Abstract
Experiments and molecular simulations have shown that the hydrophobic mismatch between proteins and membranes contributes significantly to lipid-mediated protein-protein interactions. In this article, we discuss the effect of cholesterol on lipid-mediated protein-protein interactions as function of hydrophobic mismatch, protein diameter and protein cluster size, lipid tail length, and temperature. To do so, we study a mesoscopic model of a hydrated bilayer containing lipids and cholesterol in which proteins are embedded, with a hybrid dissipative particle dynamics-Monte Carlo method. We propose a mechanism by which cholesterol affects protein interactions: protein-induced, cholesterol-enriched, or cholesterol-depleted lipid shells surrounding the proteins affect the lipid-mediated protein-protein interactions. Our calculations of the potential of mean force between proteins and protein clusters show that the addition of cholesterol dramatically reduces repulsive lipid-mediated interactions between proteins (protein clusters) with positive mismatch, but does not affect attractive interactions between proteins with negative mismatch. Cholesterol has only a modest effect on the repulsive interactions between proteins with different mismatch.
Introduction
The detection of an increasing number of transmembrane proteins in submicrometer-sized clusters (1–5) with organization that strongly depends on the membrane composition (e.g., lipid tail length (1,3,4) or cholesterol content (3,6–11)) has seriously challenged the idea that individual proteins freely diffuse in a passive biological membrane (12) and only interact via direct protein-protein interactions. Understanding the role of the membrane in the interactions between membrane proteins has become an active area of research (13,14).
In these studies, hydrophobic mismatch, i.e., the difference, d, between the length of the hydrophobic region of the protein and the bilayer hydrophobic thickness, has been frequently invoked as an important physical property that regulates lipid-protein and lipid-mediated protein-protein interactions. For example, several studies have shown that modulations of the bilayer thickness, protein tilting, protein functioning, and protein aggregation strongly depend on the protein hydrophobic mismatch, on the protein diameter, and on the lipid properties (14–34). The concept of hydrophobic mismatch has therefore significantly contributed to the insight that lipids and proteins show a collective behavior.
Theoretical studies on protein-lipid and indirect protein-protein interactions investigate mostly single-component bilayers. Several models also exist of membrane-mediated organization in lipid membranes containing two types of lipids or two phases (35–37). Examples are the lipid-annulus model (35) and the wetting and capillary condensation model (36). These models show that a preferential interaction between the proteins and a specific lipid or phase might lead to a clustering of the proteins.
In this article, we present a molecular simulation study of the effect of cholesterol on protein-lipid and lipid-mediated protein-protein interactions. Cholesterol has a significant effect on protein aggregation (3,5–11) and on a wide range of other membrane processes involving proteins (38–40). These effects are intimately related to various diseases (41).
Different mechanisms have been identified to explain why cholesterol affects the proteins. For many proteins, the exact mechanism remains unclear. In general, these mechanisms have been strongly related to the partitioning of proteins in cholesterol-rich or in cholesterol-poor regions (42,43). Three fundamentally different but not mutually exclusive mechanisms have been proposed (41–43):
-
1.
Many proteins have been observed to have a specific sterol-sensing domain (44). As a result, proteins and cholesterol interact preferentially and directly with each other, leading to a partitioning of proteins in cholesterol-rich regions.
-
2.
Indirect attractive protein-cholesterol interactions could originate from a preferential interaction between a smooth protein surface and the rigid cholesterol tetrameric ring. Proteins might be expelled from sterol-rich domains when the opposite is true (45).
-
3.
The presence of cholesterol induces changes in the bilayer material and biophysical properties, which could influence the protein-lipid and protein-protein interactions (3,8,9).
The third mechanism is the most frequently invoked. For example, in model membranes, cholesterol-rich regions might have an increased area compressibility modulus, which could lead to a partitioning of the membrane components (39,43,46). The chain ordering and bilayer thickening effect of cholesterol could lead to hydrophobic mismatch between the hydrophobic part of the protein and the bilayer or change the hydrophobic mismatch conditions such that lipid-mediated protein aggregation takes place (3,8,9).
Experiments indicate that the effect of cholesterol on the bilayer properties strongly depends on lipid type and temperature (47). For example, at physiologically relevant temperatures, the effects of cholesterol are smaller on unsaturated lipids compared to saturated lipids (47). Moreover, in real biological membranes, the addition of cholesterol does not seem to modify the membrane thickness (17). Interestingly, the increased protein aggregation in the presence of cholesterol has been observed in saturated bilayers (9), in unsaturated lipid bilayers (3,8,11), and in real membranes (6,7,10). The fact that cholesterol promotes protein clustering in different types of bilayers, but that its effect on structural and mechanical properties is not universal, indicates that there might be another mechanism. Moreover, protein clusters have been observed to be enriched in cholesterol (6), which is not accounted for by mechanisms implying a change in bilayer properties, although there might be a combination with the indirect or direct cholesterol-protein interaction mechanisms.
The mechanism implying a change of the bilayer properties focuses solely on the effect of lipids on the organization of proteins, while in reality there should also be a strong effect of proteins on the distribution of the lipids and on the properties of the bilayer, as proteins make up to 50% of the membrane. In this context the hypothetical concept of a lipid shell, a lipid domain surrounding a protein and induced by the latter, was introduced (42,48). Lipid shells might extend up to 10 nm from the protein surface, might be enriched or depleted in cholesterol, but do not need to form a separate phase and exist as mobile entities in the plane of the membrane. Lipid shells should not be confounded with the first-shell lipids (49) or the lipid annulus (35,48), which is the first layer of lipids surrounding the proteins due to direct or indirect chemical and physical interactions. Multiple ways have been proposed in which shells might form; for example, due to hydrophobic mismatch between protein and lipids (42).
Recent experiments indicate that model proteins with positive mismatch, when embedded in a bilayer containing DMPC (dimyristoylphosphatidylcholine) and cholesterol, are surrounded by a cholesterol-enriched region, while less cholesterol is observed around proteins with a smaller hydrophobic length (50). This experiment supports the shell hypothesis. Similarly, Epand and co-workers (51–53) have reported several experimental studies indicating a protein-induced formation of cholesterol-rich domains. It thus seems that there might exist another mechanism by which cholesterol affects protein-protein interactions: the lipid shell mechanism. When cholesterol is added to a bilayer, certain proteins might induce a cholesterol-enriched or cholesterol-depleted lipid shell and those lipid shells might subsequently affect the interactions between the proteins.
Experimentally it would be a difficult task to distinguish among the different mechanisms. Atomistic simulation studies identified specific cholesterol-sensing domains of certain proteins (44). However, for a systematic study of the collective behavior of lipids and proteins within a reasonable simulation time, a mesoscopic approach is required.
In this article, we build on a previously developed coarse-grained model of a hydrated bilayer containing a phospholipid and cholesterol and in which peptides are embedded (28,54–56). We systematically study the effect of cholesterol on the interactions between proteins. In particular, we use the flexibility of our model to study the effect of cholesterol at conditions where cholesterol hardly changes the properties of the bilayer and at conditions where cholesterol has a larger influence on the bilayer properties. We compute the potential of mean force (PMF) between two proteins with positive mismatch in a bilayer with and without 40 mol % cholesterol at different temperatures and provide evidence for the lipid shell mechanism.
Model and Simulation Methods
Model
In our model, groups of atoms are lumped into one mesoscopic pseudoatom. One water bead comprises three water molecules. A model lipid consists of three hydrophilic head beads to which two tails of hydrophobic beads are attached. In this article we will consider lipids with four and five beads, denoted as h3(t4)2 and h3(t5)2, respectively. The cholesterol model contains a hydrophilic bead and a hydrophobic ring to which a tail of two hydrophobic beads is attached. The h3(t4)2-cholesterol bilayer gives a reasonable representation of DMPC-cholesterol (55,56). Proteins are modeled as a bundle of NP hydrophobic chains, both ends of which are hydrophilic. By varying the number of chains and the number of hydrophobic beads per chain, one can change the diameter and the hydrophobic length of the protein, respectively. Our proteins have no specific sterol-sensing domain. The model is shown in Fig. 1.
Figure 1.
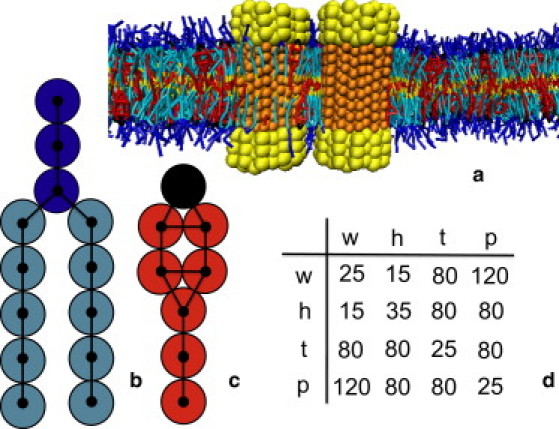
Coarse-grained model and soft-repulsive interaction parameters. (a) Snapshot of a model bilayer containing phospholipids and cholesterol and in which two proteins are embedded. Water (type w) is not shown for clarity. The protein hydrophilic (type h) and hydrophobic (type p) beads are depicted in yellow and orange, respectively. (b) Coarse-grained model of a h3(t5)2 phospholipid. Hydrophilic head beads (type h) are depicted in dark blue, while the hydrophobic tail beads (type t) are depicted in light blue. (c) Coarse-grained model of cholesterol. The hydrophilic (type h) and hydrophobic (type t) beads are depicted in black and red, respectively. (d) Table with the soft-repulsive interaction parameters aij between the four types of beads: water (w), hydrophilic (h), hydrophobic (t), and hydrophobic protein bead (p).
The nonbonded interactions between beads are described with soft-repulsive interactions,
| (1) |
where the coefficients aij > 0 represent the maximum repulsion strength, rij is the distance between beads i and j, and Rc is the cutoff radius, which gives the range of the interaction. A soft-repulsion is obtained from coarse-graining atomistic simulations (57). Contrary to atoms, beads containing groups of atoms might overlap more, and thus the interaction should be less repulsive than a classical Lennard-Jones potential. In our model, the parameters of these interactions, aij, are related to the Flory-Huggins solubility parameters (58) and are provided in Fig. 1. The intramolecular interactions include bond vibrations and bond bending (see the Supporting Material). Previous applications of this model are given in the Supporting Material.
Simulation method
The mesoscopic model was studied using a hybrid dissipative particle-dynamics Monte Carlo (DPD-MC) method. Simulations were performed in the NP⊥γT ensemble, with γ = 0. All simulations were performed in bilayers containing ∼3000 lipid or cholesterol molecules. For every cholesterol or lipid molecule, we have 25 water beads. A detailed description of the simulation method can be found in the Supporting Material and in the literature (57,59,60).
For the simulations, we use a reduced temperature. The relation with the physical temperature can, for example, be estimated from comparison with experimental main and pretransition temperatures. For h3(t4)2, one obtains T = 108.75 Tr −8.6, with Tr the reduced temperature and T the Celsius temperature (56). For h3(t5)2, one obtains T = 133 Tr −33 (28). For phospholipid-cholesterol systems, it is useful to present the results as a function of ΔT = T–Tm, with Tm the reduced temperature at which the main phase transition of the bilayer takes place:
Results
Effect of cholesterol on lipid bilayer
Before discussing the potentials of mean force (PMFs), it is useful to briefly review some aspects of the temperature-dependence of the effects of cholesterol on a phospholipid bilayer. In Fig. 2 a, the partial phase diagram is shown for a h3(t4)2-cholesterol system (56). At high temperatures, the h3(t4)2 bilayer is in a liquid-disordered phase, while below the main phase transition temperature (Tm) the bilayer is in a rippled gel phase. Addition of cholesterol above Tm results in a gradual transition (crossover) of the liquid-disordered phase to a so-called liquid-ordered phase. At sufficiently high temperatures, the bilayer remains in the liquid-disordered phase.
Figure 2.
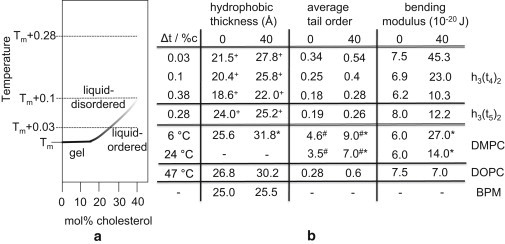
(a) Simulated partial phase diagram of h3(t4)2-cholesterol (55,56), with Tm the main phase transition temperature. (b) Effect of cholesterol on the properties of a phospholipid bilayer at different temperatures ΔT = T–Tm. Data are reported for 0 and 40 mol% cholesterol. The data for h3(t4)2 and h3(t5)2 are obtained from simulations; the data for DMPC (dimyristoylphosphatidylcholine), DOPC (dioleoylphosphatidylcholine), and basolateral plasma membrane (BPM) are experimental (17,47,67). DMPC and DOPC bilayers are examples of model saturated and unsaturated membranes, respectively, whereas BPM is an example of a real membrane. Tm,DMPC = 24°C, Tm,DOPC = −17°C. (Asterisks) Data for 30 mol % cholesterol; (plus-signs) distance between the hydrophobic beads linked to the hydrophilic ones; (pound signs) first moment (10−4 Hz) of NMR spectrum, which is proportional to the tail order parameter; and (minus-signs) no data available.
The effect of cholesterol on the bilayer hydrophobic thickness, average lipid tail order, and bilayer bending modulus is shown in Fig. 2 b for three different temperatures above Tm. For ΔT = T–Tm = 0.03, cholesterol significantly increases all three parameters due to the transition to the liquid-ordered phase. For ΔT = 0.1, the effect of cholesterol on the bending modulus is strongly reduced, and for ΔT = 0.38 the effect on all three parameters is much weaker. As shown in Fig. 2 b, similar trends have been observed experimentally (for example, for the DMPC-cholesterol system (56)).
These different temperatures allow us to separate the different effects of cholesterol on the interactions of proteins. At high temperatures, cholesterol has very little effect on the properties of the bilayer. Experimentally, this would correspond to a high-temperature DMPC bilayer—or, if we use a different mapping of our mesoscopic parameters, to those types of bilayer for which the addition of cholesterol has little effect (for example, strongly unsaturated lipid bilayers and real membranes (17)).
Protein clustering
To explore the effect of cholesterol on the interactions between small transmembrane proteins, we simulated a system consisting of 49 small (diameter: 13.5 Å) proteins inserted in a large h3(t5)2 bilayer of 100 nm2 at ΔT = 0.28. The proteins have a positive mismatch of 3.5 Å. Initially, the proteins were located at a maximal distance from each other on a square lattice. Fig. 3, a and b, shows snapshots after 2 × 106 DPD-MC cycles, which corresponds to ∼0.1–0.4 ms (61). Without cholesterol, relatively small protein clusters form (Fig. 3 a), whereas with 40 mol % cholesterol, we observed the formation of much larger protein clusters (Fig. 3 b).
Figure 3.
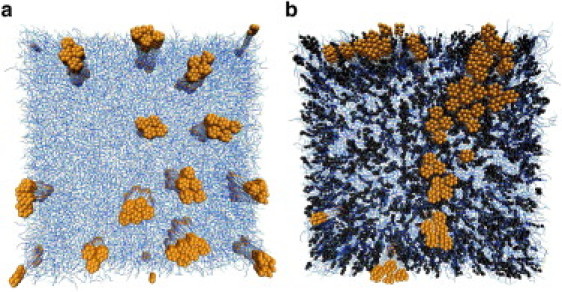
Snapshot of a top view of a lipid bilayer after 106 MC-DPD cycles. The lipids are depicted in blue, cholesterol in black. The proteins, in orange, have a small diameter of 13.5 Å and a positive mismatch of 3.5 Å in panel a and 2.3 Å in panel b. Water beads are not shown for clarity. Periodic boundary conditions apply. Initially the proteins were embedded as far as possible from each other. In panel b, the h3(t5)2 bilayer contains 40 mol% cholesterol. ΔT = 0.28.
These simulations indicate that the lipid-mediated interactions between the proteins depend on the presence of cholesterol. Ideally, one would like to use these simulations to quantitatively compare with, for example, the FRET efficiency measured during relevant protein clustering experiments (3,4). However, detailed analysis shows that these simulations are not sufficiently long and that our system size is not sufficiently big to determine reliable equilibrium cluster-size distributions from these snapshots. The simulations, however, do suggest a marked effect of cholesterol on the interactions between membrane proteins—i.e., cholesterol enhances the formation of clusters, which is also observed experimentally (3,5–11). To quantify these effects of cholesterol on the interactions between proteins, we computed the potential of mean force (see the Supporting Material). The PMF is an equilibrium property and quantifies the effective interaction between proteins (clusters), but does not give any information on protein cluster size distribution.
Interactions between two proteins
Effect of cholesterol
We computed the PMF between two large proteins (NP = 43) with diameter 32 Å at the three temperatures ΔT = 0.03, 0.1, and 0.38 in a h3(t4)2-cholesterol bilayer. In Fig. 4 a, the PMFs are shown for ΔT = 0.03 and 0.1. In a pure h3(t4)2 bilayer, the PMFs are mainly repulsive, except for the short-range direct protein-protein contact. The cut-off diameter of the soft-repulsive interactions is 6.46 Å (see the Supporting Material), and thus the repulsive interaction between the proteins is entirely lipid-mediated. In Fig. 4 a, we also show an experimental PMF (62), for a comparable system: alamethicin pores (diameter 36 Å) in DMPC at 30°C (Tm,DMPC = 24°C). It is believed that alamethicin has a slight positive mismatch in DMPC because of its tilt (62). The amplitude and the range of the simulated and experimental repulsive interaction agree very well. The experimental PMF was obtained by fitting a quadratic potential to x-ray diffraction data. Therefore, this PMF does not show the short-range oscillations.
Figure 4.
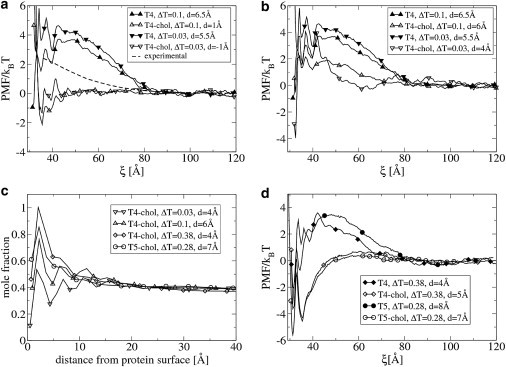
(a and b) Potential of mean force as a function of the distance ξ between two proteins with diameter 32 Å, in a h3(t4)2 bilayer with and without 40 mol % cholesterol at different temperatures ΔT = T–Tm. The value d is the positive hydrophobic mismatch between bilayer and protein. The dotted line represents the experimental PMF. In panel a, the value d changes because of the bilayer thickening effect of cholesterol. In panel b, we keep d constant by modifying the hydrophobic length of the protein. (c) Mole fraction of cholesterol as function of the distance from the protein surface. The h3(t4)2 and h3(t5)2 bilayer contains 40 mol% cholesterol. (d) Same as for panel b, but at higher values of ΔT.
At these low temperatures, cholesterol significantly increases the hydrophobic thickness of the membrane. Due to the addition of 40 mol % cholesterol, the bilayer hydrophobic thickness becomes approximately the same as the protein hydrophobic length. The hydrophobic mismatch becomes d = −1.0 and d = 1.0 Å, for ΔT = 0.03 and 0.1, respectively. As a result, we expect that the lipid-mediated interactions disappear. Indeed, this is exactly what we observe, as shown in Fig. 4 a. It is interesting to compare these results with a situation where the hydrophobic mismatch remains constant. We changed the length of the protein such that the hydrophobic mismatch is the same in a bilayer with and without cholesterol. This was achieved by increasing the protein hydrophobic length by one bead. The use of a coarse-grained model does not allow us to have exactly the same hydrophobic mismatch in every case. The results are shown in Fig. 4 b. The repulsive interactions are again strongly reduced, particularly between 45 and 80 Å. This indicates that a change in hydrophobic mismatch is not the only mechanism by which cholesterol affects the protein-protein interactions.
In Fig. 4 c, the mole fraction of cholesterol is shown around a single protein for ΔT = 0.03 and 0.1. The protein surface is enriched in cholesterol, most likely due to the preferential interaction between the rigid cholesterol ring and the smooth protein surface. However, the cholesterol-enrichment clearly extends up to a distance of 3 nm from the protein surface. This corresponds to 5–6 layers of lipids. The clear presence of maxima and minima in the curves suggests a liquid-ordered phase. In fact, the fraction of cholesterol in this domain is strongly dependent on the hydrophobic mismatch. A cholesterol-depleted region is observed around proteins with negative mismatch (see the Supporting Material).
Anderson and Jacobson (42) defined lipid shells as protein-induced domains which might extend up to 10 nm from the protein surface, and which might be enriched or depleted in cholesterol, and do not need to form a separate phase but exist as mobile entities in the plane of the membrane. This description indicates that the protein-induced domains we observe are lipid shells.
It is, however, unclear whether cholesterol reduces the repulsive protein interactions due to the presence of the cholesterol-enriched shells. Indeed, as shown in Fig. 2 b, the addition of cholesterol also significantly increases the average lipid-tail order and the bending modulus. The change in these parameters could also affect the protein interactions.
Therefore, we performed a similar simulation at a higher temperature: ΔT = 0.38. At this temperature, the effect of cholesterol on the average tail order and bending modulus is considerably less (see Fig. 2 b). The hydrophobic mismatch is again kept constant. The results are shown in Fig. 4 d. In a pure h3(t4)2 bilayer, the PMF is similar to the PMF obtained at lower temperatures. When 40 mol % cholesterol is added to the bilayer, the repulsive interaction between 45 and 80 Å is again strongly reduced. Moreover, the short distance between proteins is dramatically stabilized. In Fig. 4 c is shown that also for this case there exists a cholesterol-enriched shell around the proteins. Thus, it is very likely that the protein-induced, cholesterol-enriched domains also affect the lipid-mediated protein-protein interactions.
To check whether our results depend on the lipid tail length, we performed similar simulations in a h3(t5)2 bilayer. The results for ΔT = 0.28 are shown in Fig. 4, c and d, and are very similar to the h3(t4)2 system. Because at higher temperature the lipid shell mechanism is less superadded by effects of cholesterol on the structural and mechanical bilayer properties, we use the h3(t5)2-cholesterol system at ΔT = 0.28 to further investigate the lipid shell mechanism.
The lipid shell mechanism
The region surrounding one protein with positive mismatch is slightly enriched in cholesterol. We simulated the local membrane composition around two interacting proteins. For example, from Fig. 5, a and b (red line), one can see an increasing cholesterol concentration between two proteins interacting at 58 and 45 Å. At a distance of 58 Å, the fraction of cholesterol between both proteins is 0.5, while at 45 Å it becomes 0.8. In Fig. 6, the average cholesterol mole fraction around both proteins is shown for the entire membrane. According to the lipid shell mechanism, the effect of cholesterol on the PMF between two proteins with positive mismatch is related to the local composition around the two interacting proteins.
Figure 5.
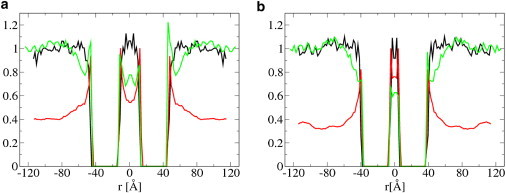
Average cholesterol mole fraction (red) and hydrophilic shielding parameters (black, green) around two large proteins. Simulations were performed in a h3(t5)2 bilayer without (green lines) and with (black lines) 40 mol % cholesterol. The proteins are at a distance of 58 Å (a) and 45 Å (b). Proteins have a diameter of 32 Å and a positive mismatch of +7 Å. ΔT = 0.28.
Figure 6.
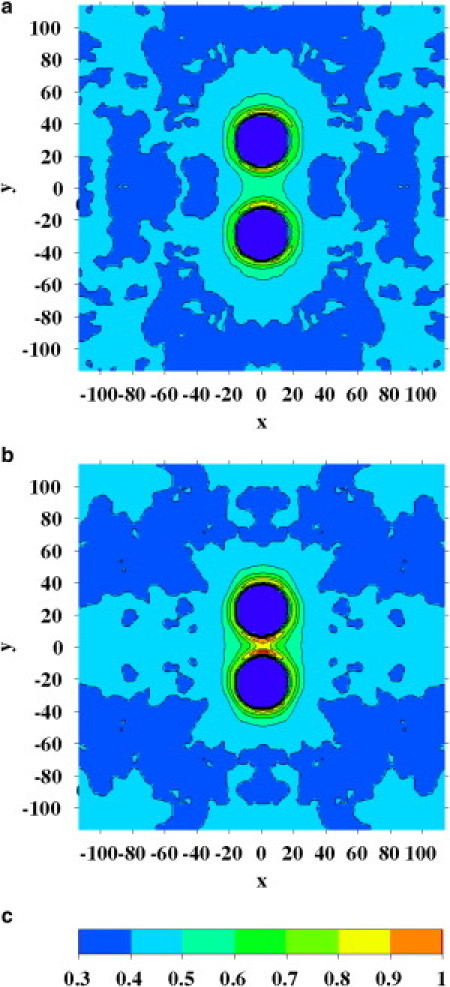
Average cholesterol mole fraction around two large proteins with diameter 32 Å with a positive mismatch of 7 Å. The proteins (dark blue circles) are at a distance of 58 Å (a) and 45 Å (b). The h3(t5)2 bilayer contains 40 mol % cholesterol. The values were averaged over both bilayer leaflets. Distances are in Å. ΔT = 0.28.
To understand how the high cholesterol fraction between the two approaching proteins affects the PMF, the concept of hydrophilic shielding is useful. In a previous article, we introduced the hydrophilic shielding parameter as a measure for the relative number of hydrophilic beads shielding the hydrophobic beads from the water, at a given position in the membrane plane (30) (see the Supporting Material).
Fig. 5, a and b, shows the hydrophilic shielding parameter for the two proteins. In a bilayer without cholesterol (green lines), at a distance sufficiently far from the proteins, the bilayer is not perturbed due to the presence of the proteins and the value of the hydrophilic shielding parameter fluctuates per definition at ∼1. In the regions close to, and in particular in between, the proteins there is a large deviation from the optimal hydrophilic shielding. When both proteins come closer, from 58 to 45 Å, the hydrophilic shielding in-between both proteins further decreases. Thus, the lipids in between the proteins can only reorganize in a way that further decreases the hydrophilic shielding in-between the proteins. This is reflected in the PMFs by the free energy increase between 58 and 45 Å (see Fig. 4 d).
The addition of cholesterol causes a completely new organization of the remaining lipids and cholesterol, with an improved shielding as result (Fig. 5, black lines). This effect is particularly strong in-between both proteins, where the shielding is now ∼1. Because the hydrophilic shielding parameter is a measure of the membrane perturbation due to the presence of the proteins, this result indicates that cholesterol naturally alleviates the unfavorable lipid perturbations due to positive hydrophobic mismatch. When both proteins come closer, from 58 to 45 Å, the average hydrophilic shielding in-between both proteins does not change. This is reflected in the PMFs by the constant free energy between 58 and 45 Å (see Fig. 4 d). In the Supporting Material, we discuss the effect of cholesterol on the hydrophobic thickness profile around both proteins.
PMFs for protein clustering
To obtain some quantitative insights into the clustering behavior, we computed the PMFs between two small (diameter 13.5 Å) proteins, between a cluster of seven proteins and a single protein, and between two clusters of seven proteins (see the Supporting Material). Using geometric arguments, one can see that a cluster of seven proteins is relatively stable under mismatch conditions.
Fig. 7 a shows the PMFs for the interaction between two single proteins, between a single protein and a cluster of seven, and between two clusters of seven proteins. In a bilayer without cholesterol, we observe two trends.
Figure 7.
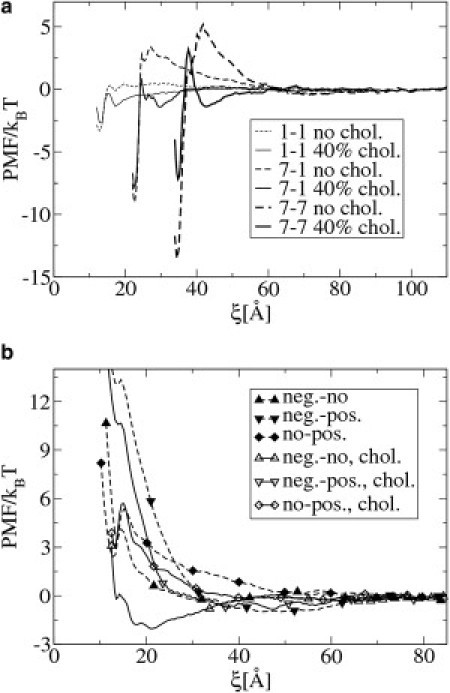
(a) Potential of mean force as a function of the distance ξ between two small (diameter 13.5 Å) proteins (1,1), between a single protein and a cluster of seven proteins (1–7), and between two clusters of seven proteins (7,7). The proteins have a positive mismatch of 2.3 Å and 3.5 Å, in the h3(t5)2 bilayer with 40 mol % (solid lines) and without (dotted lines) cholesterol, respectively. ΔT = 0.28. (b) PMF between two small proteins (1,1) with different mismatch. Positive, negligible, and negative mismatch are 8, −1, and −10 Å, respectively. The addition of cholesterol decreases the mismatch by 1.2 Å.
First, the depth of the free energy minimum, corresponding to the associated configuration, increases in the order: 1–1 < 7–1 < 7–7.
Second, the height of the repulsive barrier at intermediate distance increases in the order: 1–1 (1 kBT) < 7–1 (3 kBT) <7–7 (5.5 kBT). The presence of this repulsive barrier explains why our simulations show small clusters that were stable during the length of the simulations.
The addition of cholesterol slightly weakens the free energy minima. More importantly, cholesterol has a dramatic effect on the aggregation barriers. At 40 mol% cholesterol, the barrier is reduced from a wide barrier of 5.5 kBT to a small short-range barrier of 3 kBT. A second free energy minimum of −2 kBT, corresponding to the presence of cholesterol between the proteins (protein clusters), appears at a distance of 45 Å.
We repeated the same simulations for proteins with stronger positive mismatch (8 Å). The results are shown in the Supporting Material and are similar to the results for mismatch 3.5 Å. The height of the wide repulsive barrier between both clusters is now much higher (10.5 kBT), and the addition of cholesterol again reduces the barrier to short-range barrier of 4 kBT.
Similarly, we performed simulations for proteins with negative mismatch. The results are shown in the Supporting Material. For negative mismatch, the small aggregation barrier does not grow with increasing cluster size. Cholesterol does not affect the lipid-mediated attractive PMF between proteins with negative mismatch.
Because the aggregation barrier directly originates from the reorganization of lipids, one expects it to rise with increasing protein or protein cluster diameter. Indeed, the larger the protein or the cluster, the larger the perturbed area in between the two approaching entities. A larger membrane area with a lower hydrophilic shielding corresponds to a higher increase in the free energy. On the other hand, the effect of cholesterol should not only depend on the size of the proteins or clusters but also on the cholesterol concentration in the bulk. Indeed, the bigger the size of the protein clusters, the more cholesterol will be required in-between both clusters to alleviate the membrane perturbations.
Effect on the selectivity
We considered a system similar to the one shown in Fig. 3, but with small proteins with three different types of mismatch: negative (−10 Å), negligible (−1 Å), and positive (8 Å). We observed that proteins selectively aggregate with proteins with the same mismatch (a snapshot of the system is shown in the Supporting Material). In Fig. 7 b, the PMFs are shown between two proteins with positive and negligible, two with negative and positive, and two with negligible and positive mismatch. The lipid-mediated interactions are purely repulsive in all three cases. The PMFs between proteins with the same mismatch (shown in the Supporting Material) are attractive. These PMFs explain the selective aggregation between the proteins. We performed the same simulations again, but now in a bilayer containing 40 mol % cholesterol. The PMFs between proteins with different mismatch are shown in Fig. 7 b. Although cholesterol slightly reduces the repulsion, its effect on this type of repulsive interaction is very small. An exception is the interaction between a protein with negative and negligible mismatch, which becomes attractive due to the addition of cholesterol.
Concluding Remarks
The simulated PMFs between proteins (protein clusters) with positive mismatch indicate that there is an aggregation barrier, the size of which increases with growing cluster size. For proteins with negative mismatch, no growing aggregation barrier is observed. Thus, the cluster formation for positive mismatch will be very different from negative mismatch. For negative mismatch, one expects a phase separation between proteins and lipids, whereas for positive mismatch the aggregation barriers or the purely repulsive interactions might significantly slow down or inhibit the clustering process. This is indeed observed experimentally (4). Unfortunately, many protein clusters observed in real membranes contain a number of proteins much larger than accessible with our mesoscopic model and simulation method within a reasonable time (5–7).
Several independent experiments indicate that the addition of cholesterol further enhances the aggregation of membrane proteins (3,5–11). In this article, we provide simulation evidence for a lipid-shell mechanism by which cholesterol modifies lipid-mediated protein-protein interactions. We show that proteins with hydrophobic mismatch induce cholesterol-enriched or cholesterol-depleted shells surrounding the proteins in agreement with the shell hypothesis proposed by Anderson and Jacobson (42). The protein-induced shells then modify the lipid-mediated interactions between the proteins. Interestingly, Nyström et al. (50) recently found experimental indication for a cholesterol-enriched region around a model protein with positive mismatch in a DMPC bilayer containing a small amount of cholesterol. In agreement with our simulation results, they observed that the cholesterol-enrichment is less around proteins with no or negative mismatch. It would be very interesting to see whether all-atom simulations find evidence for the shell hypothesis.
Our results thus suggest that proteins with positive mismatch would prefer cholesterol-rich regions because cholesterol naturally replaces the perturbed lipids, and thus alleviates the perturbation. Several authors made similar observations, but explained them by using solely structural and mechanical arguments, and suggested that this sorting might play a role in the secretory pathway (39,63–65).
The interaction between two proteins with positive mismatch surrounded by cholesterol-enriched shells is significantly less repulsive. In some cases, the interaction even becomes attractive. Thus, our results suggest the formation of much larger, cholesterol-enriched, protein clusters in the presence of cholesterol. This is, for example, observed for syntaxin proteins in different real membranes (6). However, the direct comparison with experimental results is difficult, because our protein model is very general, while in reality specific protein-protein interactions exist which also determine the protein-protein interactions.
It is interesting to compare the shell mechanism to the wetting and capillary condensation mechanism proposed by Gil et al. (36) for a membrane consisting of two coexisting phases and proteins. In the shell mechanism, the two-phase coexistence is not a preexisting condition and the proteins are not surrounded by one particular phase but by a domain induced by the proteins, and with a different composition (not necessarily a different phase) than the bulk membrane. In the absence of the proteins, the two phases are present in the wetting model, while in the shell model, a single-phase homogeneous membrane exists.
We show that the origin of lipid-mediated selectivity is a lipid-mediated repulsive interaction between the different proteins. Cholesterol has only a modest effect. Experiments directly mimicking this simulation are not available in the literature. If we interpret the degree of hydrophobic mismatch as a parameter quantifying the unfavorable hydrophobic exposure of a misfolded protein, our results show how lipid-mediated interactions might play a role in the experimentally observed selective aggregation of misfolded proteins with misfolded proteins with a similar hydrophobicity (66).
In this article, we present a lipid shell mechanism by which cholesterol might affect lipid-mediated protein-protein interactions. In line with the shell hypothesis, we show that proteins induce cholesterol-enriched and cholesterol-depleted shells, which modify the interactions between the proteins. The simulation results are in line with, and might explain, several experimental observations related to an increased protein aggregation in the presence of cholesterol.
Acknowledgments
This work was supported by the Laboratory Directed Research and Development Program of the Lawrence Berkeley National Laboratory under the U.S. Department of Energy contract No. DE-AC02-05CH11231.
Supporting Material
References
- 1.Lewis B.A., Engelman D.M. Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J. Mol. Biol. 1983;166:203–210. doi: 10.1016/s0022-2836(83)80006-0. [DOI] [PubMed] [Google Scholar]
- 2.Ivanova V.P., Makarov I.M., Heimburg T. Analyzing heat capacity profiles of peptide-containing membranes: cluster formation of gramicidin A. Biophys. J. 2003;84:2427–2439. doi: 10.1016/S0006-3495(03)75047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sparr E., Ash W.L., Killian J.A. Self-association of transmembrane α-helices in model membranes: importance of helix orientation and role of hydrophobic mismatch. J. Biol. Chem. 2005;280:39324–39331. doi: 10.1074/jbc.M502810200. [DOI] [PubMed] [Google Scholar]
- 4.Botelho A.V., Huber T., Brown M.F. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sieber J.J., Willig K.I., Lang T. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 6.Lang T., Bruns D., Jahn R. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vereb G., Matkó J., Damjanovich S. Cholesterol-dependent clustering of IL-2Rα and its colocalization with HLA and CD48 on T lymphoma cells suggests their functional association with lipid rafts. Proc. Natl. Acad. Sci. USA. 2000;97:6013–6018. doi: 10.1073/pnas.97.11.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Almeida R.F., Loura L.M.S., Barrantes F.J. Cholesterol modulates the organization of the γM4 transmembrane domain of the muscle nicotinic acetylcholine receptor. Biophys. J. 2004;86:2261–2272. doi: 10.1016/S0006-3495(04)74284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristian L., Lear J.D., DeGrado W.F. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider H., Höchli M., Hackenbrock C.R. Relationship between the density distribution of intramembrane particles and electron transfer in the mitochondrial inner membrane as revealed by cholesterol incorporation. J. Cell Biol. 1982;94:387–393. doi: 10.1083/jcb.94.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray D.H., Tamm L.K. Clustering of syntaxin-1A in model membranes is modulated by phosphatidylinositol 4,5-bisphosphate and cholesterol. Biochemistry. 2009;48:4617–4625. doi: 10.1021/bi9003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer S.J., Nicolson G.L. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 13.Mouritsen O.G., Bloom M. Models of lipid-protein interactions in membranes. Annu. Rev. Biophys. Biomol. Struct. 1993;22:145–171. doi: 10.1146/annurev.bb.22.060193.001045. [DOI] [PubMed] [Google Scholar]
- 14.Killian J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 15.Harroun T.A., Heller W.T., Huang H.W. Experimental evidence for hydrophobic matching and membrane-mediated interactions in lipid bilayers containing gramicidin. Biophys. J. 1999;76:937–945. doi: 10.1016/S0006-3495(99)77257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen M.Ø., Mouritsen O.G. Lipids do influence protein function—the hydrophobic matching hypothesis revised. Biochim. Biophys. Acta: Biomembr. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Mitra K., Ubarretxena-Belandia I., Engelman D.M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Constantin D. Membrane-mediated repulsion between gramicidin pores. Biochim. Biophys. Acta. 2009;1788:1782–1789. doi: 10.1016/j.bbamem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Marcelja S. Lipid-mediated protein interaction in membranes. Biochim. Biophys. Acta. 1976;455:1–7. doi: 10.1016/0005-2736(76)90149-8. [DOI] [PubMed] [Google Scholar]
- 20.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dan N., Pincus P., Safran S.A. Membrane-induced interactions between inclusions. Langmuir. 1993;9:2768–2771. [Google Scholar]
- 22.Kralchevsky P., Paunov V., Nagayama K. Stresses in lipid-membranes and interactions between inclusions. J. Chem. Soc., Faraday Trans. 1995;91:3415–3432. [Google Scholar]
- 23.Lagüe P., Zuckermann M.J., Roux B. Lipid-mediated interactions between intrinsic membrane proteins: dependence on protein size and lipid composition. Biophys. J. 2001;81:276–284. doi: 10.1016/S0006-3495(01)75698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohinc K.V., Kralj-Igli V., May S. Interaction between two cylindrical inclusions in a symmetric lipid bilayer. J. Chem. Phys. 2003;119:7435–7444. [Google Scholar]
- 25.May S. Theories on structural perturbations of lipid bilayers. Curr. Opin. Colloid Interface Sci. 2000;5:244–249. [Google Scholar]
- 26.Wiggins P., Phillips R. Membrane-protein interactions in mechanosensitive channels. Biophys. J. 2005;88:880–902. doi: 10.1529/biophysj.104.047431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen S.O., Lopez C.F., Klein M.L. Coarse grain models and the computer simulation of soft materials. J. Phys. Condens. Matter. 2004;16:R481–R512. [Google Scholar]
- 28.Venturoli M., Smit B., Sperotto M.M. Simulation studies of protein-induced bilayer deformations, and lipid-induced protein tilting, on a mesoscopic model for lipid bilayers with embedded proteins. Biophys. J. 2005;88:1778–1798. doi: 10.1529/biophysj.104.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Periole X., Huber T., Sakmar T.P. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J. Am. Chem. Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 30.de Meyer F.J.-M., Venturoli M., Smit B. Molecular simulations of lipid-mediated protein-protein interactions. Biophys. J. 2008;95:1851–1865. doi: 10.1529/biophysj.107.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt U., Guigas G., Weiss M. Cluster formation of transmembrane proteins due to hydrophobic mismatching. Phys. Rev. Lett. 2008;101 doi: 10.1103/PhysRevLett.101.128104. 128104-1–128104-4. [DOI] [PubMed] [Google Scholar]
- 32.Psachoulia E., Fowler P.W., Sansom M.S. Helix-helix interactions in membrane proteins: coarse-grained simulations of glycophorin α-helix dimerization. Biochemistry. 2008;47:10503–10512. doi: 10.1021/bi800678t. [DOI] [PubMed] [Google Scholar]
- 33.West B., Brown F.L.H., Schmid F. Membrane-protein interactions in a generic coarse-grained model for lipid bilayers. Biophys. J. 2009;96:101–115. doi: 10.1529/biophysj.108.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janosi L., Prakash A., Doxastakis M. Lipid-modulated sequence-specific association of glycophorin A in membranes. Biophys. J. 2010;99:284–292. doi: 10.1016/j.bpj.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabra M.C., Uitdehaag J.C.M., Watts A. General model for lipid-mediated two-dimensional array formation of membrane proteins: application to bacteriorhodopsin. Biophys. J. 1998;75:1180–1188. doi: 10.1016/S0006-3495(98)74037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gil T., Ipsen J.H., Zuckermann M.J. Theoretical analysis of protein organization in lipid membranes. Biochim. Biophys. Acta. 1998;1376:245–266. doi: 10.1016/s0304-4157(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 37.Reynwar B.J., Deserno M. Membrane composition-mediated protein-protein interactions. Biointerphases. 2009;3:FA117–FA124. doi: 10.1116/1.2977492. [DOI] [PubMed] [Google Scholar]
- 38.Stüven E., Porat A., Helms J.B. Intra-Golgi protein transport depends on a cholesterol balance in the lipid membrane. J. Biol. Chem. 2003;278:53112–53122. doi: 10.1074/jbc.M300402200. [DOI] [PubMed] [Google Scholar]
- 39.Lundbaek J.A., Andersen O.S., Nielsen C. Cholesterol-induced protein sorting: an analysis of energetic feasibility. Biophys. J. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Churchward M.A., Rogasevskaia T., Coorssen J.R. Cholesterol facilitates the native mechanism of Ca2+-triggered membrane fusion. J. Cell Sci. 2005;118:4833–4848. doi: 10.1242/jcs.02601. [DOI] [PubMed] [Google Scholar]
- 41.Maxfield F.R., Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 42.Anderson R.G.W., Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 43.Epand R.M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 2006;45:279–294. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Brannigan G., Hénin J., Klein M.L. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren G.B., Houslay M.D., Birdsall N.J. Cholesterol is excluded from the phospholipid annulus surrounding an active calcium transport protein. Nature. 1975;255:684–687. doi: 10.1038/255684a0. [DOI] [PubMed] [Google Scholar]
- 46.Vidal A., McIntosh T.J. Transbilayer peptide sorting between raft and nonraft bilayers: comparisons of detergent extraction and confocal microscopy. Biophys. J. 2005;89:1106–1108. doi: 10.1529/biophysj.105.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan J., Tristram-Nagle S., Nagle J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:021931–021943. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poveda J.A., Fernández A.M., González-Ros J.M. Protein-promoted membrane domains. Biochim. Biophys. Acta. 2008;1778:1583–1590. doi: 10.1016/j.bbamem.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 49.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Nyström J.H., Lönnfors M., Nyholm T.K.M. Transmembrane peptides influence the affinity of sterols for phospholipid bilayers. Biophys. J. 2010;99:526–533. doi: 10.1016/j.bpj.2010.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epand R.M., Maekawa S., Epand R.F. Protein-induced formation of cholesterol-rich domains. Biochemistry. 2001;40:10514–10521. doi: 10.1021/bi010897s. [DOI] [PubMed] [Google Scholar]
- 52.Epand R.M., Sayer B.G., Epand R.F. Peptide-induced formation of cholesterol-rich domains. Biochemistry. 2003;42:14677–14689. doi: 10.1021/bi035587j. [DOI] [PubMed] [Google Scholar]
- 53.Epand R.M. Do proteins facilitate the formation of cholesterol-rich domains? Biochim. Biophys. Acta. 2004;1666:227–238. doi: 10.1016/j.bbamem.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Kranenburg M., Smit B. Phase behavior of model lipid bilayers. J. Phys. Chem. B. 2005;109:6553–6563. doi: 10.1021/jp0457646. [DOI] [PubMed] [Google Scholar]
- 55.de Meyer F.J.-M., Smit B. Effect of cholesterol on the structure of a phospholipid bilayer. Proc. Natl. Acad. Sci. USA. 2009;106:3654–3658. doi: 10.1073/pnas.0809959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Meyer F.J.-M., Benjamini A., Smit B. Molecular simulation of the DMPC-cholesterol phase diagram. J. Phys. Chem. B. 2010;114:10451–10461. doi: 10.1021/jp103903s. [DOI] [PubMed] [Google Scholar]
- 57.Groot R.D., Warren P.B. Dissipative particle dynamics: bridging the gap between atomistic and mesoscopic simulation. J. Chem. Phys. 1997;107:4423–4435. [Google Scholar]
- 58.Groot R.D., Rabone K.L. Mesoscopic simulation of cell membrane damage, morphology change and rupture by nonionic surfactants. Biophys. J. 2001;81:725–736. doi: 10.1016/S0006-3495(01)75737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venturoli M., Sperotto M.M., Smit B. Mesoscopic models of biological membranes. Phys. Rep. 2006;437:1–57. [Google Scholar]
- 60.Rodgers J.M., Webb M., Smit B. Alcohol solubility in a lipid bilayer: efficient grand-canonical simulation of an interfacially active molecule. J. Chem. Phys. 2010;132 doi: 10.1063/1.3314289. 064107-1–10. [DOI] [PubMed] [Google Scholar]
- 61.Venturoli, M. 2004. Mesoscopic models of lipid bilayers and bilayers with embedded proteins. PhD Thesis. University of Amsterdam, The Netherlands.
- 62.Constantin D., Brotons G., Salditt T. Interaction of alamethicin pores in DMPC bilayers. Biophys. J. 2007;92:3978–3987. doi: 10.1529/biophysj.106.101204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson C.L. Mechanisms of transport through the Golgi complex. J. Cell Sci. 2009;122:443–452. doi: 10.1242/jcs.032581. [DOI] [PubMed] [Google Scholar]
- 64.van Duyl B.Y., Meeldijk H., Killian J.A. A synergistic effect between cholesterol and tryptophan-flanked transmembrane helices modulates membrane curvature. Biochemistry. 2005;44:4526–4532. doi: 10.1021/bi047937n. [DOI] [PubMed] [Google Scholar]
- 65.Bretscher M.S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 66.Rajan R.S., Illing M.E., Kopito R.R. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. USA. 2001;98:13060–13065. doi: 10.1073/pnas.181479798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Léonard A., Dufourc E.J. Interactions of cholesterol with the membrane lipid matrix. A solid state NMR approach. Biochimie. 1991;73:1295–1302. doi: 10.1016/0300-9084(91)90092-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


