs. nonsynonymous (0.716) mutation estimated from codon usage in the CR region of the ancestral P. plagiophthalamus ventral gene and relative nucleotide substitution rates derived from the Pyrophorus sequence data]. An ω ratio >1 is a general indicator of diversifying (directional) selection (22).
QTL Sign Test. We used a modified QTL sign test (21) to further test for diversifying selection on bioluminescent color between dorsal and ventral organs. The QTL sign test examines how often one expects to see the observed number of positive effect QTL in a high phenotypic line, given all of the ways of neutrally evolving a phenotypic difference of R or more between high and low lines based on a known distribution of QTL effects. In our case, we considered R to represent the 31-nm wavelength difference between dGR and vYE luciferase alleles. These two genes were compared because they represent the likely color states for the dorsal and ventral organs, respectively, before the origins of the more recently derived dYG, vYG, and vOR alleles. Instead of QTL, our test involved using the characterized distribution of phenotypic effects of luciferase point mutations on bioluminescent color (i.e., a QTN analysis based on Orr's standard and not equal effects QTL test). The dGR and vYE genes are distinguished by nine fixed nonsynonymous substitutions affecting color (Fig. 2C and Table 1). In all nine cases, the vYE allele contained the nucleotide resulting in longer wavelength bioluminescence. Through computer simulations (10,000 repetitions), we estimated that the probability was 0.039 and that neutral evolution would generate a ≥31-nm difference in color with all nine substitutions in the vYE allele fixed for mutations having a positive effect on wavelength, supporting the diversifying selection hypothesis.
When applied in an evolutionary context, a QTL or QTN sign test does not, in itself, indicate which lineage has experienced selection. Additional information about the character state of the common ancestor is needed to assess directional change within lineages. For bioluminescence, phylogenetic analysis of luciferase gene trees can provide these data. In our test, seven of the nine mutations responsible for the color difference between dGR and vYE alleles were derived changes unique to the ventral gene, and all seven caused longer wavelength emission. Refining the analysis further, five of these seven ventral mutations were autapomorphic changes confined to Jamaican beetles (Table 1). Two additional, longer wavelength mutations in the ventral P. plagiophthalamus gene accounted for the derivation of the vOR allele. Thus, seven derived ventral mutations (five fixed differences and two polymorphisms) can be unambiguously assigned to the ventral locus of P. plagiophthalamus. The probability that all seven mutations would result in longer wavelength light, as was observed, is 0.0156 (two-tailed binomial test). These data support the occurrence of a long-term adaptive trend on Jamaica favoring more orange-colored ventral bioluminescence.
Selection on the vOR Allele. The evolutionary trend in P. plagiophthalamus toward orange light raises the question of whether the most recently derived vOR allele is undergoing a selective sweep on Jamaica. Several lines of evidence support this hypothesis. First, in the coding region of luciferase, vOR differs from vYE by three unique replacement, but zero silent, substitutions. Additional sequence data for a segment of the third intron of the ventral gene revealed a fourth difference. Second, the vOR allele class, although common at Ecclesdown (frequency = 0.201 under a single locus model), displayed lower nucleotide diversity on Jamaica (π = 0.00046 ± 0.00022, n = 10 for coding region) compared with the vYE class (π = 0.00129 ± 0.00015, n = 20). The combination of vOR divergence, moderate frequency at Ecclesdown, and limited polymorphism deviated significantly from a neutral coalescence model (P = 0.038; see Materials and Methods). Rather, the pattern implies that vOR has deterministically increased in frequency because of natural selection. Indeed, the estimated frequency of vOR was much higher at Ecclesdown than elsewhere (35/174 = 0.201 vs. 10/264 = 0.038; P < 10–7, Fisher exact test; Fig. 2 A and B), suggesting that the allele arose on the eastern side of the island and is spreading westward. It is unlikely that the pattern was caused by a population bottleneck, because allozymes indicated only modest intraisland differentiation [FST = 0.022 (0.007–0.048 95% confidence interval)]. Although we cannot rule out the possibility that the color polymorphism may stabilize because of balancing or frequency-dependent selection, our data imply that microevolutionary change on Jamaica is mirroring the history of selection for longer wavelength bioluminescence.
Discussion
We have connected several causal links in the adaptive chain (recursion) from gene to phenotype to population change for bioluminescent color in P. plagiophthalamus. We accomplished this by transforming a measurable in vivo trait (light organ bioluminescence) into a quantifiable in vitro phenotype. Combining this information with phylogenetic and population level analysis of DNA sequence variation in luciferase allowed us to identify a long-term evolutionary trend in P. plagiophthalamus favoring orange bioluminescence. Our results therefore constitute a novel example connecting the fixation of a series of selectively driven nucleotide substitutions in nature to their precisely determined phenotypic effect.
The molecular analyses we used in the study were not unique. However, the approach of working at the level of the underlying gene can be a valuable strategy for countering criticism of the adaptationist program of unwarranted acceptance of natural selection before rejection of neutrality. By discounting the neutral hypothesis for luciferase, we can now justify repeated testing for ecological causation to forge the next links in the adaptive chain for bioluminescent color from phenotype to performance to fitness. Possible explanations for the adaptive significance of more orange ventral light include: (i) sexual selection, (ii) predation from a nocturnal visual predator such as bats or birds, and (iii) character displacement caused by spectral competition from fireflies or another, now extinct, congeneric beetle.
In addition to resolving click beetle ecology, several other pertinent questions also remain concerning whether and how natural selection has affected the dorsal polymorphism, the vYG allele, and interspecific color differences among Pyrophorus species. Studies are also called for to quantify the visual acuity of beetles for different wavelengths of light to test for evidence of coevolution with bioluminescent color (26, 27). In conclusion, with the development of ever-more powerful genetic tools, we envision verification of selection at the molecular level as becoming a logical step in testing the adaptive hypothesis for a trait (28). We can only speculate on what might have been if the voyage of a young English naturalist with an amusing fondness for beetles had visited Jamaica.
Supplementary Material
Acknowledgments
We thank H. Akashi, H. Dambroski, K. Filchak, S. Kappe, M. Kreitman, N. Lobo, K. McCluskey, D. Reznick, J. Roethele, J. Wallace, and the Dominican Republic Department of Natural Resources. This work was supported by University of Notre Dame faculty and State of Indiana 21st Century grants (to J.L.F.) and Notre Dame Presidential and National Science Foundation Graduate Research Training Fellowships (to U.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: v, ventral light organ; d, dorsal; GR, green; YG, yellow–green; GY, green–yellow; YE, yellow; YO, yellow–orange; OR, orange; CR, color, intergenic recombination; NCR, no color, no intergenic recombination; QTN, quantitative trait nucleotide.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF543372, AF543378, AF543399, AF543402, AF543413, AF545853, and AF545854).
References
- 1.Darwin, C. (1988) Diary of the Voyage of H. M. S. Beagle, ed. Keynes, R. D. (Cambridge Univ. Press, New York), p. 19.
- 2.Gould, S. J. & Lewontin, R. C. (1979) Proc. R. Soc. London Ser. B 205, 581–598. [DOI] [PubMed] [Google Scholar]
- 3.Kreitman, M. & Akashi, H. (1995) Annu. Rev. Ecol. System. 26, 403–422. [Google Scholar]
- 4.Yang, Z. & Bielawski, J. P. (2000) Tree 15, 568–573. [Google Scholar]
- 5.Ford, M. J. (2002) Mol. Ecol. 11, 125–1262.11903910 [Google Scholar]
- 6.Seliger, H. H., Buck, J. B., Fastie, W. G. & McElroy, W. D. (1964) J. Gen. Physiol. 48, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biggley, W. H., Lloyd, J. E. & Seliger, H. H. (1967) J. Gen. Physiol. 50, 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colepicolo-Neto, P., Costa, C. & Bechara, E. J. H. (1986) Insect Biochem. 5, 803–810. [Google Scholar]
- 9.Bechara, E. J. H. (1988) Adv. Oxygen. Process. 1, 123–178. [Google Scholar]
- 10.Wood, K. V., Lam, Y. A., Seliger, H. H. & McElroy, W. D. (1989) Science 244, 700–702. [DOI] [PubMed] [Google Scholar]
- 11.Wood, K. V. (1990) J. Biolumin. Chemilum. 5, 107–114. [DOI] [PubMed] [Google Scholar]
- 12.Wood, K. V. (1995) Photochem. Photobiol. 62, 662–673. [Google Scholar]
- 13.DeWet, J. R., Wood, K. V., Helinski, D. R. & DeLuca, M. (1985) Proc. Natl. Acad. Sci. USA 82, 7870–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conti, E., Franks, N. P. & Brick, P. (1996) Structure (London) 4, 287–298. [DOI] [PubMed] [Google Scholar]
- 15.Swofford, D. L. (2000) PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b8.
- 16.Costa, C. (1976) Papeis Avulsos de Zoologia 29, 141–154. [Google Scholar]
- 17.Viviani, V. R, Silva, A. C. R., Perez, G. L. O., Santelli, R. V., Bechara, E. J. H. & Reinach, F. C. (1999) Photochem. Photobiol. 70, 254–260. [DOI] [PubMed] [Google Scholar]
- 18.Betran, E., Rozas, J., Navarro, A. & Barbadilla, A. (1997) Genetics 146, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimodaira, H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 1114–1116. [Google Scholar]
- 20.McDonald, J. H. & Kreitman, M. E. (1991) Nature 351, 652–654. [DOI] [PubMed] [Google Scholar]
- 21.Orr, H. A. (1998) Genetics 149, 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, Z. & Swanson, W. J. (2002) Mol. Biol. Evol. 19, 49–57. [DOI] [PubMed] [Google Scholar]
- 23.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- 24.Weir, B. S. & Cockerham, C. C. (1984) Evolution (Lawrence, Kans.) 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- 25.Hudson, R. R., Bailey K., Skarecky D., Kwiatowski J. & Ayala, F. J. (1994) Genetics 136, 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cronin, T. W., Jarvilehto, M., Weckstrom, M. & Lall, A. B. (2000) Comp. Physiol. 186, 1–12. [DOI] [PubMed] [Google Scholar]
- 27.Lall, A. B., Ventura, D. S. F., Bechara, E. J. H., de Souza, J. M., Colepicolo-Nito, P. & Viviani, V. R. (2000) J. Insect Physiol. 46, 1137–1141. [DOI] [PubMed] [Google Scholar]
- 28.Sabeti, P. C., Reich, D. E., Higgins, J. M., Levine, H. Z., Richter, D. J., Schaffner, S. F., Gabriel, S. B., Platko, J. V., Patterson, N. J., McDonald, G. J., et al. (2002) Nature 419, 832–837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.