Abstract
The age distribution of female mosquitoes in the field is a critical component of vectorial capacity because of the extrinsic incubation period of mosquito-borne pathogens. However this parameter has not been well characterized in malaria vectors because of methodological difficulties; transcriptional profiling provides a potential new approach for age determination. In Anopheles gambiae, microarrays were used to examine global gene expression over adult life. Nine genes were selected from the 2714 gene transcripts that displayed age-related transcription patterns, and quantitative reverse transcription PCR used to select the four best performing genes. The resulting age estimation assay was able to predict female age from lab-reared samples with sufficient accuracy to provide a potentially useful tool for studies of malaria epidemiology and control.
Keywords: malaria, mosquito, vector, microarray, lifespan
Introduction
One of the most sensitive parameters contributing to the vectorial capacity of malaria-transmitting Anopheles mosquitoes is the age distribution of wild females. After picking up the Plasmodium parasite there is an incubation period before transmission can occur – during which time exflagination, fertilization, zygote elongation, ookinete migration, oocyst formation and the migration of thousands of sporozoites to the salivary glands have to occur. Only then is the mosquito infectious and able to transmit malaria the next time it takes a bloodmeal. Timing of the extrinsic incubation period depends on temperature and the species of Plasmodium/Anopheles, but typically at least 9–15 days are needed; for example around 11 days for Plasmodium falciparum in Anopheles gambiae at 24 °C (Garrett-Jones, 1964; Gilles & Warrell, 2002; Smith & McKenzie, 2004), plus there is typically at least 2 days from eclosion until females take their first bloodmeal. Adult mosquitoes experience a high daily mortality rate such that only a small percentage of the total population actually survive long enough to transmit malaria (Brownstein et al., 2003; Cook et al., 2008).
Despite the critical importance of estimating Anopheles age in field-caught specimens to the understanding of malaria transmission dynamics and epidemiology, and the impact of control interventions, age remains one of the least understood components of vectorial capacity. Ovary dissection and microscopic examination have traditionally been used to estimate the number of gonotrophic cycles a female mosquito has gone through (Beklemishev et al., 1959; Detinova, 1962; Tyndale-Biscoe, 1984; Lehane, 1985; Hayes & Wall, 1999; Hugo et al., 2008), but this is a relatively difficult and inaccurate method and is not an absolute indicator of age because time between blood feeds can vary according to conditions and availability of human hosts for biting.
Novel control strategies to shorten mosquito lifespan are currently in development. Life-shortening Wolbachia bacteria from Drosophila melanogaster (Min & Benzer, 1997) have been successfully transferred into mosquitoes (McMeniman et al., 2009) with the same phenotype, thus providing an attractive control methodology. Testing of life-shortening fungal biopesticides against malaria vectors is underway (Blanford et al., 2005; Scholte et al., 2005; Bell et al., 2009). In addition, population age structure can be skewed toward younger individuals by current insecticidal malaria control programmes through cumulative uptake of insecticides, and it has been proposed that different insecticides should be specifically selected for delayed rather than immediate killing effects as a strategy to delay the evolution of insecticide resistance in malaria vectors (Read et al., 2009). Thus, the ability to estimate the age structure of natural Anopheles populations before and after interventions has acquired considerable importance, and there is a need to develop additional methodologies with which to do so.
A new approach developed in Aedes aegypti mosquitoes utilizes changes in the levels of expression of three genes in the adult mosquito (Cook et al., 2006, 2007). Expression changes were measured by quantitative reverse transcription PCR (qRT-PCR) on caged mosquitoes of known age and then used to build a multivariate calibration model to predict the age of field caught specimens from the same locality. This technique has been used successfully to predict age of Ae. aegypti from Cairns, Australia (Cook et al., 2006, Hugo et al., 2010). Here we set out to develop comparable methodologies for An. gambiae mosquitoes, the most important malaria vector in Africa. In order to select the most informative genes for the assay, we characterized global transcription in adult An. gambiae throughout adult life for both sexes. We used the An. gambiae genome database (Holt et al., 2002) to design and employ a microarray to examine global gene transcription profiles up to 30 days post-eclosion; a previous array study had examined transcription up to 15 days post-eclosion (Marinotti et al., 2006) but it was necessary to extend this period in order to capture expression profiles in older insects.
Results
Microarray results and selection of candidate genes
Whole genome microarrays were used to identify An. gambiae genes that displayed age-dependent transcription levels in both male and female adult mosquitoes. A custom microarray was designed with a single probe per An. gambiae gene transcript (13254 probes). One-colour microarray hybridizations were run with total RNA extracted from pools of male and female mosquitoes at 0, 10, 20 and 30 days post-eclosion. Fluorescence data were quantified from the microarray scan files using Agilent Technologies' Feature Extraction software. Data processing and analysis were carried out with GeneSpring GX. Two-way analysis of variance (P≤ 0.01; Benjamini–Hochberg multiple test correction) identified 2713 gene transcripts that displayed significant differential expression across adult mosquito age and no significant difference resulting from sex or age–sex interaction (Fig. 1). From these, candidate genes were selected for showing a strong correlation between transcript levels and adult age/a greater than fourfold change in transcription across multiple age classes. Five transcripts, AGAP004115-RA, AGAP006187-RA, AGAP006985-RA, AGAP010398-RA and AGAP012396-RA (Fig. S1A–E, respectively), showed a strong positive correlation between transcript levels and both male and female mosquito age. Two transcripts, AGAP006829-RA and AGAP008447-RA (Fig. S1F, H, respectively), displayed a negative correlation between transcript levels and age. The final two down-regulated gene transcripts, AGAP007963-RA and AGAP009871-RA (Fig. S1G, H, respectively), were selected because they are putative orthologues of the two most important genes used in the Ae. aegypti transcriptional age-grading protocol (Cook et al., 2006). AGAP007963-RA was selected from the 2714 gene transcripts that displayed age-related transcription (ie no significant effect of sex or age-sex interaction). AGAP009871-RA had significant age- and sex-related transcription. The reference gene, 40S ribosomal protein S7 (RS7_ANOGA; AGAP10592-RA), was not differentially regulated with adult male or female mosquito age (Fig. S1J).
Figure 1.
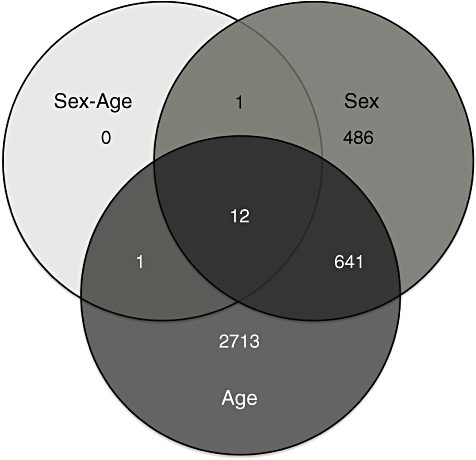
Venn diagram showing the distribution of gene transcripts that have significant differential expression with respect to age, sex and/or a sex-age interaction. A total of 3367 gene transcripts displayed significant differential expression with age; however, only 2713 transcripts did not show any significant effects with respect to sex or sex-age interaction.
The transcriptional profiles of the nine candidate genes were investigated in individual adult mosquitoes using qRT-PCR. Primers were designed to amplify a 100–150 base pair region of each gene transcript (Table 1). Transcript abundance of each candidate gene was determined relative to RS7_ANOGA as a logcontrast of cycle threshold (Ct) values. A low Ct value indicates high transcript abundance and a high Ct value corresponds to a low transcript copy number. All nine candidate genes showed age-related transcriptional profiles and results were generally consistent with that observed in the microarray experiment. All up-regulated candidate genes, AGAP004115, AGAP006187, AGAP006985, AGAP010398-RA and AGAP012396, showed negative, linear associations between logcontrast Ct values and age, in both male and female mosquitoes (Fig. S2A–E, respectively). Three of the four down-regulated candidate genes, AGAP006829, AGAP007963 and AGAP009871 (Fig. S2F, G and I, respectively), displayed positive associations with logcontrast Ct values and age, in females and males. The remaining candidate gene, AGAP008447 (Fig. S2H), had a positive association across female age and an unexpected negative association with male age. Transcript AGAP008447 was subsequently excluded from the analysis because of the possible confounding effects of insecticide resistance status (see Discussion).
Table 1.
Candidate age-grading genes selected from the microarray analysis
| Transcript identifier | Gene name | Putative function |
|---|---|---|
| AGAP004115-RA | Cystinosin, putative | |
| AGAP006187-RA | G12_ANOGA | Protein G12 precursor (ANG12) |
| AGAP006985-RA | Conserved hypothetical protein | |
| AGAP010398-RA | Dimethylaniline monooxygenase, putative | |
| AGAP012936-RA | Cystinosin, putative | |
| AGAP006829-RA | CPR59 | Cuticular protein 59 |
| AGAP007963-RA* | Calcium-binding protein, putative | |
| AGAP008447-RA† | CPLCG4 | Adult cuticle protein |
| AGAP009871-RA* | CPR75 | Cuticular protein 75 (RR-1 family) |
| AGAP010592-RA | RS7_ANOGA | 40S ribosomal protein S7 |
Transcript identifiers and gene function originate from Vectorbase annotations. When gene function has not been annotated putative gene function has been inferred from Drosophila melanogaster and/or Aedes aegypti orthologues. The four best performing gene transcripts used for the age estimation analysis shown in Fig. 3 are in bold.
Orthologues of genes selected for use in the Aedes aegypti age estimation assay (Cook et al., 2006).
Gene excluded from the analysis because of possible confounding effects of insecticide resistance status (see Discussion).
Redundancy analysis was used to reduce multiple gene expression measures into a redundancy variate, a linear combination of gene expression measures, which maximized correlation with mosquito age. This analysis was initially carried out using all genes and from there an optimal set of genes was selected based on the canonical coefficient loadings from the redundancy analysis; more informative genes have a larger absolute loading (either positive or negative depending on the relationship between a particular gene expression profile and age). Based on these criteria, four transcripts were selected for the age estimation assay: AGAP006187, AGAP007963, AGAP010398 and AGAP012936. These genes performed very well in females generating a calibration model that was highly correlated with age (Fig. 2; R2= 0.8206, n= 34, P < 0.0001). However in males, these genes generated a redundancy variate that was not as strongly correlated with age (Fig. S3; R2= 0.6099, n= 35, P < 0.0001); the reasons for this difference are unclear.
Figure 2.
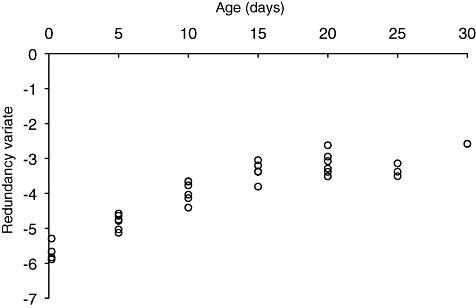
Female Anopheles gambiae calibration model generated from the expression profiles of the four best candidate genes. Logcontrast normalized cycle threshold values for AGAP006187, AGAP006985, AGAP007963 and AGAP010398 were entered into a canonical correlation analysis. This analysis generates a redundancy variate, a linear combination of the four gene expression measures, which maximizes correlation with mosquito age. The redundancy variate score for each individual is plotted and the linear regression of these data on age provides the calibration model.
Age predictions were derived for females using inverse regression of the calibration model constructed from the four most informative genes (Fig. 3). A leave-one-out cross validation methodology was used to generate a large number of age predictions from the available data. The precision of each age prediction was estimated using a nonparametric bootstrapping procedure that derived 95% confidence intervals (CIs). The median age within these 95% CIs was considered to be the predicted age. Predicted ages had a mean 95% CI of ± 4.2 days across all ages. The mean residual value (the absolute difference between predicted age and actual age) for females is 4.3 days. Negative ages are predicted but this was to be expected when predicted ages are close to zero simply as a result of error within the calibration model.
Figure 3.
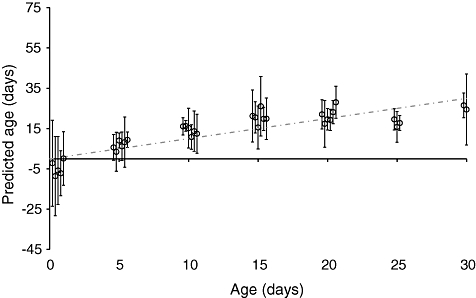
Age predictions for individual female mosquitoes using four gene expression measures. Age predictions were made using a leave-one-out cross validation approach, whereby each data point was sequentially removed from the training data before the age of the excluded individual was predicted. Age was predicted by inverse regression of the female calibration model. Error bars indicate a 95% confidence interval derived by a nonparametric bootstrap procedure.
For males, predicted ages had a mean 95% CI of ±9.1 days across all ages (Fig. S4), and a mean residual value of 10.1 days. This higher variability was expected given the poorer fit observed in the calibration model. Four samples were notable outliers in the age predictions; however, this could not be attributed to a single disparate gene expression measure across samples – that is, it was a different gene for each specimen that caused the inaccuracy in age prediction. The distribution of the residual values when plotted against predicted age suggests that there was no bias associated with the age predictions for either sex (Fig. S5).
Discussion
The primary purpose of these experiments was to develop an assay capable of detecting differences in female population age structure in order to be able to compare populations with and without control interventions, and the effects of age structure on malaria epidemiology. We consider the data presented to provide the baseline for an assay that would be a very useful addition to the field as the confidence intervals in individual age predictions are acceptable for making population-level comparisons. The assay was less successful for males, but the requirement for age prediction methodologies is in any case much smaller in males, which do not blood feed or transmit malaria. In addition, male age-grading methodologies are now available that use the number of spermatocysts in An. gambiae s.s. male testes and the relative size of their sperm reservoirs (Huho et al., 2006).
Three of the genes examined encoded cuticular proteins. Cuticular protein genes of low complexity (CPLC) constitute a family of at least 210 genes, including 27 in a cluster encoding family CPLCG (Cornman et al., 2008; Cornman & Willis, 2009). Several adult cuticular protein genes have recently been reported to show increased levels of expression associated with pyrethroid insecticide resistance (Vontas et al., 2007; Djouaka et al., 2008; Awolola et al., 2009), including CPLCG4 (previously named CPLC#; AGAP008447). This appears to constitute a mechanism of resistance based on cuticular thickening to reduce insecticide penetration, but understanding of this mechanism is in its early stages. Because of the potential for variation in the expression of cuticular proteins associated with insecticide resistance status of populations under investigation, and because this factor is not yet well understood, the adult cuticular protein gene was excluded from the current assay. In addition variation in cuticular hydrocarbons amongst and within species in the An. gambiae complex (Carlson & Service, 1979, 1980; Anyanwu et al., 2000, 2001) could act as a further confounding variable. This variation is possibly associated with selection for mate choice, as has been shown in Drosophila for example (Foley et al., 2007).
The analysis of cuticular proteins for age estimation has been assayed in An. gambiae and other anophelines with varying degrees of success (Randimby et al., 2002; Caputo et al., 2005; Hugo et al., 2006; Mayagaya et al., 2009). A recently reported assay using near -infrared spectroscopy (Mayagaya et al., 2009) showed promise as, in the authors' words, a ‘quick and dirty’ assay with minimal sample processing costs once the initial outlay on a spectrophotometer has been made, allowing large numbers of samples to be assayed. This assay does not appear sensitive enough to estimate age differences beyond about 15 days, which is the most epidemiologically significant class. However, it is possible that a combination of assays could be used to improve throughput and reduce costs, with near-infrared spectroscopy used to assign females to ‘younger’ and ‘older’ classes, and then qRT-PCR based on the methodologies described here could be employed on the older class (which would be a minority subset). In this case, however, it would be important to examine and control for any insecticide resistance effects on cuticle composition, for the reasons outlined above.
Future experiments will examine the robustness and utility of the assay on natural populations from different geographical locations, molecular forms of An. gambiae s.s., and sibling species within the An. gambiae complex such as Anopheles arabiensis. The effects on the accuracy of the age predictions of temperature variation and larval rearing conditions will also be examined; it is possible that other genes in Table 1 than those selected may prove to be more robust under different conditions. However we consider that the assay presented here provides a solid foundation for future refinement, and represents a new tool that can assist a variety of studies related to malaria epidemiology and control.
Experimental procedures
Mosquito rearing and collection
An. gambiae G3 colonies were maintained under standard insectary conditions (∼28 °C; ∼70% relative humidity; 12:2 h light : dark). Adults were given 10% sucrose solution ad libitum; however no bloodmeals were provided. Adults emerging within a 24 h period were considered a single age class and each age class allocated to separate cages. Adult male and female mosquitoes were collected at 0 (≤24 h), 5, 10, 15, 20, 25 and 30 days post-eclosion. Collected individuals were knocked down with ∼2 min exposure to −20 °C before the abdomen was dissected and discarded. The head and thorax were transferred directly into Trizol reagent (Invitrogen, Carlsbad, CA, USA) and stored at −80 °C.
Microarray design
Custom gene expression arrays (8 × 15 k format) were designed using Agilent Technologies' eArray (https://earray.chem.agilent.com/earray). The microarray contained 13 254 custom probes, a single 60-mer probe per An. gambiae transcript (Vectorbase genebuild AgamP3.4 transcripts; http://www.vectorbase.org) and a standard set of 536 Agilent control features.
Total RNA isolation and labelling protocol
Global transcription was quantified at 0, 10, 20 and 30 days of adult age in both male and female An. gambiae. Total RNA was extracted from a pool of five individuals (heads and thoraces only) in 500 µl Trizol reagent. The manufacturer's standard RNA extraction protocol was used, with the exception that RNA was precipitated by a ∼16 h incubation at −20 °C. Array experiments were performed according to Agilent Technologies' One-colour Microarray-based Gene Expression Analysis protocol (v. 5.7, Agilent Technologies, Palo Alto, CA, USA). All consumables and equipment were from Agilent Technologies unless otherwise stated. Briefly, 1 µg total RNA and 5 µl of 1:5000 dilution Agilent One-colour Spike-in mix were amplified and labelled using the Quick Amp Kit protocols (Agilent Technologies). Labelled cRNA was then purified and quantified before hybridization to the arrays using Agilent Technologies' Gene Expression Hybridization Kit, SureHyb chambers, Microarray Hybridization oven and standard protocols. Hybridized arrays were washed with Gene Expression Wash Buffers (without the addition of Triton X102) and Stabilization and Drying Solution to protect against ozone degradation.
Microarray scanning, feature extraction and data analysis
Microarrays were scanned and feature extraction undertaken by technicians at Oxford Gene Technology (Oxford, UK). Scans were at 5 µm resolution using the extended dynamic range feature of the Agilent Technologies' DNA microarray scanner (G2505B). Feature Extraction software (v. 9.5.3.1; Agilent Technologies) was used to quantify and process fluorescence data from the array image files. This process was carried out according to Agilent's One-colour Gene Expression protocol (GE1-v5-95_Feb07). Data files were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) according to Minimum Information About a Microarray Experiment (MIAME) requirements (accession GSE18194).
Array analysis was undertaken with Genespring GX software (v. 10.0.2; Agilent Technologies). The array technology file was created directly from the eArray website (see Microarray design section) using the corresponding microarray design files. Signal intensity data were processed as follows; threshold to 1.0, log2 transformation, 75th percentile shift normalization and baseline transformation to the median of all samples. Array data were filtered by raw signal values (excluding data ≤20th percentile) and flags generated during feature extraction. Flags indicated whether a feature signal was uniform, saturated and significant above background. Differentially expressed transcripts were identified by a two-way analysis of variance at P≤ 0.01 (asymptotic P-value computation) with Benjamini–Hochberg multiple testing correction. Gene transcripts that were differentially expressed only with respect to adult mosquito age were selected. These transcripts were then filtered for transcriptional profiles that were either positively (r > 0.8) or negatively (r < −0.4) correlated with mosquito age. Attention was also given to transcripts with fourfold or greater changes in transcription between the various age classes.
Quantitative RT-PCR age-grading assay
Candidate gene and reference gene expression were measured in adult male and female mosquitoes at 0, 5, 10, 15, 20, 25 and 30 days post-eclosion using qRT-PCR.
Total RNA was extracted from the head and thorax of individual mosquitoes using 250 µl Trizol reagent. Extractions were carried out according to the manufacturer's protocol with the exception of RNA precipitation, which was a ∼16 h incubation at −20 °C. RNA was re-suspended in 20 µl RNase-free water. Total RNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). 500 ng total RNA was treated with 1 unit of DNase I (amplification grade, Invitrogen) mixed with supplied 10× DNase buffer solution. The reaction was incubated at room temperature for 15 min before being terminated with 25 mM ethylenediaminetetraacetic acid and incubated at 65 °C for 10 min.
Reverse transcription was performed using the Superscript VILO cDNA synthesis kit (Invitrogen) according to the manufacturer's protocol. cDNA samples were diluted fivefold prior to qRT-PCR amplification to eliminate PCR inhibitors. Quantitative RT-PCRs were performed in triplicate on a Lightcycler 480 (Roche, Indianapolis, IN, USA) using Express SYBR® Greener qPCR supermix Universal (Invitrogen), 1 µl diluted cDNA template and 400 nM of each primer. Thermal cycling conditions were 95 °C for 5 min; then 45 cycles of 95 °C for 10 s, 58 °C for 10 s, 72 °C for 15 s and a single fluorescence acquisition; and then a melt curve of 95 °C for 5 s, 65 °C for 1 min, then ramp to 97 °C with continuous fluorescence acquisition (5 acquisitions/°C). Ct values were determined using the second-derivative maximum method (Roche).
Multivariate calibration and age predictions
The statistical methods employed to predict mosquito age followed the multivariate calibration approach as previously outlined (Cook et al., 2006, 2007). Briefly, mean Ct values were calculated and normalized to the reference gene (RS7_ANOGA) by logcontrast ratios. Redundancy analysis, implemented through a canonical correlation analysis in SAS (SAS Institute Inc., Cary, NC, USA), was used to calculate a linear combination of gene expression measures (a redundancy variate) that maximized the correlation with mosquito age. The linear regression of the first redundancy variate on age was used to construct a calibration model. Age predictions were estimated from this calibration model by inverse regression techniques. The error associated with age predictions was estimated using a nonparametric bootstrapping procedure. In this study, calibration models were evaluated by generating age predictions using a leave-one-out cross validation approach.
Acknowledgments
This study was supported by an EMBO Long term fellowship to P. E. C., the EC FP7 award ‘AnoPopAge’, and the Wellcome Trust. We thank C. Godfray and M. Blagrove for comments on the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article under the DOI reference: DOI 10.1111/j.1365-2583.2010.01034.x
Figure S1. Age-dependent transcription levels in both male and female adult mosquitoes. Transcription profiles of the nine candidate genes (A–I) and reference gene (J) were measured using one-colour microarrays and displayed as normalized signal intensity (log2) across adult female and male mosquito lifespan.
Figure S2. Logcontrast gene expression measures across adult male and female mosquito lifespan. Transcript abundance of each candidate gene (A–I) was determined relative to RS7_ANOGA as a logcontrast of cycle threshold (Ct) values.
Figure S3. Male Anopheles gambiae calibration model generated from the expression profiles of the four best candidate genes. Logcontrast normalized cycle threshold (Ct) values for AGAP006187, AGAP006985, AGAP007963 and AGAP010398 were entered into a canonical correlation analysis. This analysis generates a redundancy variate, a linear combination of the four gene expression measures, which maximizes correlation with mosquito age. The redundancy variate score for each individual is plotted and the linear regression of these data on age provides the calibration model.
Figure S4. Age predictions for individual male mosquitoes using four gene expression measures. Age predictions were made using a leave-one-out cross validation approach, whereby each data point was sequentially removed from the training data before the age of the excluded individual was predicted. Age was predicted by inverse regression of the female calibration model. Error bars indicate a 95% confidence interval derived by a nonparametric bootstrap procedure.
Figure S5. Residuals, the absolute differences between predicted age and actual age, for individual female (A) and male (B) mosquitoes plotted against predicted age. The distribution of the residuals suggests that there was no bias associated with age predictions.
Please note: Neither the Editors nor Wiley-Blackwell are responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Anyanwu GI, Molyneux DH, Phillips A. Variation in cuticular hydrocarbons among strains of the Anopheles gambiae sensu stricto by analysis of cuticular hydrocarbons using gas liquid chromatography of larvae. Mem Inst Oswaldo Cruz. 2000;95:295–300. doi: 10.1590/s0074-02762000000300003. [DOI] [PubMed] [Google Scholar]
- Anyanwu GI, Davies DH, Molyneux DH, Priestman A. Cuticular-hydrocarbon discrimination between Anopheles gambiae s.s. and An. arabiensis larval karyotypes. Ann Trop Med Parasitol. 2001;95:843–852. doi: 10.1080/00034980120112270. [DOI] [PubMed] [Google Scholar]
- Awolola TS, Oduola OA, Strode C, Koekemoer LL, Brooke B, Ranson H. Evidence of multiple pyrethroid resistance mechanisms in the malaria vector Anopheles gambiae sensu stricto from Nigeria. Trans R Soc Trop Med Hyg. 2009;103:1139–1145. doi: 10.1016/j.trstmh.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Beklemishev WN, Detinova TS, Polovodova VP. Determination of physiological age in anophelines and of age distribution in anopheline populations in the USSR. Bull WHO. 1959;21:223–232. [PMC free article] [PubMed] [Google Scholar]
- Bell AS, Blanford S, Jenkins N, Thomas MB, Read AF. Real-time quantitative PCR for analysis of candidate fungal biopesticides against malaria: technique validation and first applications. J Invert Pathol. 2009;100:160–168. doi: 10.1016/j.jip.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanford S, Chan BH, Jenkins N, Sim D, Turner RJ, Read AF, et al. Fungal pathogen reduces potential for malaria transmission. Science. 2005;308:1638–1641. doi: 10.1126/science.1108423. [DOI] [PubMed] [Google Scholar]
- Brownstein JS, Hett E, O'Neill SL. The potential of virulent Wolbachia to modulate disease transmission by insects. J Invert Pathol. 2003;84:24–29. doi: 10.1016/s0022-2011(03)00082-x. [DOI] [PubMed] [Google Scholar]
- Caputo B, Dani FR, Horne GL, Petrarca V, Turillazzi S, Coluzzi M, et al. Identification and composition of cuticular hydrocarbons of the major Afrotropical malaria vector An. gambiae s.s. (Diptera: Cilicidae): analysis of sexual dimorphism and age-related changes. J Mass Spectrom. 2005;40:1595–1604. doi: 10.1002/jms.961. [DOI] [PubMed] [Google Scholar]
- Carlson DA, Service MW. Differentiation between species of the Anopheles gambiae Giles complex (Diptera: Culicidae) by analysis of cuticular hydrocarbons. Ann Trop Med Parasitol. 1979;73:589–592. doi: 10.1080/00034983.1979.11687301. [DOI] [PubMed] [Google Scholar]
- Carlson DA, Service MW. Identification of mosquitoes of Anopheles gambiae species complex A and B by analysis of cuticular components. Science. 1980;207:1089–1091. doi: 10.1126/science.7355276. [DOI] [PubMed] [Google Scholar]
- Cook PE, Hugo LE, Iturbe-Ormaetxe I, Williams CR, Chenoweth SF, Ritchie SA, et al. The use of transcriptional profiles to predict adult mosquito age under field conditions. Proc Natl Acad Sci USA. 2006;103:18060–18065. doi: 10.1073/pnas.0604875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PE, Hugo LE, Iturbe-Ormaetxe I, Williams CR, Chenoweth SF, Ritchie SA, et al. Predicting the age of mosquitoes using transcriptional profiles. Nat Protoc. 2007;2:2796–2806. doi: 10.1038/nprot.2007.396. [DOI] [PubMed] [Google Scholar]
- Cook PE, McMeniman CJ, O'Neill SL. Modifying insect population age structure to control vector-borne disease. Adv Exp Med Biol. 2008;627:126–140. doi: 10.1007/978-0-387-78225-6_11. [DOI] [PubMed] [Google Scholar]
- Cornman RS, Willis JH. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Mol Biol. 2009;18:607–622. doi: 10.1111/j.1365-2583.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH. Annotation and analysis of a large cuticular protein family with the R & R Consensus in Anopheles gambiae. BMC Genomics. 2008;9:22. doi: 10.1186/1471-2164-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detinova TS. Age grading methods in Diptera of medical importance with special reference to some vectors of malaria. Mono Ser WHO. 1962;47:1–216. [PubMed] [Google Scholar]
- Djouaka RF, Bakare AA, Coulibaly ON, Akogbeto MC, Ranson H, Hemingway J, et al. Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics. 2008;9:538. doi: 10.1186/1471-2164-9-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B, Chenoweth SF, Nuzhdin SV, Blows MW. Natural genetic variation in cuticular hydrocarbon expression in male and female Drosophila melanogaster. Genetics. 2007;175:1465–1477. doi: 10.1534/genetics.106.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Jones C. Prognosis for interruption of malaria transmission through assessment of the mosquito's vectorial capacity. Nature. 1964;204:1173–1175. doi: 10.1038/2041173a0. [DOI] [PubMed] [Google Scholar]
- Gilles HM, Warrell DA. Essential Malariology. London: Arnold; 2002. [Google Scholar]
- Hayes EJ, Wall R. Age-grading adult insects: a review of techniques. Physiol Entomol. 1999;24:1–10. [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Hugo LE, Kay BH, Eaglesham GK, Holling N, Ryan PA. Investigation of cuticular hydrocarbons for determining the age and survivorship of Australasian mosquitoes. Am J Trop Med Hyg. 2006;74:462–474. [PubMed] [Google Scholar]
- Hugo LE, Quick-Miles S, Kay BH, Ryan PA. Evaluations of mosquito age grading techniques based on morphological changes. J Med Entomol. 2008;45:353–369. doi: 10.1603/0022-2585(2008)45[353:eomagt]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hugo LE, Cook PE, Johnson PH, Rapley LP, Kay BH, Ryan PA, et al. Field validation of a transcriptional assay for the prediction of age of uncaged Aedes aegypti mosquitoes in Northern Australia. PLoS Negl Trop Dis. 2010;4:e608. doi: 10.1371/journal.pntd.0000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huho BJ, Ng'habi KR, Killeen GF, Nkwengulila G, Knols BGJ, Ferguson HM. A reliable morphological method to assess the age of male An. gambiae. Malar J. 2006;5:62. doi: 10.1186/1475-2875-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane MJ. Determining the age of an insect. Parasitol Today. 1985;1:81–85. doi: 10.1016/0169-4758(85)90048-1. [DOI] [PubMed] [Google Scholar]
- Marinotti O, Calvo E, Nguyen QK, Dissanayake S, Ribeiro JM, James AA. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- Mayagaya VS, Michel K, Benedict MQ, Killeen GF, Wirtz RA, Ferguson HM, et al. Non-destructive determination of age and species of Anopheles gambiae s.l. using near-infrared spectroscopy. Am J Trop Med Hyg. 2009;81:622–630. doi: 10.4269/ajtmh.2009.09-0192. [DOI] [PubMed] [Google Scholar]
- McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–144. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- Min KT, Benzer S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci USA. 1997;94:10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randimby FM, Gorham J, Duchemin JB, Robert V, Lehane MJ. Value of HPLC and fluorescent pteridines in estimating the age of Anopheles gambiae adults. Arch Inst Pasteur Madagascar. 2002;68:86–89. [PubMed] [Google Scholar]
- Read AF, Lynch PA, Thomas MB. How to make evolution-proof insecticides for malaria control. PLoS Biol. 2009;7:e1000058. doi: 10.1371/journal.pbio.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte E-J, Ng'habi K, Kihonda J, Takken W, Paaijmans K, Abdulla S, et al. An entomopathogenic fungus for control of adult African malaria mosquitoes. Science. 2005;308:1641–1642. doi: 10.1126/science.1108639. [DOI] [PubMed] [Google Scholar]
- Smith DL, McKenzie FE. Statics and dynamics of malaria infection in Anopheles mosquitoes. Malar J. 2004;3:13. doi: 10.1186/1475-2875-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndale-Biscoe M. Age-grading methods in adult insects: a review. Bull Entomol Res. 1984;74:341–377. [Google Scholar]
- Vontas J, David JP, Nikou D, Hemingway J, Christophides GK, Louis C, et al. Transcriptional analysis of insecticide resistance in Anopheles stephensi using cross-species microarray hybridization. Insect Mol Biol. 2007;16:315–324. doi: 10.1111/j.1365-2583.2007.00728.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


