Abstract
We have found that genomic diversity is generally positively correlated with abiotic and biotic stress levels (1–3). However, beyond a high-threshold level of stress, the diversity declines to a few adapted genotypes. The Dead Sea is the harshest planetary hypersaline environment (340 g·liter–1 total dissolved salts, ≈10 times sea water). Hence, the Dead Sea is an excellent natural laboratory for testing the “rise and fall” pattern of genetic diversity with stress proposed in this article. Here, we examined genomic diversity of the ascomycete fungus Aspergillus versicolor from saline, nonsaline, and hypersaline Dead Sea environments. We screened the coding and noncoding genomes of A. versicolor isolates by using >600 AFLP (amplified fragment length polymorphism) markers (equal to loci). Genomic diversity was positively correlated with stress, culminating in the Dead Sea surface but dropped drastically in 50- to 280-m-deep seawater. The genomic diversity pattern paralleled the pattern of sexual reproduction of fungal species across the same southward gradient of increasing stress in Israel. This parallel may suggest that diversity and sex are intertwined intimately according to the rise and fall pattern and adaptively selected by natural selection in fungal genome evolution. Future large-scale verification in micromycetes will define further the trajectories of diversity and sex in the rise and fall pattern.
Genomic diversity in nature across all forms of life at global, regional, and local scales has been investigated widely since 1975 at the Institute of Evolution at the University of Haifa (Haifa, Israel) (1–4). The major problem investigated was how much of the genomic diversity in nature is adaptive. Our results indicated that genomic diversity is positively correlated with, and partially predictable by, ecological diversity and environmental stress (3). Theoretically, spatial and temporal ecological variation is of prime importance in maintaining genomic diversity in nature. Even in small isolated populations, genomic diversity is influenced strongly by natural selection, including diversifying, balancing, cyclical, and purifying selective regimes, interacting with but ultimately overriding the effects of mutation, migration, and stochasticity (3). The genomic era dramatically reinforced insights into genomic diversity as a factor in the twin evolutionary processes of adaptation and speciation. Molecular methods such as amplified fragment length polymorphism (AFLP) (5) can screen diversity effectively in both coding and noncoding regions of individuals and populations, making DNA-based studies excellent monitors of evolution. Here, we examined in filamentous fungi the relation between increasing ecological salinity stress and genome diversity from mild to extreme stress, culminating in the extreme hypersaline Dead Sea.
The Dead Sea, located in the Syrian–African Rift Valley between Israel and Jordan, is a unique ecological theater. Its waters are hypersaline (340 g·liter–1 total dissolved salts) and dominated by divalent cations (46 g·liter–1 Mg2+ and 17 g·liter–1 Ca2+ in addition to 36.5 g·liter–1 Na+, 7.8 g·liter–1 K+, and 225 g·liter–1 Cl–) (6). The low water activity (<0.669) (7) and the acidity (pH 5.9) make it an extreme and hostile environment to most forms of life (8). Currently, conditions continue to deteriorate as the water level of the Dead Sea declines constantly, dropping annually ≈100 cm (9).
The extremely impoverished biota of the Dead Sea is dominated by the unicellular green alga Dunaliella parva, as the primary producer, and by various species of halophilic Archaea of the family Halobacteriaceae, as main consumers (8). Blooms of these microorganisms occur only in exceptionally rainy years when extensive rain and floods cause a significant dilution of the upper layers of the water column (10–13).
Filamentous fungi in the Dead Sea water (DSW) were discovered in 1998 (14). To date, 77 fungal species have been recovered from DSW samples, including Gymnascella marismortui, an endemic species not previously described (15–20). Most of the species are sporadic or rare, but a spatiotemporal study revealed a steady species core including Aspergillus versicolor and Eurotium herbariorum (18). Some of the fungi isolated from the Dead Sea grew in DSW that was diluted to half its original salinity, and their spores remained viable after incubation in undiluted DSW for extended periods (21). Live mycelium of nine fungal species, including Eurotium amstelodami, E. herbariorum, and Eurotium rubrum, were found growing on driftwood that submerged near the aquatic shore (20, 22).
Here, we examine the relationship between increasing environmental stress and genomic (coding and noncoding) diversity in the ascomycete fungus A. versicolor. We substantiate the general pattern: genomic diversity is positively correlated with stress up to a high-threshold level but then drops to a minimum at extreme stress levels at the edge of life, where only a few fit genotypes can survive. Likewise, we demonstrate parallelism between genomic diversity and sex trajectories, rising with stress and falling near the hypersalinity edge of life.
Materials and Methods
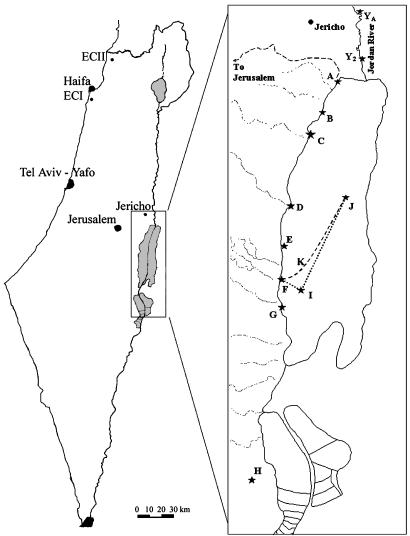
Fungal Isolation and Growth. The examined species A. versicolor (Vuill.) Tirab. was recovered from the shoreline of the hypersaline DSW up to a depth of 280 m (18); the Dead Sea terrestrial shore (DSTS) (23), including the Arubotaim salt cave (Mount Sedom) (24); the freshwater Jordan River;§ and the nonsaline soil of “Evolution Canyon” II (ECII) in Lower Nahal Keziv (western Upper Galilee) (ref. 25; Fig. 1).
Fig. 1.
Map of Israel containing isolation localities and a detailed Dead Sea area. J, Dead Sea deep water (DSDW; 50–280 m); K and I, Dead Sea surface water (DSSW); A–G, Dead Sea aquatic shore (DSAS); A–G, DSTS; H, Dead Sea Arubotaim salt cave; YA and Y2, southern Jordan River (JRS).
Liquid cultures were grown in 100 ml of medium containing 10 g·liter–1 glucose and 1 g·liter–1 yeast extract, prepared in distilled water, and mixed by a shaker (150 rpm) in 250-ml Erlenmeyer flasks to provide mycelia for DNA extraction. In the case of irregular (osmophilic) strains, the medium was prepared with sterile DSW (sampled from a 200-m depth in the center of the sea), which was diluted with sterile distilled water to 30% (vol/vol) of its original salinity. Mycelia were harvested by filtration through cellulose filter paper and kept at –80°C until DNA extraction.
DNA Extraction and AFLP Analysis. DNA was extracted from mycelia according to the method of Lee and Taylor (26) with an additional incubation with 5 μl of RNase 5 at 37°C for 30 min, followed by a second extraction with 700 μl of phenol/chloroform/isoamyl alcohol (25:24:1) and centrifugation as in the first extraction phase. The AFLP technique (5) was modified for a nonradioactive method and detection with an ALFexpress automated sequencer (Amersham Biosciences) as described (27). Primer sequences for preselective PCR were GACTGCGTACCAATTC (Eco+0) and GATGAGTCCTGAGTAA (Mse+0). For selective PCR, the primer combinations Eco+ACT:Mse+0, Eco+ACA:Mse+C, Eco+AGT:Mse+C, and Eco+C, combined with Mse+AG, Mse+CCG, Mse+CGC, or Mse+CGG, were chosen according to the resolution of their PCR product on a sequencing gel. In total, 605 AFLP sites (considered gene loci; 86 ± 17.7 per primer pair) were obtained.
Data Analysis. Reproducible bands with sizes of 40–500 bp in different samples across the gel were scored. A binary matrix was constructed for all isolates by scoring the presence (1) or absence (0) of AFLP bands.
For analysis of genomic diversity within populations, the unbiased estimate of expected genetic diversity Ĥe or mean unbiased gene diversity was applied 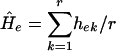 , where hek is the value of he for kth locus, and r is the total number of loci studied; he is an unbiased estimate of genetic diversity for a single locus given by
, where hek is the value of he for kth locus, and r is the total number of loci studied; he is an unbiased estimate of genetic diversity for a single locus given by 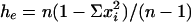 , where n is the number of haploid individuals and xi is the corresponding frequency of the ith allele at a locus in a sample from the population (28, 29). Additional genetic indices included percent of polymorphic loci estimates (95% criterion) and genetic distance calculated as Nei's unbiased minimum distance (30), represented by dendrograms. An exact test for population differentiation (28) was performed also. The test analyzes each locus in a data set to determine whether differences in allele frequencies exist. Fisher's combined probability test was used also as a global test over loci to determine the overall significance. The test was calculated with 10 batches, 2,000 permutations per batch, and 1,000 dememorization steps for estimation of probability value. All population statistics and dendrograms were calculated with tfpga software, Version 1.3 (31).
, where n is the number of haploid individuals and xi is the corresponding frequency of the ith allele at a locus in a sample from the population (28, 29). Additional genetic indices included percent of polymorphic loci estimates (95% criterion) and genetic distance calculated as Nei's unbiased minimum distance (30), represented by dendrograms. An exact test for population differentiation (28) was performed also. The test analyzes each locus in a data set to determine whether differences in allele frequencies exist. Fisher's combined probability test was used also as a global test over loci to determine the overall significance. The test was calculated with 10 batches, 2,000 permutations per batch, and 1,000 dememorization steps for estimation of probability value. All population statistics and dendrograms were calculated with tfpga software, Version 1.3 (31).
Correlation was examined between the polymorphism levels of A. versicolor populations and the ratio of morphologically sexual fungal species at sites in Israel (32), both displaying positive association with ecological stress. Polymorphism was estimated on a random sample of half (302 loci) of all loci. For each sample, the appropriate coefficient (Nei's Ĥe) (28) for each of the six (full set) or five (only the linear rise) considered populations was calculated. The permutation procedure was designed to calculate 100,000 random samples from a population. Thus, we received 100,000 variants of the original data. The second data set, the percentage of morphologically sexual species, had 100,000 random samples generated from the corresponding probability distribution. For each sample, Spearman's rank coefficient correlation was calculated (33). We then calculated the significance of each value of the coefficient correlation on the basis of every possible rearrangement of the data (33).
Results
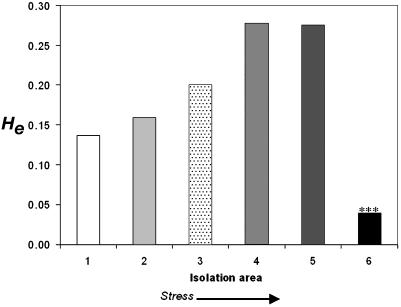
Population Structure and Division. The analysis of A. versicolor involves 605 bands (equal to loci). The Ĥe is presented graphically in Fig. 2. Ĥe increases with stress (bars 1, 2 < 3 < 4, 5) with a sharp drop seen in bar 6, i.e., from the freshwater population of the JRS (Fig. 2, bar 1) and the northern Mediterranean terrestrial population of ECII → DSTS → to DSAS → DSSW → DSDW (genetic diversity Ĥe = 0.137 < 0.159 < 0.201 < 0.278, 0.275 > 0.039; and polymorphism P = 25.62, 23.97 < 37.69 < 61.16, 51.57 > 10.58; Fig. 2, bars 1–6, respectively). Stress can be measured by a combination of salinity and pressure. Salinity levels are given in Table 1. The Dead Sea brine's salinity is relatively uniform throughout the whole body of water, but the increased stress level results from bathymetric pressure. The brines are ≈1.236 g·liter–1; therefore, pressure in the DSDW (>50 m) spans >6 atmospheres. The highest levels of Ĥe were in the DSAS and DSSW (Fig. 2, bars 4 and 5, respectively) and the lowest Ĥe occurred in the DSDW (50–280 m deep). The Ĥe pattern displays a “rise” with stress and a “fall” with extreme stress at the hypersaline edge of life (Fig. 2).
Fig. 2.
Ĥe of populations of the filamentous fungus A. versicolor based on 605 AFLP bands from random gene loci across the coding and noncoding genome. 1, JRS; 2, “African” south-facing slope (ECII); 3, DSTS; 4, DSAS; 5, DSSW; 6, DSDW. ***, A population significantly different from the one that precedes it (p > 0.001).
Table 1. A. versicolor (filamentous fungus) populations subjected to AFLP analysis.
| Symbol (Fig. 1)
|
No. of isolates
|
Salinity
|
||||
|---|---|---|---|---|---|---|
| Area | Population | Isolate identification | % of weight | % of sea water | ||
| ECII, Lower | ECII canyon bottom | ECIIB | 1 | Av ECII1 | >1.5 | ≈43 |
| Nahal Keziv | ECII “African” south-facing slope | ECIIA | 2 | Av ECII2-3 | >1.5 | ≈43 |
| JRS | Abdallah Bridge (≈13 km north of the Dead Sea) | YA | 1 | Av 6509 | >1 | ≈29 |
| Baptism church (≈8 km north of the Dead Sea) | YB | 2 | Av 6510-11 | >1 | ≈29 | |
| DSTS | DSTS | A-G | 2 | Av DSsl-2 | 29-5 | 143-829 |
| Dead Sea Arubotaim salt cave | H | 1 | Av DSSC | Unknown | Unknown | |
| DSW | DSDW (depth 50-280 m) | J | 5 | Av 7011-12/14/27, A 6001 | >34 | ≈970 |
| DSSW | K, I | 3 | Av 6007, Av6057-58 | ≈34 | ≈970 | |
| DSAS | A-G | 3 | Av 5001/4, Av 6026* | ≈34 | ≈970 | |
Fisher's Exact Test for Population Differentiation. We used Fisher's combined probability test to determine the overall significance of differences between populations based on the testing of interallelic differences of each of the 605 loci. The DSDW population (50–280 m deep) was significantly different from both the DSAS population and the DSSW population (p < 0.0001, df = 950). The two nonsaline populations of ECII and the freshwater isolates of the JRS did not differ significantly. By contrast, the A. versicolor metapopulation of the DSW diverged significantly from the JRS and ECII (both p < 0.0000). No significant difference was found between the DSW metapopulation and the DSTS population.
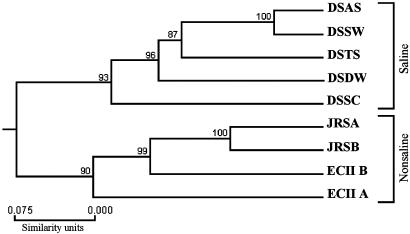
Genetic Distance and Cluster Analysis. Genetic distance (28) was calculated between the populations as presented in Table 2 and graphically represented as a dendrogram in Fig. 4. A. versicolor had high polymorphism correlated with distribution. The populations have a clear (100% bootstrap support) division to saline and nonsaline environments.
Table 2.
Genetic distances between populations of A. versicolor calculated as Nei's (28) unbiased minimum distance
Statistically significant differentiation of populations.
Fig. 4.
Dendrogram of the ascomycete A. versicolor populations. Numbers near a node indicate the percentage of bootstrap values that support the branch. DSSC, Dead Sea Arubotaim salt cave; JRSA, Abdallah Bridge; JRSB, Baptism church; ECII A, ECII “African” south-facing slope; ECII B, ECII canyon bottom.
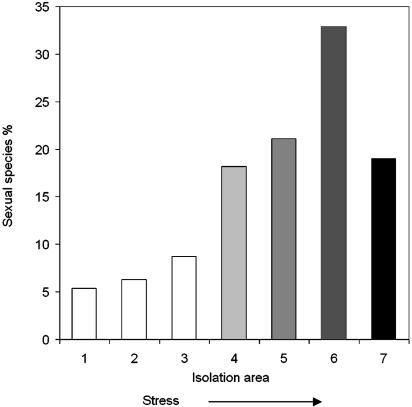
Correlation Between Genetic Diversity and Sexual Reproduction. We correlated the pattern of genetic polymorphism P of A. versicolor (Fig. 2) with the pattern of sexual reproduction S in Israeli fungi (Fig. 3). We conducted two correlations: (i) solely of the rise of P and S with stress based on the first five points, and (ii) the “rise and fall” of P and S with stress based on six points. For correlation i in 92% of the samples, the significance level between P and S was <5%. For correlation ii in 70% of the samples, the significance level between P and S was also <5%.
Fig. 3.
Proportion of sexual species in the mycobiota of different Israeli regions. 1, ECI (Lower Nahal Oren, Mt. Carmel); 2, ECII; 3, central coastal plain; 4, northern Negev Desert; 5, Dead Sea area; 6, DSTS (hypersaline mud); 7, DSW (hypersaline water). In DSW, the y axis represents the percent of sexual species from a total of 70 species identified. Sites 4–6 are characterized by increasing stress intensity, followed by increasing proportion of sexual reproduction and a sharp decline in sea water (site 7).
Discussion
Stress and Evolution: Insights from Studies of Fungi. Here, we presented genomic evidence in the cosmopolitan fungus A. versicolor, which lives in the Dead Sea and displays positive correlation with ecological stress, climaxing in the surface of the hypersaline Dead Sea but then significantly declining into only a few adapted genotypes in the deep-sea water column. Complementarily, we showed that this rise and fall trajectory of genomic diversity significantly parallels that of fungal sexual reproduction in multiple species across Israel. Thus, the filamentous fungus living in the extremely hypersaline Dead Sea highlights two major unresolved problems of evolutionary biology. (i) Does genetic diversity in extreme environments near the edge of life continue the upward expected trajectory of positive correlation with stress? (ii) How does genetic diversity relate to sexual reproduction? These questions are intimately related to the basic questions that we have explored across life since 1975 at the Institute of Evolution. How much of the genomic and phenomic diversity in nature is adaptive and processed by natural selection? What are the origin and evolution of adaptation and speciation processes in spatiotemporal macro- and microgeographic stressful environments?
Genetic Diversity in Nature at Climatic, Thermal, and Chemical Edges of Life. Since 1975, our research program at the Institute of Evolution at the University of Haifa, on the evolution of genome–phenome diversity under environmental stress (3), has revealed abundant genotypic and phenotypic diversity in nature. In general, the structure and evolution of this diversity, at both the molecular and organismal levels and at all geographical scales (global, regional, and local), are nonrandom and structured; they display regularities across life and are largely positively correlated with, and partly predictable by, abiotic and biotic environmental heterogeneity and stress. Biodiversity evolution, apparently even in small isolated populations, is strongly influenced by natural selection, including diversifying, balancing, cyclical, and purifying selective regimes, interacting with, but ultimately overriding, the effects of mutation, migration, and genetic drift (3). These results, derived from examining numerous species and thousands of plant and animal populations in Israel and across the planet (refs. 1–4, 34, and references therein, including higher basidiomycetes, ref. 35), supported the environmental theory of genetic diversity and, specifically, the niche-width variation hypothesis (36). This theory was validated here in our current critical test of the soil ascomycete fungus A. versicolor across Israel. Genomic diversity of A. versicolor culminated in the shores and surface water of the Dead Sea, one of the harshest planetary environments at the hypersaline edge of life, but then drastically declined in the hypersaline deep-sea water (50–280 m) with >34% salinity and high pressure. Can this proposed rise and fall pattern of genomic diversity be generalized? Several lines of evidence (i.e., climatic, thermal, and chemical of different model organisms at the edge of life discussed below) support this pattern in our research program of genetic diversity in nature.
Climatic Stress. We used Israel as our genetic laboratory to examine the association of increasing aridity stress southward from the northern mesic to the southern xeric deserts (37). We first tested protein polymorphism for 27 enzymatic loci in 13 unrelated genera of plants, invertebrates, and vertebrates, involving 21 species, 142 populations, and 5,474 individuals. The species, including fungi (35), varied biologically but shared a short (260 km) ecological gradient of increasing aridity stress both eastward and southward (but primarily southward). We found genetic parallelism across most taxa and loci. Heterozygosity (H) and Ĥe were positively and overall significantly correlated with rainfall variation, i.e., with increasing drought resistance corroborating the environmental theory of genetic diversity, primarily the niche-width variation hypothesis (36), over both space and time. We demonstrated further that H increases in Negev Desert and Sinai Desert populations (38) in five species of Sphincterochila landsnails, the lizard species Agama stellio, and the spiny mouse species Acomys cahirinus. This rise in diversity parallels a rise in stress levels, as reflected by a southward decrease in annual rainfall and an increase in rainfall variation and unpredictability. Not all Mediterranean species (e.g., Gryllotalpa gryllotalpa mole crickets) show a rise in H; a decline in H is evident when populations penetrate into the deep desert and the Dead Sea basin (39). Because protein heterozygosity is presumably associated with developmental homeostasis and physiological function, the fitness of individuals in ecologically uncertain and fluctuating stressful environments, such as deserts (but excluding extreme situations), is selected for higher protein heterozygosity. Later, we showed that this finding is true also for DNA diversity of noncoding genomes typed for diverse molecular markers (e.g., ref. 40). Theory predicts that at stable conditions, viability maintained multilocus polymorphism, the fitness of a genotype tends to increase with the number of heterozygous loci that it contains (41, 42). But does this trajectory continue to the edge of life?
Thermal Stress. A remarkable and more drastic decline of genomic diversity at the thermal edge of life was found in barnacles (43). In critical tests in barnacles in nearby cool (≈25°C) and warm (≈35°C) inflow and outflow canals, respectively, we showed strong temperature selection on allozymes and size of the acorn barnacle Balanus amphitrite. Allozymic diversity dropped from H = 0.095, 0114, and 0.111 to H = 0.026, 0.055, and 0.041 in three repetitive experiments. Corroborating evidence that strong thermal selection caused the sharp drop in H was evidenced by the 4-fold decrease in numbers and 3-fold decrease in body size of B. amphitrite, the total elimination of temperate Balanus eburneus, and the exclusion of most other species in the warm canal that abound in the cool canal, as is true also for other organisms under extreme thermal stress (ref. 43 and references therein).
Mercury Pollution Stress. Genetic diversity declined dramatically at extreme chemical stress in controlled laboratory experiments of mercury pollution on allozymic diversity of 15 phosphoglucomutase (Pgm) genotypes in the Mediterranean shrimp Palaemon elegans, involving 2,765 shrimps with 1,560 survivors (44). Selection for the MS (Pgm) heterozygote increased from 23% to 39% within the range 0.00–0.18 ppm HgCl2. By contrast, its frequency decreased in high HgCl2 concentrations (0.26–0.40 ppm), and the frequency of the MM (Pgm) homozygote increased and prevailed in those high HgCl2 concentrations. Homozygosity was clearly selected by purifying selection at the mercury edge of shrimp survival, and 1 homozygote was superior to all of the other 5 homozygotes and 10 heterozygotes.
Hypersaline Stress. Mole crickets. A drastic decline was indicated in our mole cricket studies of allozyme heterozygosity in a highly saline environment in Israel (39). Heterozygosity, based on 21 allozyme loci, dropped from 0.119 in Gryllotalpa tali living across Israel to 0.039 in Gryllotalpa marismortui living in saline habitats (the margin of the Dead Sea) at an extreme environment for mole crickets (39).
Filamentous fungi. The results obtained here for A. versicolor are even more dramatic than those obtained for Gryllotalpa marismortui. A. versicolor is a cosmopolitan species (45), distributed from the Antarctic (46) to the Egyptian desert (47), that is adapted to cope with low water activity (aw = 0.71–0.79) (48) and high radiation (49). Here, we unraveled a steady southward increase of overall genomic diversity in both the coding and noncoding regions from Ĥe = 0.137 (Jordan River) to a high of 0.278 at the DSSW, dropping precipitously to a low of Ĥe = 0.039 in the deep sea in variable depths 50–280 m. The DSDW population diverged significantly from the DSSW population and DSAS populations. Furthermore, the DSW metapopulation (comprising DSAS, DSSW, and DSDW populations) had diverged significantly from the nonsaline populations of the JRS and ECII (Table 2 and Fig. 4). This divergence corroborates the proposed rise and fall pattern discussed above. The DSDW provides extreme combined stress conditions, in both salinity and pressure at the edge of life, consistent with the selection of only a few adapted homozygotes. The combined hypersalinity and pressure is similar to the state found in deep-sea basins where “living fossils,” such as stromatolites, may be encountered.
Adaptive Salt Tolerance. Phenotypic expression of salt tolerance corroborates the genetic evidence. Dead Sea shore and sea isolates of A. versicolor have common adaptive properties (primarily, high-salt tolerance). In physiological viability trials (21), fungal strains from the saline DSTS with 5–29% salinity showed an intermediate survival pattern between that of the sea (DSW) and nonsaline (ECII) isolates in both spore and mycelium testing. They were less tolerant to incubation in undiluted DSW than the isolates obtained from the sea brines. On the other hand, in diluted DSW (80%), simulating a diluted upper layer formed after rains and flooding, spores of shore isolates (DSTS) remained viable to the same extent as the DSW isolates (21).
Ecological Stress and Sex Evolution in Soil Microfungi. If ecological stress shapes diversity levels, then how does it affect sexuality? The elucidation of the origin and maintenance of sex is a major unresolved problem in evolutionary biology. More than two dozen theoretical models have been proposed to explain the maintenance of sexual recombination (32). Recently, we reported estimates of the distribution of morphologically sexual and asexual soil microfungi in nature, indicating a positive correlation between increased sexuality (by means of meiospores) with ecological stress (ref. 32, data adapted here as Fig. 3 with the addition of the estimate from the DSW). Most remarkably, the genomic diversity and sexuality trajectories of soil fungi based on the present study and the studies of Grishkan et al. (32) are significantly parallel (Figs. 2 and 3). This parallelism is exciting because it may suggest that ecological stress affects diversity and sex similarly and that they are intimately intertwined and shaped by similar ecological stresses. Our present results strongly support a positive association among ecological stress, genomic diversity, and sexual reproduction in micromycetes and call for additional tests to substantiate a general pattern. Both genomic diversity and sex seem to follow the rise and fall pattern, increasing with stress toward the hypersaline edge of life, where both appear to decline drastically.
Indeed, stressful conditions are known to increase genetic polymorphism, recombination, mutation, gene conversion, and sex (50), as we have shown also at the EC microscale model of life in the soil fungus Sordaria fimicola (51, 52) and in Drosophila (ref. 3 and references therein). Increase in sexual reproduction with increasing stress has been demonstrated, not only regionally in Israeli fungi (32) but also locally in the “African” stressful slope in ECI and ECII (25, 53). Clearly, increases in mutation, recombination, gene conversion, and sex ensure higher levels of genetic diversity, providing a higher potential for genetic adaptation. But when the edge of life is approached climatically, thermally, or chemically and the niche becomes narrower, homogeneous, and extremely stressful, all of the diversity-generating mechanisms decline, and natural selection seems to turn from diversifying and balancing selective regimes to a purifying selective regime (i.e., selection for highly adaptive but few homozygous clones).
Conclusions and Prospects
The long-term research program of genetic diversity in natural populations of microorganisms, plants, animals, and humans at global, regional, and local scales at both protein (1–4) and DNA (40) levels, conducted since 1975 at the Institute of Evolution at the University of Haifa, showed similar adaptive strategies linking genetic diversity with stress and now also with sexual reproduction (32) (Fig. 3). The results indicate that genetic diversity in nature is nonrandom and positively correlated with abiotic and biotic diverse stresses and is oriented by balancing, diversifying, and cyclical selection regimes. However, at the edge of life, diversity appears to decline to a few adapted genotypes and seems to be oriented by purifying selection. Sexual reproduction displays patterns parallel to those of genetic diversity but calls for additional verification tests.
The extreme, harsh ecological theater of the Dead Sea represents the hypersaline edge of life and, hence, provides an extraordinary test case of the environmental theories on genetic diversity and sex. We have demonstrated that the cosmopolitan soil fungus A. versicolor corroborates the niche-width environmental theory of genetic diversity. It supports our theory that genomic diversity and sex levels are coupled adaptively and that both coding and noncoding genomes are influenced by diverse forms of natural selection in accordance with the patterns and levels of stress generating evolutionary adaptive patterns. Future fungal tests in the Dead Sea could further explore and test this evolutionary generalization.
Acknowledgments
We thank Professor A. Oren from the Division of Microbial and Molecular Ecology at the Hebrew University of Jerusalem for his remarks and insights, and Dr. I. Grishkan from the Institute of Evolution at the University of Haifa for contributing fungal strains from the Dead Sea shore and ECII in Lower Nahal Keziv, in western Upper Galilee, Israel. This study was supported in part by the Authority of Graduate Studies of the University of Haifa, the Wolf Foundation, the Israel Discount Bank Chair of Evolutionary Biology, and the Ancell–Teicher Research Foundation for Genetics and Molecular Evolution.
Abbreviations: AFLP, amplified fragment length polymorphism; DSDW, Dead Sea deep water; DSSW, Dead Sea surface water; DSAS, Dead Sea aquatic shore; DSW, Dead Sea water (in general); DSTS, Dead Sea terrestrial shore; ECI/II, “Evolution Canyon” I and II; JRS, southern Jordan River.
Footnotes
Kis-Papo, T., Grishkan, I., Oren, A. & Nevo, E. (2001) Isr. J. Plant Sci. 49, 160 (abstr.).
References
- 1.Nevo, E. (1988) Evol. Biol. 23, 217–247. [Google Scholar]
- 2.Nevo, E. (1998) J. Exp. Zool. 282, 95–119. [Google Scholar]
- 3.Nevo, E. (2001) Proc. Natl. Acad. Sci. USA 98, 6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nevo, E. (2001) in Encyclopedia of Biodiversity, ed. Levin, S. A. (Academic, San Diego), Vol. 3, pp. 195–213. [Google Scholar]
- 5.Vos, P., Hogers, R., Bleeker, M., Reijans, M., Lee, T. V. D., Hornes, M., Frijters, A., Pot, J., Kuiper, J. M. & Zabeaur, M. (1995) Nucleic Acids Res. 23, 4407–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stiller, M. & Nissenbaum, A. (1999) Geochim. Cosmochim. Acta 63, 3467–3475. [Google Scholar]
- 7.Krumgalz, B. S. & Millero, F. J. (1982) Mar. Chem. 11, 209–222. [Google Scholar]
- 8.Oren, A. (2003) in Fungal Life in the Dead Sea, eds. Nevo, E., Oren, A. & Wasser, S. P. (Gantner, Ruggell, Liechtenstein), pp. 117–140.
- 9.Gertman, I. & Hecht, A. (2002) J. Mar. Syst. 35, 169–181. [Google Scholar]
- 10.Oren, A. (1988) in Advances in Microbial Ecology, ed. Marshall, K. C. (Plenum, New York), Vol. 10, pp. 193–229. [Google Scholar]
- 11.Oren, A. (1999) Hydrobiologia 405, 1–9. [Google Scholar]
- 12.Oren, A. (2000) Arch. Hydrobiol. Spec. Issues Advanc. Limnol. 55, 531–542. [Google Scholar]
- 13.Oren, A., Gurevich, P., Anati, D. A., Barkan, E. & Luz, B. (1995) Hydrobiologia 297, 173–185. [Google Scholar]
- 14.Buchalo, A. S., Nevo, E., Wasser, S. P., Oren, A. & Molitoris, H. P. (1998) Proc. R. Soc. London Ser. B 265, 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchalo, A. S., Wasser, S. P., Molitoris, H. P., Volz, P. A., Kurchenko, I., Lauer, I. & Rawal, B. (1999) in Evolutionary Theory and Processes: Modern Perspectives. Papers in Honour of Eviatar Nevo, ed. Wasser, S. P. (Kluwer, Dordrecht, The Netherlands), pp. 293–300.
- 16.Buchalo, A. S., Nevo, E., Wasser, S. P., Oren, A., Molitoris, H. P. & Volz, P. A. (2000) Mycol. Res. 104, 132–133. [Google Scholar]
- 17.Buchalo, A. S., Nevo, E., Wasser, S. P. & Volz, P. A. (2000) in Journey to Diverse Microbial Worlds, ed. Seckbach, J. (Kluwer, Dordrecht, The Netherlands), pp. 239–252.
- 18.Kis-Papo, T., Grishkan, I., Oren, A., Wasser, S. P. & Nevo, E. (2001) Mycol. Res. 105, 749–756. [Google Scholar]
- 19.Kis-Papo, T. (2003) Ph.D. dissertation (University of Haifa, Haifa, Israel).
- 20.Kis-Papo, T., Grishkan, I., Gunde-Cimerman, N., Oren, A., Wasser, S. P. & Nevo, E. (2003) in Fungal Life in the Dead Sea, eds. Oren, A., Nevo, E. & Wasser, S. P. (Gantner, Ruggell, Liechtenstein), pp. 271–292.
- 21.Kis-Papo, T., Oren, A., Wasser, S. P. & Nevo, E. (2003) Microb. Ecol. 45, 183–190. [DOI] [PubMed] [Google Scholar]
- 22.Buchalo, A. (2003) in Fungal Life in the Dead Sea, eds. Nevo, E., Oren, A. & Wasser, S. P. (Gantner, Ruggell, Liechtenstein, Koeltz Scientific, Königstein), pp. 141–161.
- 23.Grishkan, I., Nevo, E. & Wasser, S. P. (2003) Mycol. Prog. 2, 19–28. [Google Scholar]
- 24.Grishkan, I., Nevo, E. & Wasser, S. P. (2003) J. Arid Environ., in press.
- 25.Grishkan, I., Nevo, E., Wasser, S. P. & Beharav, A. (2003) Biol. J. Linn. Soc. 78, 527–540. [Google Scholar]
- 26.Lee, S. B. & Taylor, J. W. (1990) in PCR Protocols: A Guide to Methods and Applications, eds. Innis, M. A., Gelfand, D. H. & Sni, J. J. (Academic, London), pp. 282–287.
- 27.Satish, N., Krugman, T., Vinogradova, O. N., Nevo, E. & Kashi, Y. (2001) Microb. Ecol. 42, 306–316. [DOI] [PubMed] [Google Scholar]
- 28.Nei, M. (1978) Genetics 89, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei, M. (1987) Molecular Evolutionary Genomics (Columbia Univ. Press, New York).
- 30.Raymond, M. L. & Rousset, F. (1995) Evolution (Lawrence, Kans.) 49, 1280–1283. [DOI] [PubMed] [Google Scholar]
- 31.Miller, M. P. (1997) Tools for Population Genomic Analyses (tfpga): A Windows Program for Analysis of Allozyme and Molecular Population Genomic Data, Version 1.3. Available at http://herb.bio.nau.edu/~miller. Accessed June 17, 2002.
- 32.Grishkan, I., Korol, A. B., Nevo, E. & Wasser, S. P. (2003) Proc. R. Soc. London Ser. B 270, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendall, M. G. & Gibbons, J. D. (1990) Rank Correlation Methods (Edward Arnold, London).
- 34.Nevo, E., Beiles, A. & Ben-Shlomo, R. (1984) Lect. Notes Biomath. 53, 13–213. [Google Scholar]
- 35.Lewinsohn, D., Nevo, E., Hadar, Y., Wasser, S. W. & Beharav, A. (2000) Mycol. Res. 104, 1184–1190. [Google Scholar]
- 36.Van Valen, L. (1965) Am. Nat. 99, 377–390. [Google Scholar]
- 37.Nevo, E. & Beiles, A. (1988) Biol. J. Linn. Soc. 35, 229–245. [Google Scholar]
- 38.Nevo, E. & Beiles, A. (1989) J. Arid Environ. 17, 241–244. [Google Scholar]
- 39.Nevo, E., Beiles, A., Korol, A. B., Ronin, Y. I., Pavlicek T. & Hamilton, W. D. (2000) Evolution (Lawrence, Kans.) 54, 586–605. [DOI] [PubMed] [Google Scholar]
- 40.Nevo, E., Ben-Shlomo, R., Beiles, A., Ronin, Y., Blum, S. & Hillel, J. (1996) in Gene Families: Structure, Function, Genetics and Evolution, eds. Holmes, R. S. & Lim, H. A. (World Scientific, Singapore), pp. 55–70.
- 41.Karlin, S. & Feldman, M. W. (1981) Genetics 97, 475–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turelli, M. & Ginzburg, L. R. (1983) Genetics 104, 191–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nevo, E., Shimony, T. & Libni, M. (1977) Nature 267, 699–701. [DOI] [PubMed] [Google Scholar]
- 44.Nevo, E., Perl, T., Beiles, A. & Wool, D. (1981) Experientia 37, 1152–1154. [Google Scholar]
- 45.Klich, M. A. (2002) Mycologia 94, 21–27. [PubMed] [Google Scholar]
- 46.Fenice, M., Selbmann, L., Zucconi, L. & Onofri, S. (1997) Polar Biol. 17, 275–280. [Google Scholar]
- 47.el-Gindy, A. A. & Saad, R. R. (1990) Zentralbl. Mikrobiol. 145, 547–551. [PubMed] [Google Scholar]
- 48.Lacey, J. & Magan, N. (1991) in Cereal Grain: Mycotoxins, Fungi Quality in Drying and Storage, ed. Chelkowski, J. (Elsevier, Amsterdam), pp. 77–118.
- 49.Zhdanova, N. N., Zakharchenko, V. A., Vember, V. V. & Nakonechnaya, L. T. (2000) Mycol. Res. 104, 1421–1426. [Google Scholar]
- 50.Korol, A. B., Preygel, I. A. & Preygal, S. I. (1994) Recombination Variability and Evolution (Chapman & Hall, London).
- 51.Lamb, B. C., Saleem, M., Scott, W., Thapa, N. & Nevo, E. (1998) Genetics 149, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saleem, M., Lamb, B. C. & Nevo, E. (2001) Genetics 159, 1573–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grishkan, I., Nevo, E., Wasser, S. P. & Pavlicek, T. (2000) Isr. J. Plant Sci. 48, 297–308. [Google Scholar]