Abstract
Blood pressure (BP) is a major cardiovascular disease risk factor. To date, few variants associated with inter-individual BP variation have been identified. A genome-wide association study of systolic (SBP), diastolic BP (DBP), and hypertension in the CHARGE Consortium (n=29,136) identified 13 SNPs for SBP, 20 for DBP, and 10 for hypertension at p <4×10-7. The top 10 loci for SBP and DBP were incorporated into a risk score; mean BP and prevalence of hypertension increased in relation to number of risk alleles carried. When 10 CHARGE SNPs for each trait were meta-analyzed jointly with the Global BPgen Consortium (n=34,433), four CHARGE loci attained genome-wide significance (p<5×10-8) for SBP (ATP2B1, CYP17A1, PLEKHA7, SH2B3), six for DBP (ATP2B1, CACNB2, CSK/ULK3, SH2B3, TBX3/TBX5, ULK4), and one for hypertension (ATP2B1). Identifying novel BP genes advances our understanding of BP regulation and highlights potential drug targets for the prevention or treatment of hypertension.
High blood pressure affects about one third of adults and contributes to 13.5 million deaths worldwide each year and about half the global risk for stroke and ischemic heart disease. 1,2 Clinical trials, dating back more than forty years, have proven that drug treatment to lower blood pressure dramatically reduces the risk of cardiovascular events in people with hypertension.3,4
The substantial (30-60 percent)5 heritability of blood pressure has prompted extensive efforts to identify its genetic underpinnings. The search for genes associated with inter-individual variation in blood pressure in the general population has used a variety of complementary approaches, which have yielded relatively few clues. Linkage and candidate gene studies, despite considerable knowledge about pathways that are critical to blood pressure homeostasis, have provided limited consistent evidence of blood pressure quantitative trait loci.6,7,8 The study of families with rare Mendelian high or low blood pressure syndromes has identified mutations with gain or loss of function in about a dozen renal sodium regulatory genes.9 Common variants in two renal sodium regulatory genes have been found to be associated with blood pressure in the general population.10 The vast majority of the genetic contribution to variation in blood pressure, however, remains unexplained.
Large-scale genome-wide association studies (GWAS), in which hundreds of thousands of common genetic variants are genotyped and analyzed for disease association, have shown great success in identifying genes associated with common diseases and traits.11,12 The fact that six GWAS published to date, however, have not identified loci associated with blood pressure or hypertension at p<5×10-8, has raised concerns about the utility of this approach for these traits.13,14,15,16,17,18
If blood pressure variation in the general population is due to multiple variants with small effects, very large study samples are needed to identify them. We established the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium19 to identify common genetic variation associated with complex traits. The CHARGE Consortium consists of 29,136 participants of European descent who had undergone standardized blood pressure measurements in six population-based cohort studies: the Age, Gene/Environment Susceptibility Reykjavik Study (AGES), Atherosclerosis Risk in Communities (ARIC), Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), Rotterdam Study (RS) and the Rotterdam Extension Study (RES). We report the top findings of our GWAS of systolic blood pressure, diastolic blood pressure, and hypertension and provide replication results for our most promising loci in the Global BPgen Consortium,20 another GWAS consortium of similar size, and report combined meta-analysis findings of the two consortia for the most promising CHARGE loci.
Results
Study samples
The total sample size for this analysis was 29,136 (AGES, n=3,219; ARIC, n=8,047; CHS, n=3,277; FHS, n=8,096; RS n=4,737; RES, n=1760). Characteristics of the study sample are presented in Supplementary Table 1. The mean age of the study participants at the initial examination varied from 38 years (FHS) to 72 years (CHS). The mean observed (and treatment corrected) systolic blood pressures across the six cohorts ranged from 118 (120) mm Hg (ARIC) to 143 (145) mm Hg (RES); the mean diastolic blood pressure ranged from 72 (73) mm Hg (ARIC) to 83 (84) mm Hg (AGES). The proportion of participants taking antihypertensive medication ranged from 5 percent (FHS) to 38 percent (CHS) and the proportion with hypertension ranged from 17 percent (FHS) to 60 percent (RES).
Meta-analysis of CHARGE cohort results
Within cohort analyses were combined by meta-analysis and the results for all SNPs with p value <1×10-6 are presented in Table 1 for systolic blood pressure, Table 2 for diastolic blood pressure, and Table 3 for hypertension; within each of these tables the results for the other two blood pressure phenotypes are also provided. Supplementary Table 2 provides summary data for the top SNPs for each phenotype for each of the 6 cohorts. QQ and −log10(p) genome-wide association plots are presented in Supplementary Figure 1 for systolic blood pressure, diastolic blood pressure, and hypertension. The QQ plots show departure from the line of identity at approximately 1×10-3 for systolic and diastolic blood pressure. The number of SNPs with p values <1×10-3 was 3433 for systolic and 3558 for diastolic blood pressure vs. 2540 expected. The proportion of SNPs with p <1×10-3 that are intragenic was 47 percent for systolic and diastolic blood pressure vs. an average of 37 percent for all imputed SNPs. For systolic blood pressure the meta-analysis identified 13 SNPs with p <4×10-7 (stage 1 threshold). The strongest signal for systolic pressure was for rs2681492 (p=3.0×10-11) in ATP2B1 on chromosome 12q21-23. A low minor allele frequency variant in C18orf1 (rs8096897; p=3.2×10-7) showed evidence of association with systolic blood pressure as did CASZ1 (rs880315; p=2.1×10-7). A signal was identified on chromosome 12q24 for SH2B3 (rs3184504; p=5.7×10-7) and for nearby ATXN2 (rs653178; p=8.5×10-7). PLEKHA7 (chromosome 11p15.1, rs381815, p=5.8×10-7) and a locus on chromosome 2q31-33 adjacent to PMS1 and MSTN (rs7571613, p=7.2×10-7) revealed suggestive evidence of association. Of note, many of the top systolic blood pressure SNPs were also associated with other blood pressure phenotypes (Table 1). The top allelic associations were generally consistent in size and direction across the 6 individual cohorts (Supplementary Table 2).
Table 1.
Genome-wide Association Results for Systolic Blood Pressure SNPs with P Value <1×10-6 Sorted by Systolic Blood Pressure Meta-analysis P Value
| SNP identifier | Chr | Position | Gene | MAF | CHARGE Meta-analysis SBP | CHARGE Meta-analysis DBP | CHARGE Meta-analysis Hypertension | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p | Beta | SE | p | Beta | SE | p | |||||
| rs2681492 | 12 | 88537220 | ATP2B1 | 0.20 | -1.26 | 0.19 | 3.0E-11 | -0.62 | 0.11 | 4.6E-08 | -0.14 | 0.03 | 8.4E-08 |
| rs2681472 | 12 | 88533090 | ATP2B1 | 0.18 | -1.29 | 0.19 | 3.5E-11 | -0.64 | 0.11 | 3.7E-08 | -0.16 | 0.03 | 1.7E-08 |
| rs11105354 | 12 | 88550654 | ATP2B1 | 0.18 | -1.30 | 0.20 | 3.7E-11 | -0.63 | 0.11 | 5.8E-08 | -0.16 | 0.03 | 1.8E-08 |
| rs11105364 | 12 | 88593407 | 0.18 | -1.30 | 0.20 | 4.8E-11 | -0.63 | 0.12 | 1.2E-07 | -0.16 | 0.03 | 2.1E-08 | |
| rs17249754 | 12 | 88584717 | 0.18 | -1.30 | 0.20 | 5.2E-11 | -0.63 | 0.12 | 1.0E-07 | -0.16 | 0.03 | 2.2E-08 | |
| rs11105368 | 12 | 88598572 | 0.18 | -1.30 | 0.20 | 5.3E-11 | -0.63 | 0.12 | 1.3E-07 | -0.16 | 0.03 | 2.2E-08 | |
| rs12579302 | 12 | 88574634 | 0.18 | -1.29 | 0.20 | 6.2E-11 | -0.62 | 0.12 | 1.3E-07 | -0.16 | 0.03 | 2.2E-08 | |
| rs12230074 | 12 | 88614998 | 0.17 | -1.31 | 0.20 | 9.1E-11 | -0.62 | 0.12 | 3.4E-07 | -0.17 | 0.03 | 2.9E-08 | |
| rs11105378 | 12 | 88614872 | 0.17 | -1.31 | 0.20 | 9.1E-11 | -0.62 | 0.12 | 3.1E-07 | -0.17 | 0.03 | 2.8E-08 | |
| rs4842666 | 12 | 88465680 | 0.17 | -1.20 | 0.21 | 6.5E-09 | -0.62 | 0.12 | 4.5E-07 | -0.15 | 0.03 | 3.4E-07 | |
| rs8096897 | 18 | 13428905 | C18orf1 | 0.01 | -12.87 | 2.33 | 3.2E-08 | -4.07 | 1.33 | 2.9E-03 | -0.73 | 0.35 | 0.04 |
| rs11105328 | 12 | 88466521 | 0.18 | -1.11 | 0.20 | 4.2E-08 | -0.61 | 0.12 | 5.1E-07 | -0.15 | 0.03 | 7.1E-07 | |
| rs880315 | 1 | 10719453 | CASZ1 | 0.35 | 0.89 | 0.17 | 2.1E-07 | 0.30 | 0.10 | 2.9E-03 | 0.09 | 0.02 | 6.2E-05 |
| rs3184504 | 12 | 110368991 | SH2B3 | 0.48 | 0.75 | 0.15 | 5.7E-07 | 0.50 | 0.09 | 1.7E-08 | 0.07 | 0.02 | 7.4E-04 |
| rs381815 | 11 | 16858844 | PLEKHA7 | 0.26 | 0.84 | 0.17 | 5.8E-07 | 0.51 | 0.10 | 4.3E-07 | 0.09 | 0.02 | 1.7E-04 |
| rs7926335 | 11 | 16874445 | PLEKHA7 | 0.26 | 0.85 | 0.17 | 5.8E-07 | 0.51 | 0.10 | 4.8E-07 | 0.09 | 0.02 | 1.9E-04 |
| rs7571613 | 2 | 190513907 | PMS1 | 0.18 | 0.96 | 0.19 | 7.2E-07 | 0.55 | 0.11 | 2.2E-06 | 0.09 | 0.03 | 5.2E-04 |
| rs11895934 | 2 | 190510498 | 0.18 | 0.96 | 0.19 | 7.3E-07 | 0.55 | 0.11 | 2.2E-06 | 0.09 | 0.03 | 5.5E-04 | |
| rs7564968 | 2 | 190520217 | 0.18 | 0.96 | 0.19 | 8.0E-07 | 0.55 | 0.11 | 2.3E-06 | 0.09 | 0.03 | 4.9E-04 | |
| rs653178 | 12 | 110492139 | ATXN2 | 0.48 | 0.74 | 0.15 | 8.5E-07 | 0.50 | 0.09 | 2.0E-08 | 0.07 | 0.02 | 7.8E-04 |
| rs284277 | 1 | 10713384 | CASZ1 | 0.35 | 0.79 | 0.16 | 9.4E-07 | 0.24 | 0.09 | 0.01 | 0.09 | 0.02 | 6.9E-05 |
Chr=chromosome; MAF=minor allele frequency; NA=not available DBP=diastolic blood pressure; SBP=systolic blood pressure
Beta is the effect size on blood pressure, in mm Hg, per allele based on the additive genetic model
Table 2.
Genome-wide Association Results for Diastolic Blood Pressure SNPs with P Value <1×10-6 Sorted by Diastolic Blood Pressure Meta-analysis P Value
| SNP identifier | Chr | Position | Gene | MAF | CHARGE Meta-analysis DBP | CHARGE Meta-analysis SBP | CHARGE Meta-analysis Hypertension | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | Beta | SE | p | Beta | SE | p | |||||
| rs3184504 | 12 | 110368991 | SH2B3 | 0.48 | 0.50 | 0.09 | 1.7E-08 | 0.75 | 0.15 | 5.7E-07 | 0.07 | 0.02 | 7.4E-04 |
| rs653178 | 12 | 110492139 | ATXN2 | 0.48 | 0.50 | 0.09 | 2.0E-08 | 0.74 | 0.15 | 8.5E-07 | 0.07 | 0.02 | 7.7E-04 |
| rs2681472 | 12 | 88533090 | ATP2B1 | 0.17 | -0.64 | 0.12 | 3.7E-08 | -1.29 | 0.19 | 3.5E-11 | -0.16 | 0.03 | 1.7E-08 |
| rs4766578 | 12 | 110388754 | ATXN2 | 0.49 | 0.49 | 0.09 | 4.2E-08 | 0.73 | 0.15 | 1.2E-06 | 0.06 | 0.02 | 1.9E-03 |
| rs10774625 | 12 | 110394602 | ATXN2 | 0.49 | 0.49 | 0.09 | 4.2E-08 | 0.73 | 0.15 | 1.1E-06 | 0.06 | 0.02 | 1.8E-03 |
| rs2681492 | 12 | 88537220 | ATP2B1 | 0.19 | -0.62 | 0.11 | 4.6E-08 | -1.26 | 0.18 | 3.0E-11 | -0.14 | 0.03 | 8.4E-08 |
| rs11105354 | 12 | 88550654 | ATP2B1 | 0.17 | -0.63 | 0.12 | 5.8E-08 | -1.30 | 0.19 | 3.7E-11 | -0.16 | 0.03 | 1.8E-08 |
| rs17630235 | 12 | 111076069 | TRAFD1 | 0.43 | 0.50 | 0.09 | 1.0E-07 | 0.69 | 0.15 | 1.1E-05 | 0.06 | 0.02 | 4.3E-03 |
| rs17249754 | 12 | 88584717 | 0.17 | -0.63 | 0.12 | 1.0E-07 | -1.30 | 0.19 | 5.2E-11 | -0.16 | 0.03 | 2.2E-08 | |
| rs11066188 | 12 | 111095097 | C12orf51 | 0.43 | 0.50 | 0.09 | 1.1E-07 | 0.68 | 0.15 | 1.3E-05 | 0.06 | 0.02 | 4.2E-03 |
| rs11105364 | 12 | 88593407 | 0.17 | -0.63 | 0.12 | 1.2E-07 | -1.30 | 0.19 | 4.8E-11 | -0.16 | 0.03 | 2.1E-08 | |
| rs11105368 | 12 | 88598572 | 0.17 | -0.63 | 0.12 | 1.2E-07 | -1.30 | 0.19 | 5.3E-11 | -0.16 | 0.03 | 2.2E-08 | |
| rs12579302 | 12 | 88574634 | 0.17 | -0.62 | 0.12 | 1.2E-07 | -1.29 | 0.19 | 6.2E-11 | -0.16 | 0.03 | 2.2E-08 | |
| rs2384550 | 12 | 113837114 | TBX3/TBX5 | 0.35 | -0.48 | 0.09 | 1.3E-07 | -0.71 | 0.15 | 4.3E-06 | -0.08 | 0.02 | 5.6E-05 |
| rs1991391 | 12 | 113837049 | 0.35 | -0.48 | 0.09 | 1.4E-07 | -0.71 | 0.15 | 3.8E-06 | -0.09 | 0.02 | 5.6E-05 | |
| rs6489992 | 12 | 113837152 | 0.37 | -0.48 | 0.09 | 2.0E-07 | -0.71 | 0.15 | 4.7E-06 | -0.08 | 0.02 | 1.9E-04 | |
| rs11065987 | 12 | 110556807 | 0.42 | 0.48 | 0.09 | 2.2E-07 | 0.70 | 0.15 | 9.4E-06 | 0.06 | 0.02 | 4.1E-03 | |
| rs11024074 | 11 | 16873795 | PLEKHA7 | 0.28 | 0.50 | 0.10 | 2.8E-07 | 0.79 | 0.16 | 1.6E-06 | 0.09 | 0.02 | 5.2E-05 |
| rs11105378 | 12 | 88614872 | 0.17 | -0.62 | 0.12 | 3.1E-07 | -1.31 | 0.20 | 9.1E-11 | -0.17 | 0.03 | 2.8E-08 | |
| rs12230074 | 12 | 88614998 | 0.17 | -0.62 | 0.12 | 3.4E-07 | -1.31 | 0.20 | 9.1E-11 | -0.17 | 0.03 | 2.9E-08 | |
| rs7963771 | 12 | 113827875 | 0.31 | -0.53 | 0.10 | 4.3E-07 | -0.73 | 0.17 | 4.7E-05 | -0.07 | 0.02 | 3.8E-03 | |
| rs381815 | 11 | 16858844 | PLEKHA7 | 0.26 | 0.51 | 0.10 | 4.3E-07 | 0.84 | 0.16 | 5.8E-07 | 0.09 | 0.02 | 1.7E-04 |
| rs4842666 | 12 | 88465680 | 0.17 | -0.62 | 0.12 | 4.5E-07 | -1.20 | 0.20 | 6.5E-09 | -0.15 | 0.03 | 3.4E-07 | |
| rs7926335 | 11 | 16874445 | PLEKHA7 | 0.26 | 0.51 | 0.10 | 4.8E-07 | 0.85 | 0.16 | 5.8E-07 | 0.09 | 0.02 | 1.9E-04 |
| rs11105328 | 12 | 88466521 | 0.18 | -0.61 | 0.12 | 5.1E-07 | -1.11 | 0.20 | 4.2E-08 | -0.15 | 0.03 | 7.1E-07 | |
| rs17696736 | 12 | 110971201 | C12orf30 | 0.44 | 0.46 | 0.09 | 5.1E-07 | 0.64 | 0.15 | 3.5E-05 | 0.05 | 0.02 | 0.015 |
| rs10744835 | 12 | 113838232 | 0.30 | -0.49 | 0.10 | 7.1E-07 | -0.68 | 0.16 | 3.9E-05 | -0.07 | 0.02 | 1.5E-03 | |
| rs7977406 | 12 | 113843807 | 0.30 | -0.49 | 0.10 | 7.6E-07 | -0.69 | 0.16 | 2.9E-05 | -0.08 | 0.02 | 1.2E-03 | |
| rs9815354 | 3 | 41887655 | ULK4 | 0.17 | 0.60 | 0.12 | 7.8E-07 | 0.08 | 0.20 | 6.9E-01 | -0.01 | 0.03 | 0.83 |
| rs6495122 | 15 | 72912698 | CSK | 0.42 | 0.45 | 0.09 | 8.0E-07 | 0.64 | 0.15 | 2.7E-05 | 0.07 | 0.02 | 4.0E-03 |
| rs11014166 | 10 | 18748804 | CACNB2 | 0.34 | -0.46 | 0.09 | 8.7E-07 | -0.74 | 0.15 | 2.1E-06 | -0.11 | 0.02 | 7.8E-07 |
| rs6768438 | 3 | 41840359 | ULK4 | 0.16 | 0.59 | 0.12 | 9.7E-07 | 0.11 | 0.20 | 5.9E-01 | 0.01 | 0.03 | 0.84 |
| rs9852991 | 3 | 41850459 | ULK4 | 0.16 | 0.59 | 0.12 | 9.7E-07 | 0.11 | 0.20 | 5.9E-01 | 0.01 | 0.03 | 0.85 |
| rs13401889 | 2 | 190618804 | MSTN | 0.21 | 0.54 | 0.11 | 9.7E-07 | 0.88 | 0.18 | 2.7E-06 | 0.10 | 0.03 | 1.6E-04 |
| rs9816772 | 3 | 41847881 | ULK4 | 0.16 | 0.59 | 0.12 | 9.7E-07 | 0.11 | 0.20 | 5.9E-01 | 0.01 | 0.03 | 0.85 |
Chr=chromosome; MAF=minor allele frequency; NA=not available DBP=diastolic blood pressure; SBP=systolic blood pressure
Beta is the effect size on blood pressure, in mm Hg, per allele based on the additive genetic model
Table 3.
Genome-wide Association Results for Hypertension SNPs with P Value <1×10-6 Sorted by Hypertension Meta-analysis P Value
| SNP identifier | Chr | Position | Gene | MAF | CHARGE Meta-analysis Hypertension | CHARGE Meta-analysis SBP | CHARGE Meta-analysis DBP | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p | Beta | SE | p | Beta | SE | p | |||||
| rs2681472 | 12 | 88533090 | ATP2B1 | 0.17 | -0.16 | 0.03 | 1.7E-08 | -1.29 | 0.19 | 3.5E-11 | -0.64 | 0.11 | 3.7E-08 |
| rs11105354 | 12 | 88550654 | ATP2B1 | 0.17 | -0.16 | 0.03 | 1.8E-08 | -1.30 | 0.19 | 3.7E-11 | -0.63 | 0.11 | 5.8E-08 |
| rs11105364 | 12 | 88593407 | 0.17 | -0.16 | 0.03 | 2.1E-08 | -1.30 | 0.19 | 4.8E-11 | -0.63 | 0.12 | 1.2E-07 | |
| rs17249754 | 12 | 88584717 | 0.17 | -0.16 | 0.03 | 2.2E-08 | -1.30 | 0.19 | 5.2E-11 | -0.63 | 0.12 | 1.0E-07 | |
| rs11105368 | 12 | 88598572 | 0.17 | -0.16 | 0.03 | 2.2E-08 | -1.30 | 0.19 | 5.3E-11 | -0.63 | 0.12 | 1.2E-07 | |
| rs12579302 | 12 | 88574634 | 0.17 | -0.16 | 0.03 | 2.2E-08 | -1.29 | 0.19 | 6.2E-11 | -0.62 | 0.12 | 1.2E-07 | |
| rs11105378 | 12 | 88614872 | 0.16 | -0.17 | 0.03 | 2.8E-08 | -1.31 | 0.20 | 9.1E-11 | -0.62 | 0.12 | 3.1E-07 | |
| rs12230074 | 12 | 88614998 | 0.16 | -0.17 | 0.03 | 2.8E-08 | -1.31 | 0.20 | 9.1E-11 | -0.62 | 0.12 | 3.4E-07 | |
| rs2681492 | 12 | 88537220 | ATP2B1 | 0.19 | -0.14 | 0.03 | 8.4E-08 | -1.26 | 0.18 | 3.0E-11 | -0.62 | 0.11 | 4.6E-08 |
| rs4842666 | 12 | 88465680 | 0.17 | -0.15 | 0.03 | 3.4E-07 | -1.20 | 0.20 | 6.5E-09 | -0.62 | 0.12 | 4.5E-07 | |
| rs7640747 | 3 | 37571809 | ITGA9 | 0.38 | 0.12 | 0.02 | 4.8E-07 | 0.56 | 0.16 | 5.9E-04 | 0.32 | 0.09 | 9.5E-04 |
| rs11105328 | 12 | 88466521 | 0.18 | -0.15 | 0.03 | 7.1E-07 | -1.11 | 0.20 | 4.2E-08 | -0.61 | 0.12 | 5.1E-07 | |
| rs743395 | 3 | 37573386 | ITGA9 | 0.38 | 0.12 | 0.02 | 7.5E-07 | 0.58 | 0.16 | 4.4E-04 | 0.33 | 0.10 | 8.0E-04 |
| rs11014166 | 10 | 18748804 | CACNB2 | 0.17 | -0.11 | 0.02 | 7.8E-07 | -0.74 | 0.15 | 2.1E-06 | -0.46 | 0.09 | 8.7E-07 |
DBP=diastolic blood pressure; SBP=systolic blood pressure
For diastolic blood pressure (Table 2) there were 20 SNPs with p <4×10-7. Significant association signals were detected in a large 1-megabase block of linkage disequilibrium on chromosome 12q24 that includes SH2B3 (rs3184504, p=1.7×10-8), ATXN2 (rs653178, p=2.0×10-8; r2=1.0 with rs3184504), and TRAFD1 (rs17630235, p=1.0×10-7; r2=0.66 with rs3184504). This block encompasses CUTL2, FAM109A, SH2B3, ATXN2, BRAP, ACAD10, ALDH2, MAPKAPK5, TMEM116, ERP29, C12orf30, TRAFD1, C12orf51, RPL6, and PTPN11 (Supplementary Figure 2). In addition, ATP2B1 (chromosome 12q21, rs2681472, p=3.7×10-8), TBX3/TBX5 (chromosome 12q24, rs2384550, p=1.3×10-7), and PLEKHA7 (chromosome 11p15, rs11024074, p=2.8×10-7) showed association with diastolic blood pressure. Suggestive evidence of association was found for loci in or adjacent to ULK4 (chromosome 3p22.1), CSK/ULK3 (chromosome 15q24), and CACNB2 (chromosome 10p12). Multiple diastolic blood pressure SNPs were also associated with other blood pressure phenotypes (Table 2). The top allelic associations were generally consistent in size and direction across studies (Supplementary Table 2).
For the dichotomous trait of hypertension (Table 3), one significant association was detected for ATP2B1 (rs2681472, p=1.7×10-8), with an odds ratio for hypertension of 1.17 per risk allele. Suggestive evidence of association was detected for ITGA9 (chromosome 3p22.2, rs7640747, p=4.8×10-7) and CACNB2 (rs11014166, p=8.7×10-7).
Independent replication in Global BPgen and combined meta-analysis of top CHARGE SNPs
Thirty SNPs representing the top ten CHARGE Consortium loci for systolic pressure, diastolic pressure, and hypertension were exchanged for lookup within the Global BPgen Consortium GWAS results. One SNP for systolic blood pressure, four for diastolic blood pressure, and one for hypertension that attained stage 1 p <4×10-7 in CHARGE were assessed for evidence of independent replication in Global BPgen (Table 4). Five of these six associations fulfilled criteria for external replication in Global BPgen of p <0.008 (0.05/6, one tailed test). The replicated loci included ATP2B1 (for systolic blood pressure, diastolic blood pressure and hypertension), SH2B3 (diastolic blood pressure), and TBX3/TBX5 (diastolic blood pressure). PLEKHA7 did not replicate for diastolic blood pressure (rs11024074, p=0.03 in Global BPgen), however, another SNP in PLEKHA7 was genome-wide significant (at p <5×10-8, stage 2) for systolic blood pressure (rs381815) in the joint meta-analysis of CHARGE and Global BPgen. Of note, for 29 of 30 CHARGE SNPs that were exchanged, the directional association (sign of beta) was identical in both consortia.
Table 4.
Meta-analysis of CHARGE and Global BPgen of Top 10 Loci for Systolic and Diastolic Blood Pressure and Hypertension in CHARGE
| CHARGE meta-analysis results | Global BPgen meta-analysis results | CHARGE + Global BPgen meta-analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP identifier | Chr | Position | Nearest Gene | Alleles (coded / other) | Freq. of coded allele | Beta | SE | p-value | Beta | SE | p-value | Beta | SE | p-value |
|
Systolic blood pressure | ||||||||||||||
| rs12046278 | 1 | 10,722,164 | CASZ1 | T/C | 0.64 | -0.84 | 0.18 | 1.84E-06 | -0.29 | 0.15 | 5.71E-02 | -0.53 | 0.12 | 4.77E-06 |
| rs7571613 | 2 | 190,513,907 | PMS1 | A/G | 0.82 | -0.96 | 0.19 | 7.28E-07 | -0.23 | 0.16 | 1.59E-01 | -0.54 | 0.13 | 1.90E-05 |
| rs448378 | 3 | 170,583,593 | MDS1 | A/G | 0.52 | -0.71 | 0.15 | 1.28E-06 | -0.36 | 0.13 | 4.76E-03 | -0.51 | 0.10 | 1.18E-07 |
| rs2736376 | 8 | 11,155,175 | MTMR9 | C/G | 0.13 | -1.08 | 0.23 | 1.90E-06 | -0.06 | 0.19 | 7.36E-01 | -0.48 | 0.15 | 9.15E-04 |
| rs1910252 | 8 | 49,569,915 | EFCAB1 | T/C | 0.18 | -0.93 | 0.19 | 1.70E-06 | -0.07 | 0.17 | 6.80E-01 | -0.43 | 0.13 | 6.13E-04 |
| rs11014166 | 10 | 18,748,804 | CACNB2 | A/T | 0.66 | 0.74 | 0.16 | 2.11E-06 | 0.33 | 0.13 | 1.31E-02 | 0.50 | 0.10 | 7.03E-07 |
| rs1004467 | 10 | 104,584,497 | CYP17A1 | A/G | 0.90 | 1.20 | 0.25 | 1.99E-06 | 0.94 | 0.21 | 1.08E-05 | 1.05 | 0.16 | 1.28E-10 |
| rs381815 | 11 | 16,858,844 | PLEKHA7 | T/C | 0.26 | 0.84 | 0.17 | 5.76E-07 | 0.52 | 0.14 | 2.72E-04 | 0.65 | 0.11 | 1.89E-09 |
| rs2681492 | 12 | 88,537,220 | ATP2B1 | T/C | 0.80 | 1.26 | 0.19 | 3.01E-11 | 0.50 | 0.17 | 4.07E-03 | 0.85 | 0.13 | 3.76E-11 |
| rs3184504 | 12 | 110,368,991 | SH2B3 | T/C | 0.48 | 0.75 | 0.15 | 5.73E-07 | 0.45 | 0.13 | 6.36E-04 | 0.58 | 0.10 | 4.52E-09 |
|
Diastolic blood pressure | ||||||||||||||
| rs13423988 | 2 | 68,764,770 | GPR73/ARHGAP25 | T/C | 0.17 | 0.59 | 0.12 | 1.09E-06 | 0.11 | 0.11 | 3.22E-01 | 0.33 | 0.08 | 5.00E-05 |
| rs13401889 | 2 | 190,618,804 | MSTN | T/C | 0.79 | -0.54 | 0.11 | 9.58E-07 | -0.10 | 0.11 | 3.55E-01 | -0.31 | 0.08 | 4.82E-05 |
| rs9815354 | 3 | 41,887,655 | ULK4 | A/G | 0.17 | 0.60 | 0.12 | 7.88E-07 | 0.40 | 0.11 | 3.79E-04 | 0.49 | 0.08 | 2.54E-09 |
| rs7016759 | 8 | 49,574,969 | EFCAB1 | T/C | 0.83 | 0.57 | 0.12 | 1.87E-06 | 0.06 | 0.11 | 5.79E-01 | 0.30 | 0.08 | 2.29E-04 |
| rs11014166 | 10 | 18,748,804 | CACNB2 | A/T | 0.66 | 0.46 | 0.09 | 8.82E-07 | 0.28 | 0.09 | 1.46E-03 | 0.37 | 0.06 | 1.24E-08 |
| rs11024074 | 11 | 16,873,795 | PLEKHA7 | T/C | 0.72 | -0.50 | 0.10 | 2.83E-07 | -0.17 | 0.09 | 6.82E-02 | -0.33 | 0.07 | 1.20E-06 |
| rs2681472 | 12 | 88,533,090 | ATP2B1 | A/G | 0.83 | 0.64 | 0.12 | 3.74E-08 | 0.36 | 0.12 | 2.43E-03 | 0.50 | 0.08 | 1.47E-09 |
| rs3184504 | 12 | 110,368,991 | SH2B3 | T/C | 0.48 | 0.50 | 0.09 | 1.68E-08 | 0.45 | 0.09 | 2.83E-07 | 0.48 | 0.06 | 2.58E-14 |
| rs2384550 | 12 | 113,837,114 | TBX3/TBX5 | A/G | 0.35 | -0.48 | 0.09 | 1.32E-07 | -0.23 | 0.09 | 1.06E-02 | -0.35 | 0.06 | 3.75E-08 |
| rs6495122 | 15 | 72,912,698 | CSK/ULK3 | A/C | 0.42 | 0.45 | 0.09 | 8.10E-07 | 0.35 | 0.09 | 3.98E-05 | 0.40 | 0.06 | 1.84E-10 |
|
Hypertension | ||||||||||||||
| rs17806132 | 2 | 190,416,532 | PMS1 | A/G | 0.16 | 0.14 | 0.03 | 1.14E-05 | 0.04 | 0.04 | 2.87E-01 | 0.10 | 0.02 | 4.70E-05 |
| rs305489 | 3 | 11,986,163 | SYN2 | A/G | 0.55 | 0.10 | 0.02 | 4.20E-06 | 0.01 | 0.03 | 7.75E-01 | 0.06 | 0.02 | 1.70E-04 |
| rs7640747 | 3 | 37,571,809 | ITGA9 | C/G | 0.62 | -0.12 | 0.02 | 4.53E-07 | -0.02 | 0.03 | 4.12E-01 | -0.08 | 0.02 | 1.24E-05 |
| rs11775334 | 8 | 10,109,030 | MSRA | A/G | 0.32 | 0.10 | 0.02 | 1.03E-05 | 0.05 | 0.03 | 5.86E-02 | 0.08 | 0.02 | 4.05E-06 |
| rs899364 | 8 | 11,366,954 | FAM167A/BLK | T/G | 0.32 | -0.10 | 0.02 | 6.95E-06 | -0.04 | 0.03 | 1.32E-01 | -0.08 | 0.02 | 1.01E-05 |
| rs11014166 | 10 | 18,748,804 | CACNB2 | A/T | 0.66 | 0.11 | 0.02 | 7.96E-07 | 0.07 | 0.03 | 1.06E-02 | 0.09 | 0.02 | 5.72E-08 |
| rs2681472 | 12 | 88,533,090 | ATP2B1 | A/G | 0.83 | 0.16 | 0.03 | 1.65E-08 | 0.13 | 0.04 | 2.15E-04 | 0.15 | 0.02 | 1.75E-11 |
| rs278126 | 12 | 118,620,100 | CIT | T/G | 0.28 | 0.11 | 0.02 | 8.34E-06 | -0.01 | 0.03 | 6.72E-01 | 0.06 | 0.02 | 1.74E-03 |
| rs11612893 | 12 | 129,290,572 | FZD10/PIWIL1 | T/G | 0.10 | -0.19 | 0.04 | 7.62E-06 | -0.04 | 0.06 | 4.19E-01 | -0.14 | 0.03 | 5.50E-05 |
| rs16982520 | 20 | 57,192,115 | ZNF831/ EDN3 | A/G | 0.88 | -0.14 | 0.03 | 4.95E-06 | -0.11 | 0.04 | 7.44E-03 | -0.13 | 0.02 | 1.59E-07 |
Chr=chromosome;
SNPs in bold attained p <5×10-8 in meta-analysis of CHARGE and Global BPgen
Table 4 provides results of joint meta-analysis of CHARGE and Global BPgen for the top 10 CHARGE SNPs for systolic blood pressure, diastolic blood pressure, and hypertension. Four genome-wide significant (stage 2 p <5×10-8) associations emerged for systolic blood pressure (CYP17A1, PLEKHA7, ATP2B1, and SH2B3), 6 for diastolic blood pressure (ULK4, CACNB2, ATP2B1, SH2B3, TBX3/TBX5, and a locus adjacent to CSK/ULK3), and 1 for hypertension (ATP2B1). Three additional associations attained p <4×10-7 (MDS1 for systolic blood pressure; CACNB2 and a region near EDN3 for hypertension). Plots of association results across each of the genome-wide significant loci are presented in Figures 1 and 2 using the SNAP tool.21 Forest plots (Supplementary Figures 3 and 4) reveal modest effect sizes (beta coefficients) of approximately 1 mm Hg systolic and 0.5 mm Hg diastolic blood pressure for each risk allele attaining genome-wide significance in the combined analysis with Global BPgen.
Figure 1. Locus-specific association maps for systolic blood pressure.
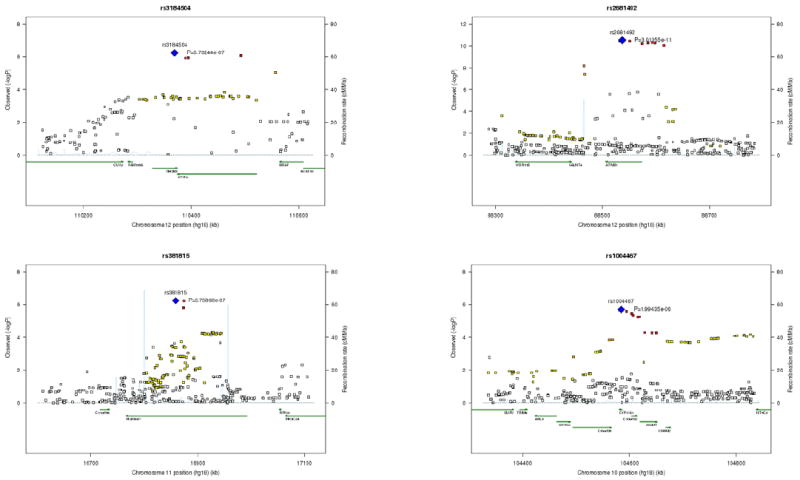
Locus-specific (-log10p-values) maps in the CHARGE meta-analysis of systolic blood pressure for four loci with genome-wide significance in the joint analysis of CHARGE with Global BPgen. The four loci are represented by rs3184504 (SH2B3), rs2681492 (ATP2B1), rs381815 (PLEKHA7), and rs1004467 (CYP17A1). For each locus the sentinel SNP (lowest p value) is depicted in blue, SNPs in red have r2 ≥0.8 with the sentinel SNP; SNPs in orange have r2 ≥0.5; those in yellow have r2 ≥0.2; those in white have r2 < 0.2 with the sentinel SNP. Superimposed on the plot are gene locations (green) and recombination rates (blue).
Figure 2. Locus-specific association maps for diastolic blood pressure.
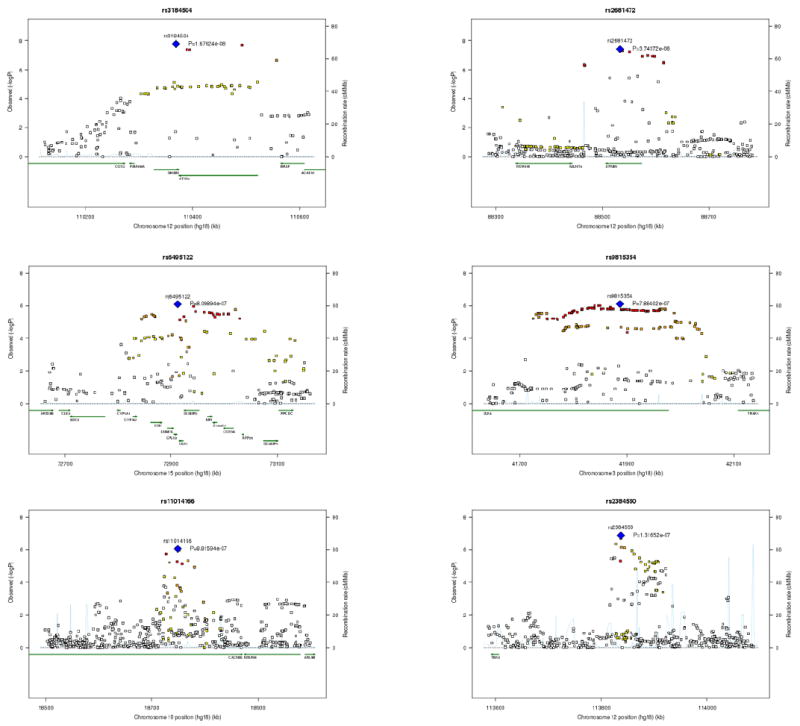
Locus-specific (-log10p-values) maps in the CHARGE meta-analysis of diastolic blood pressure for six loci with genome-wide significance in the joint analysis of CHARGE with Global BPgen. The six loci are represented by rs3184504 (SH2B3), rs2681472 (ATP2B1), rs6495122 (CSK), rs9815354 (ULK4), rs11014166 (CACNB2), and rs2384550 (TBX3/TBX5). For each locus the sentinel SNP (lowest p value) is depicted in blue, SNPs in red have r2 ≥0.8 with the sentinel SNP; SNPs in orange have r2 ≥0.5; those in yellow have r2 ≥0.2; those in white have r2 < 0.2 with the sentinel SNP. Superimposed on the plot are gene locations (green) and recombination rates (blue).
Blood pressure risk score
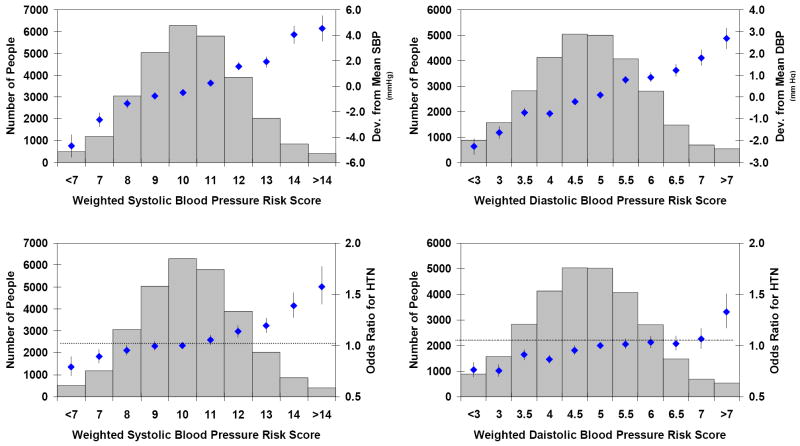
Weighted risk scores, incorporating the top 10 CHARGE loci for systolic and diastolic blood pressure, were applied to the study results to examine the influence of risk alleles in aggregate on deviation from mean blood pressure levels and odds ratios for hypertension. Figure 3 reveals a continuous and graded relation of risk score on blood pressure levels and odds ratios for hypertension. Inverse-variance weighted regression estimates of slope (beta) and its standard error (SE) were obtained across risk score groups for deviation from mean blood pressure and odds ratios for hypertension. To summarize these findings, two-tailed p values (from Z = beta / SE[beta]) were obtained from testing the null hypothesis of a zero slope across risk score groups. The p values across risk score groups were: 1.8×10-27 (systolic blood pressure vs. systolic blood pressure risk score), 1.7×10-56 (diatolic blood pressure vs. diastolic blood pressure risk score), 1.4×10-17 (hypertension vs. systolic blood pressure risk score), and 8.4×10-10 (hypertension vs. diastolic blood pressure risk score).
Figure 3. Systolic and Diastolic Blood Pressure Risk Scores.
This figure shows deviation in blood pressure or odds ratio for hypertension as solid diamonds with whiskers extending to ±1 standard error. Sample sizes for blood pressure risk score groups are shown by the blue bars. The top panels present deviation from mean systolic (left panel) and diastolic blood pressure (right panel) in mm Hg according to weighted risk score. The bottom panels show odds ratios for hypertension in relation to systolic (left panel) and diastolic blood pressure (right panel) weighted risk score. DBP=diastolic blood pressure; HTN=hypertension; SBP=systolic blood pressure. The p values for slope across risk score groups were all highly significant: 1.8×10-27 (systolic blood pressure vs. systolic blood pressure risk score), 1.7×10-56 (diatolic blood pressure vs. diastolic blood pressure risk score), 1.4×10-17 (hypertension vs. systolic blood pressure risk score), and 8.4×10-10 (hypertension vs. diastolic blood pressure risk score).
Putative functional variation
A search for nonsynonymous SNPs among our blood pressure association results identified five such variants including rs3184504 in SH2B3 (stage 2 p value for diastolic blood pressure 2.6×10-14), rs267561 in ITGA9 (stage 1 p value for hypertension 2.6×10-6), and three linked non-synonymous SNPs in ULK4 (rs2272007, rs3774372, and rs1716975; pairwise r2 0.82-1.0; lowest stage 1 p value 1.5×10-6 for diastolic blood pressure). To further identify putative functional associations within our GWAS results, we culled from the 2.5 million HapMap SNPs in our analysis those that were previously reported from GWAS to be associated with altered gene expression in liver22 (n=3,322) or lymphoblastoid cell lines23 (n=10,823). These expression-associated SNPs (eSNPs or eQTLs) were then interrogated for association with blood pressure phenotypes within our GWAS results (Table 5). Of note, three of our genome-wide significant loci were captured through the analysis eSNPs including: rs739496 in SH2B3, which is associated with altered expression of nearby HSS00340376 in liver; rs6495126 near CSK/ULK3, which is associated with altered expression of ULK3 in liver; and non-synonymous SNPs rs1716975 and rs2272007 in ULK4, which are associated with altered expression of ULK4 in lymphoblastoid cell lines. In addition, rs7571613 near PMS1 and MSTN is associated with altered expression of ORMDL1 and PMS1 in lymphoblastoid cell lines. Additional eSNPs with suggestive evidence of association with blood pressure phenotypes were: rs7537765 near MTHFR/NPPA (expressed gene CLCN6); and several SNPs in JARID1A that are associated with expression of JARID1A, SLC6A12, and CCDC77.
Table 5.
SNPs association with blood pressure and altered tissue-specific gene expression
| SNP | Chr | Position | Nearest gene(s) | SBP pval | DBP pval | HTN pval | Expressed Gene(s) | eSNP pval‡ |
|---|---|---|---|---|---|---|---|---|
| Liver eSNPs | ||||||||
| rs7537765 | 1 | 11809889 | NPPA; NPPB; CLCN6; MTHFR | 1.8E-05 | 9.2E-04 | 1.5E-04 | CLCN6 | 1.4E-07 |
| rs249209 | 5 | 79902964 | DHFR; DP58; UNQ9217 | 8.5E-05 | 0.07 | 1.2E-03 | HSS00169533 | 4.4E-05 |
| rs10963072 | 9 | 17368775 | C9orf39 | 0.03 | 0.11 | 6.3E-05 | C9orf39 | 2.0E-06 |
| rs525381 | 12 | 255052 | JARID1A; SLC6A13 | 9.3E-05 | 0.14 | 5.3E-04 | CCDC77; SLC6A12 | 2.1E-10 |
| rs739496 | 12 | 110372041 | SH2B3; ATXN2 | 2.9E-04 | 1.3E-05 | 0.01 | HSS00340376 | 1.1E-06 |
| rs7312321 | 12 | 118520617 | CCDC60 | 9.0E-04 | 0.03 | 6.0E-05 | Contig30372_RC | 1.1E-30 |
| rs6495126 | 15 | 72962078 | MPI/SCAMP2/ULK3 | 3.0E-04 | 3.6E-05 | 1.2E-04 | ULK3; AK001918; RPP25 | 8.6E-06 |
| Lymphoblastoid cell line eSNPs | ||||||||
| rs1384883 | 1 | 74274065 | FPGT | 9.9E-03 | 7.2E-05 | 2.5E-03 | LRRC44;BC042056 | 1.4E-21 |
| rs12466395 | 2 | 190488943 | PMS1 | 8.8E-04 | 5.3E-05 | 8.8E-05 | ORMDL1 | 2.4E-15 |
| rs7571613 | 2 | 190513907 | PMS1 | 7.3E-07 | 2.2E-06 | 5.3E-04 | ORMDL1;PMS1 | 1.5E-08 |
| rs1454301 | 2 | 190518307 | PMS1 | 1.1E-06 | 2.2E-06 | 1.1E-03 | PMS1;ORMDL1 | 2.5E-09 |
| rs2053163 | 2 | 190535268 | PMS1 | 1.7E-05 | 5.8E-06 | 5.8E-03 | ORMDL1 | 2.1E-12 |
| rs6749643 | 2 | 190543718 | MSTN | 1.6E-06 | 2.2E-06 | 7.3E-04 | ORMDL1;PMS1 | 5.5E-09 |
| rs7575810 | 2 | 190560410 | MSTN | 2.5E-05 | 2.7E-05 | 0.03 | ORMDL1 | 3.6E-10 |
| rs1474359 | 2 | 190641251 | MSTN | 1.9E-05 | 1.2E-05 | 1.0E-03 | ORMDL1;PMS1 | 1.7E-11 |
| rs1052501 | 3 | 41900402 | ULK4 | 0.84 | 4.2E-05 | 0.64 | ULK4 | 3.9E-08 |
| rs1716975 | 3 | 41935010 | ULK4 | 0.94 | 2.2E-06 | 0.96 | ULK4 | 7.9E-08 |
| rs2272007 | 3 | 41971140 | ULK4 | 0.87 | 1.5E-06 | 0.83 | ULK4 | 2.7E-08 |
| rs4572871 | 4 | 83979911 | SEC31A; SCD5 | 2.3E-05 | 9.7E-03 | 3.5E-04 | SCD5 | 6.4E-41 |
| rs6601414 | 8 | 10014158 | MSRA | 3.0E-04 | 3.4E-05 | 4.9E-03 | C8orf5 | 8.2E-09 |
| rs13254942 | 8 | 10295088 | MSRA | 5.6E-05 | 1.9E-03 | 1.5E-04 | C8orf5;FAM167A | 1.0E-08 |
| rs2898290 | 8 | 11471318 | BLK | 2.3E-05 | 7.0E-03 | 7.9E-05 | C8orf5;FAM167A;BLK | 3.7E-12 |
| rs4980878 | 12 | 297338 | JARID1A; SLC6A13 | 4.8E-05 | 0.12 | 1.6E-04 | JARID1A | 2.7E-09 |
| rs1860360 | 12 | 364161 | JARID1A; CCDC77 | 6.2E-05 | 0.10 | 1.6E-04 | JARID1A | 2.7E-09 |
Highlighted p values are < 1/n, where n is the number of eSNPs interrogated.
p value for association of eSNP with gene expression in liver or lymphoblastoid cell lines.
DBP=diastolic blood pressure; HTN=hypertension; SBP=systolic blood pressure
Discussion
In this meta-analysis of results from 29,136 participants from six large prospective observational studies in the CHARGE Consortium, we identified multiple loci with evidence of association with levels of systolic and diastolic blood pressure and hypertension. We further replicated genome-wide significant SNPs in 34,433 independent subjects from the Global BPgen Consortium, and the joint analysis of results from the two consortia identified 11 genome-wide significant associations: four loci for systolic blood pressure (ATP2B1, p=3.8×10-11; CYP17A1, p=1.3×10-10; PLEKHA7, p= 1.9×10-9; SH2B3, p= 4.5×10-9), six loci for diastolic blood pressure (ATP2B1, p=1.5×10-9; CACNB2, p=1.2×10-8; CSK/ULK3 p=1.8×10-10; SH2B3, p= 2.6×10-14; TBX3/TBX5, p=3.8×10-8; ULK4, p=2.5×10-9), and one locus for hypertension (ATP2B1, p=1.8×10-11). There was considerable concordance among top loci across all three phenotypes; ATP2B1 showed significant association will systolic blood pressure, diastolic blood pressure, and hypertension, CACNB2 showed strong evidence of association with all three traits, and SH2B3 demonstrated significant association with systolic and diastolic blood pressure. Of note, rs1004467, a common intronic variant in CYP17A1, a gene associated with a rare Mendelian form of hypertension, emerged as a genome-wide significant locus in the meta-analysis of results from both consortia. Several additional loci showed suggestive association results including MDS1, ITGA9, EDN3, and PMS1/MSTN. The top 10 risk alleles for systolic and diastolic blood pressure within CHARGE were each associated with about a 1 and 0.5 mm Hg increase in systolic and diastolc blood pressure, respectively; there was a continuous and graded relation of the number of risk alleles to mean levels of SBP and DBP and odds ratios for hypertension. Last, analysis of gene expression associated SNPs within our GWAS provided additional promising blood pressure candidates (by virtue of the identified expressed genes) including JARID1A/SLC6A12/CCDC77, ORMDL1 and CLCN6.
We identified genome-wide significant association of ATP2B1 with systolic and diastolic blood pressure and with hypertension (17 percent increase in odds per risk allele and 37 percent increase for two risk alleles). This gene encodes PMCA1, a plasma membrane calcium/calmodulin dependent ATPase that is expressed in vascular endothelium and is involved in calcium pumping from the cytosol to the extracellular compartment.24 An investigation of cultured rat aortic smooth muscle cells found elevated PMCA1 mRNA levels in spontaneously hypertensive rats compared to nonhypertensive controls, consistent with a role of ATP2B1 in blood pressure regulation.25
Genetic variation can contribute to altered blood pressure regulation by altering the structure of coded proteins or by altering gene expression levels (i.e. protein quantity). For SH2B3 we have strong evidence to suport both mechanisms; a missense SNP (altered protein structure) and an eSNP (altered expression) were associated with blood pressure. Our most highly significant SNP for diastolic blood pressure (and our second strongest signal for systolic blood pressure) was the nonsynonymous SNP rs3184504 in SH2B3 (Tables 1, 2, and 4), which introduces amino the acid substitution W262R in a plekstrin homology domain on exon 3. This coding variant is predicted by PolyPhen26 to be probably damaging to the coded protein. This SNP has recently also been found to be reproducibly associated with type 1 diabetes mellitus and celiac disease.27,28 The association of this SNP with two autoimmune diseases suggests that immune response pathways may influence blood pressure by mechanisms previously not appreciated. SH2B3 knockout mice are viable but show increased sensitivity to cytokines and abnormal growth factor signaling.29 In addition, eSNP rs739496 (Table 5) was associated with blood pressure levels and with liver expression of a transcript adjacent to SH2B3. SH2B3 is located in a large block of linkage disequilibrium on chromosome 12 that contained multiple association signals across 700 kb from rs3184504 in SH2B3 to rs11066188 in C12orf51 for systolic and diastolic blood pressure and contains many genes (Figure 1, Figure 2, and Supplementary Figure 2). Located near the midpoint between SH2B3 and C12orf51 is ALDH2, encoding acetaldehyde dehydrogenase 2, a critical enzyme in alcohol metabolism. A recent meta-analysis found that male homozygotes for the Lys671Glu variant (rs671) in ALDH2, had an increased odds of hypertension (odds ratio 2.42, p=4.8×10-6) and 7 mm Hg higher mean systolic blood pressure (p = 1.1×10-12) when compared with major allele homozygotes.30 Although rs671 is absent in whites of European descent in HapMap and was not included in our GWAS, our intriguing findings in the region encircling ALDH2 are consistent with a role of this gene in blood pressure regulation in people of European descent.
A SNP (rs1004467) attaining genome-wide significance is in CYP17A1, encoding steroid 17-alpha-hydroxylase, an enzyme necessary for steroidogenesis. Mutations in CYP17A1 are found in patients with 17α-hydroxylase deficiency, which is characterized by congenital adrenal hyperplasia with apparent mineralocorticoid excess, salt retention, hypokalemia, and hypertension.31 Numerous mutations in CYP17A1 have been identified in patients with 17α-hydroxylase deficiency leading to a spectrum of phenotypic severity.32 Although mutations in CYP17A1 causing phenotypic 17α-hydroxylase deficiency are rare, our data suggest that common variants in CYP17A1 may also be associated with blood pressure by promoting mild forms of enzyme deficiency or dysfunction.
CACNB2, encoding the beta-2 subunit of a voltage-gated calcium channel, was associated with diastolic blood pressure and showed suggestive evidence of association with systolic pressure and hypertension. The gene is expressed in the heart and a nonsynomymous variant in CACNB2 was identified in affected individuals with Brugada syndrome.33 CACNB2 is one member of a family of voltage-gated calcium channel genes, several of which have effects on blood pressure regulation and serve as target of calcium channel blockers. The beta-2 subunit interacts with alpha-1 calcium channels (CaV1.2) and this is a mechanism by which variation in CACNB2 may alter blood pressure.34
The joint meta-analysis of CHARGE and Global BPgen (Table 4) also identified PLEKHA7, ULK4, TBX3/TBX5, and a region adjacent to CSK/ULK3/CYP1A2 as genome-wide significant loci. Mutations in TBX5 (T-box transcription factor 5) cause structural cardiac malformations and can be associated with altered expression of NPPA,35 which also was a locus of interest in our eSNP analysis. CSK encodes cytoplasmic tyrosine kinase, which is involved in angiotensin II dependent vascular smoot muscle cell proliferation.36 Little is know about ULK3 or ULK4 and how variation in these genes might affect blood pressure. Three CHARGE loci that were identified as genome-wide significant in this analysis were also found to be genome-wide significant in the Global BPgen Consortium meta-analysis. They were CYP17A1 (rs1004467 in CYP17A1 in CHARGE vs. rs11191548 in Global BPgen, respectively; r2=0.42), SH2B3/ATXN2 (rs3184504 vs. rs653178; r2=1.0), and a locus containing CSK/ULK3/CYP1A2 (rs6495122 vs. rs4886606; r2=0.56). In addition, both consortia identified MDS1 as a locus of interest (rs448378 vs. rs1918974; r2=1.0). The region containing MTHFR/NPPA, which attained genome-wide significance in Global BPgen, was identified as a region of interest in the CHARGE analysis of eSNPs ([Table 5] rs7537765 in CHARGE vs. rs17367504 in Global BPgen; r2=0.94). Other loci of interest (5×10-8 < p < 4×10-7) in the joint analysis of CHARGE and Global BPgen were MDS1 (rs448378, p=1.2×10-7) and a region adjacent to EDN3 (rs16982520, p=1.6×10-7). Endothelin-3 may play a role in renal-mediated hypertension in the rat.37
A search for putative functional variation within our GWAS identified five nonsynonymous SNPs. In addition rs3184504 in SH2B3 (discussed above), rs267561 in ITGA9, which showed suggestive evidence of association with hypertension (p=2.6×10-6), produces an E507G substitution that is predicted by PolyPhen to have possibly damaging effects.26 Three linked non-synonymous SNPs in ULK4 showed suggestive evidence of association with diastolic blood pressure (rs2272007, p=1.5×10-6; rs3774372, p=1.6×10-6; rs1716975, p=2.2×10-6; pairwise r2 0.82-1.0); these amino acid substitutions are predicted to be benign individually, but their conjoint effects on protein function is unknown. Interrogation of our GWAS results for SNPs that are associated with blood pressure phenotypes and altered gene expression confirmed SH2B3, ULK4 and ULK3 as loci of interest (Table 5). Another locus detected via eSNP associations with blood pressure was rs7537765 near NPPA, which encodes atrial natriuretic peptide and which was in linkage disequilibrium with rs198358 (r2=0.58), a SNP that has been shown to be associated with higher circulating natriuretic peptide levels and lower systolic blood pressure.38 Other promising candidates by virtue of the expressed genes in our eSNP analysis are JARID1A/SLC6A12/CCDC77, ORMDL1/PMS1, and CLCN6.
Although the conjoint effect of multiple risk alleles on blood pressure can be substantial, our findings underscore the small effect size of individual common allelic variants -- about 1 mm Hg each for systolic and 0.5 mm Hg each for diastolic blood pressure per variant allele -- and the necessity of very large sample sizes for detection of robust and significant results. The combined analysis of CHARGE and Global BPgen for our top SNPs reflects a sample size of 63,569 individuals and illustrates the advantage of large consortia for meta-analysis of genome-wide data to identify new clues to the genetic underpinnings of common complex traits. Given the small effect sizes detected, it is not surprising that previous blood pressure GWAS failed to identify genome-wide significant results at p <5×10-8. 13,14,15,16,17,18
Current understanding of allelic variation affecting blood pressure in the general population is in its infancy; until recently there have been few genetic variants reproducibly associated with blood pressure variation in the community. Our CHARGE findings, in conjunction with those of the Global BPgen Consortium,38 establish the utility of genome-wide association approaches to identify common allelic variants pertaining to blood pressure physiology and pathophysiology. Our findings are consistent with the hypothesis that variation in scores, if not hundreds, of genes contribute to blood pressure variation. This hypothesis is supported by the excess number of SNPs showing association at p <1×10-3with blood pressure phenotypes. Future efforts to identify additional alleles associated with blood pressure will require complementary strategies including larger genome-wide studies to identify additional common alleles and resequencing efforts in large samples to identify rare variants.
In aggregate, the proportion of blood pressure variation explained by the top 10 CHARGE systolic and diastolic blood pressure SNPs across the six cohorts is 1 percent (increment in r2) after accounting for the major non-genetic determinants of blood pressure: age, age squared, sex, and body mass index. The conjoint effect of multiple risk alleles on blood pressure levels, however, amounts to several mm Hg (Figure 3), which is sufficient to increase cardiovascular disease risk. Observational data indicate that a prolonged increase in diastolic blood pressure of 5 mm Hg is associated with a 34 percent increase in risk for stroke and a 21 percent increase in risk of coronary events.39
Future analyses using larger samples can benefit from specific features of our study design. First, the vast majority of blood pressure values used in our analyses were obtained more than 15 years ago, when blood pressure treatment, which confounds genetic analyses, was less widely used; contemporary blood pressure data might be less likely to detect genetic associations. Second, because allelic variation may affect both the low and high ends of the blood pressure distribution, we used the more powerful approach of analyzing blood pressure as a continuous trait, yet we also identified a genome-wide significant locus for hypertension. At the same time, one should recognize that this study, utilizing participants of European descent only, can’t be applied to other populations. Although our analysis of eSNPs indicates that some of the genome-wide significant blood pressure loci we identified are associated with altered gene expression, the relevance of these findings to blood pressure is speculative. A similar approach, however, has been used to identify putative disease genes for childhood asthma,40 Crohn’s disease,41 and a network of genes implicated in obesity.42
In conclusion, we have identified multiple genome-wide significant blood pressure loci that can be used to guide fine mapping efforts to pinpoint causal variants and to understand how the implicated genes alter blood pressure physiology and contribute to hypertension. The characterization of new blood pressure loci can serve as a basis for future approaches to early detection of high risk individuals and for the development of novel therapies for the prevention or treatment of hypertension.
Methods
Consortium Organization
The CHARGE Consortium19 includes 6 cohort studies that completed genome-wide genotyping and had extensive data on multiple phenotypes including blood pressure. Each study adopted collaboration guidelines and established a consensus on phenotype harmonization, covariate selection, and an analytical plan for within-study genome-wide association and prospective meta-analysis of results across studies. Each study received instittutional review board approval of its consent procedures, examination and surveillance components, data security measures, and DNA collection and its use for genetic research. All participants in each study gave written informed consent for participation in the study and the conduct of genetic research. Details of each study, blood pressure measurement protocols, inclusion and exclusion criteria, and genotyping are provided in the Supplementary Methods and Supplementary Tables 1 and 3.
Genotype Imputation
For imputation of genotypes to the HapMap set of approximately 2.5 million SNPs, ARIC, FHS, and RS used a Hidden Markov Model as implemented in MACH,43 and CHS used BIMBAM10 v0.9944 (Supplementary Table 3). SNP imputation combined genotype data from each sample with the HapMap CEU samples and then inferred genotypes probabilistically based on shared haplotype stretches between study samples and HapMap release 22 build 36. Imputation results are summarized as an ‘allele dosage’ defined as the expected number of copies of the minor allele at that SNP (a fractional value between 0.0 and 2.0) for each genotype.
Statistical analyses
Cross-sectional analyses were conducted within each cohort using an additive genetic model, and within-study associations were combined by prospective meta-analysis. The phenotypes for meta-analysis were systolic and diastolic blood pressure and hypertension at the first examination attended. For participants who were taking antihypertensive medication we added 10 mm Hg to observed systolic blood pressure values and 5 mm Hg to diastolic values.45 Hypertension was defined as systolic blood pressure ≥140 or diastolic blood pressure ≥90 mm Hg or drug treatment for hypertension at time of assessment. Within each cohort, regression models were fitted for systolic and diastolic blood pressure (separately) and allele dosage, adjusting for sex, age, age squared, and BMI.
Meta-analysis of results was performed using inverse-variance weighting. Prior to meta-analysis, results were filtered for minor allele frequency <0.005 and the genomic control parameter was calculated to adjust each study. After meta-analysis, the genomic control parameter was re-calculated to adjust for between-study heterogeneity.46 A pre-determined threshold of 4×10-7 (stage 1) was used to indicate genome-wide significance within CHARGE. For 2.5×106 tests (the total number of imputed SNPs), this threshold means that the expected number of false positive results is ≤1; the validity of this bound is not affected by correlation between test statistics.47 Ten leading SNPs for systolic, 10 for diastolic blood pressure, and 10 for hypertension were exchanged between CHARGE and Global BPgen, a consortium with a sample size of 34,433 whites of European ancestry with analogous genome-wide data.38 SNP selection was limited to one SNP per locus of interest, defined by an r2 ≤ 0.2. For rs880315, imputation in Global BPgen was suboptimal and this SNP was replaced with rs12046272. rs8096897 and rs10972206 were not selected for exchange due to low minor allele frequencies (defined as <0.01 for continuous traits, <0.05 for hypertension). rs5761405 was selected for exchange, but was not available in the imputed results from Global BPgen, so the next most highly significant locus was selected in its place. For all 30 exchanged SNPs we performed meta-analysis of CHARGE and Global BPgen results using inverse variance weighting and considered a p value in the joint analysis (stage 2) significant at p=5×10-8. Significant replication of a genome-wide significant SNP in CHARGE was defined as a p value <0.008 for the same SNP in Global BPgen (0.05/6 genome-wide significant SNPs submitted for replication). One-sided tests were used to assess replication when the alignment of an allele and its directional effect were identical between CHARGE and Global BPgen.
Analysis of hypertension was conducted within each cohort, and the within-study associations were combined by meta-analysis. Within each cohort, regression models were fitted for hypertension, adjusting for sex, age, age squared, and BMI. Meta-analysis of results was performed using inverse-variance weighting. Prior to meta-analysis, results were filtered for low minor allele frequency <0.01 and the genomic control parameter calculated to adjust each study. After meta-analysis, the genomic control parameter was calculated again to adjust for between-study heterogeneity. In the meta-analysis of CHARGE and Global BPgen, the analytical approach used in Global BPgen was different from that of CHARGE; in Global BPgen non-hypertensive controls were defined as individuals not taking any hypertensive medications and having a SBP ≤120 mm Hg and a DBP ≤85 mm Hg.
Blood pressure risk score was a weighted sum across 10 top SNPs (separately for systolic and diastolic blood pressure) combining beta coefficients and doses of risk alleles, rounded to 1 mm Hg for systolic blood pressure (groups ≤ 6 to ≥15) and 0.5 mm Hg for diastolic blood pressure (groups ≤ 2.5 to ≥7.5). Within a study, for each risk score group we calculated deviations of empirical blood pressure from the study mean. Across studies, we estimated mean deviation and standard error within risk score group, weighted by group-and study-specific sample sizes. For hypertension, odds ratios (and standard errors) were the corresponding summary statistics, with the reference group being those with a weighted systolic risk score of 10 or a diastolic score of 5.
SNP associations with altered gene expression
To assess putative functional associations in our GWAS, we used bioinformatics tools to query existing GWAS databases of SNPs associated with cis-gene expression levels in immortalized liver (n=3,322) 22 and lymphoblastoid cell lines (n=10,823).23 These expression-associated SNPs were then explored for association with blood pressure in the fully imputed HapMap blood pressure results for CHARGE. Statistical significance was defined by a p value of 1/n (where n is the number of tissue-specific cis eSNPs interrogated); this threshold will yield on average 1 false positive per tissue examined. The p value thresholds for significance of eSNP associations for liver and lymphoblastoid cell lines were 3.0×10-4 and 9.2×10-5, respectively.
Supplementary Material
Acknowledgments
The authors acknowledge the essential role of the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium in development and support of this manuscript. CHARGE members include the Netherland’s Rotterdam Study (RS), Framingham Heart Study (FHS), Cardiovascular Health Study (CHS), the NHLBI’s Atherosclerosis Risk in Communities (ARIC) Study, and the NIA’s Iceland Age, Gene/Environment Susceptibility (AGES) Study.
The Age, Gene/Environment Susceptibility Reykjavik Study is funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament).
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, and grants R01HL087641, R01HL59367, R37HL051021, R01HL086694 and U10HL054512; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. AK is supported by a German Research Foundation Fellowship.
The Cardiovascular Health Study was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
The National Heart, Lung, and Blood Institute’s Framingham Heart Study is a joint project of the National Institutes of Health and Boston University School of Medicine, and was supported by the National Heart, Lung, and Blood Institute’s Framingham Heart Study (contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (contract No. N02-HL-6-4278). Analyses reflect the efforts and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly(RIDE); The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission(DG XII); the Municipality of Rotterdam and the ErasmusMC translational research fund (2004-44). Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO Groot, 175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (014.93.015; RIDE2). This study was supported by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Michael Moorhouse, Marijn Verkerk and Sander Bervoets for their help in creating the database and Maxim Struchalin for his contributions to the imputations of the data. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practioners and pharmacists.
References
- 1.Fields LE, Burt VL, Cutler JA, Hughes J, Roccella EJ, Sorlie P. The burden of adult hypertension in the United States, 1999–2000: a rising tide. Hypertension. 2004;44:398–404. doi: 10.1161/01.HYP.0000142248.54761.56. [DOI] [PubMed] [Google Scholar]
- 2.Lawes CM, Vander Hoorn S, Rodgers A. International Society of Hypertension. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371(9623):1513–8. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 3.VA Cooperative Study Group. Effects of treatment on morbidity in hypertension. Results in patients with diastolic blood pressures averaging 115 through 129 mm Hg. JAMA. 1967;202(11):1028–34. [PubMed] [Google Scholar]
- 4.SHEP Cooperative Research Group. Prevention of stroke by antihypertensive drug treatment in older persons with isolated systolic hypertension. Final results of the Systolic Hypertension in the Elderly Program (SHEP) JAMA. 1991;265(24):3255–64. [PubMed] [Google Scholar]
- 5.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a Blood Pressure Gene on Chromosome 17: Genome Scan Results for longitudinal blood pressure phenotypes in subjects from the Framingham Heart Study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 6.Koivukoski L, Fisher SA, Kanninen T, Lewis CM, von Wowern F, Hunt S, Kardia SL, Levy D, Perola M, Rankinen T, Rao DC, Rice T, Thiel BA, Melander O. Meta-analysis of genome-wide scans for hypertension and blood pressure in Caucasians shows evidence of susceptibility regions on chromosomes 2 and 3. Hum Mol Genet. 2004;13:2325–32. doi: 10.1093/hmg/ddh237. [DOI] [PubMed] [Google Scholar]
- 7.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Chang YP, Liu X, Kim JD, Ikeda MA, Layton MR, Weder AB, Cooper RS, Kardia SL, Rao DC, Hunt SC, Luke A, Boerwinkle E, Chakravarti A. Multiple genes for essential-hypertension susceptibility on chromosome 1q. Am J Hum Genet. 2007;80:253–64. doi: 10.1086/510918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lifton RP. Genetic determinants of human hypertension. Proc Natl Acad Sci. 1995;92(19):8545–51. doi: 10.1073/pnas.92.19.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani SJ. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hyperension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 12.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 15.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, Mano H, Yasunami M, Kimura A, Kita Y, Ueshima H, Nakayama T, Soma M, Hata A, Fujioka A, Kawano Y, Nakao K, Sekine A, Yoshida T, Nakamura Y, Saruta T, Ogihara T, Sugano S, Miki T, Tomoike H. High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet. 2008;17(4):617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 17.Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nature Genetics 2008. 2008 doi: 10.1038/ng.271. online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Whole-genome association study identifies STK39 as a hypertension susceptibility gene. PNAS. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Psaty BM, O’Donnell CJ, Gudnason VL, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from five cohorts. Circ Cardiovasc Genet. 2008 doi: 10.1161/CIRCGENETICS.108.829747. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Global BPgen Consortium manuscript. Submitted jointly to Nature Genetics. 2008 [Google Scholar]
- 21.Johnson AD, Handsaker RE, Pulit S, Nizzari M, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 24.Pande J, Mallhi KK, Sawh A, Szewczyk MM, Simson F, Grover A. Aortic smooth muscle and endothelial plasma membrane CA2+ pump isoforms are inhibited differently by the extracellular inhibitor caloxin 1b1. Am J Physiol. 2006;290:1341–1349. doi: 10.1152/ajpcell.00573.2005. [DOI] [PubMed] [Google Scholar]
- 25.Monteith GR, Kable EP, Kuo TH, Roufogalis BD. Elevated plasma membrane and sarcoplasmic reticulum Ca2+ pump mRNA levels in cultured aortic smooth muscle cells from spontaneously hypertensive rats. Biochem Biophys Res Commun. 1997;230(2):344–6. doi: 10.1006/bbrc.1996.5956. [DOI] [PubMed] [Google Scholar]
- 26.Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork Peer. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 27.Hunt KA, Zhernakova A, Turner G, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velazquez L, Cheng AM, Fleming HE, Furlonger C, Vesely S, Bernstein A, Paige CJ, Pawson T. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med. 2002;195(12):1599–611. doi: 10.1084/jem.20011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Davey Smith G, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5(3):e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa-Santos M, Kater CE, Auchus RJ. The Brazilian CAH Multicenter Study Group Two prevalent CYP17 mutations and genotype-phenotype correlations in 24 Brazilian patients with 17-hydroxylase deficiency. J Clin Endocrinol Metab. 2004;89:49–60. doi: 10.1210/jc.2003-031021. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Cui B, Sun S, Shi T, Zheng S, Bi Y, Liu J, Zhao Y, Chen J, Ning G, Li X. Phenotype-genotype correlation in eight Chinese 17alpha-hydroxylase/17,20 lyase-deficiency patients with five novel mutations of CYP17A1 gene. J Clin Endocrinol Metab. 2006;91(9):3619–25. doi: 10.1210/jc.2005-2283. [DOI] [PubMed] [Google Scholar]
- 33.Antzelevitch C, Pollevick GD, Cordeiro JM, et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. 2007;115:442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lao QZ, Kobrinsky E, Harry JB, Ravindran A, Soldatov NM. New determinant for the CaVbeta2 subunit modulation of the CaV1.2 calcium channel. J Biol Chem. 2008;283:15577–88. doi: 10.1074/jbc.M802035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, Lam J, Wilde AA, Lekanne Deprez RH, Moorman AF. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102(11):1433–42. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- 36.Sayeski PP, Showkat-Ali M. The critical role of c-Src and the Shc/Grb2/ERK2 signaling pathway in angiotensin II-dependent VSMC proliferation. Experimental Cell Research. 2003;287:339–349. doi: 10.1016/s0014-4827(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 37.Vogel V, Bäcker A, Heller J, Kramer HJ. The renal endothelin system in the Prague hypertensive rat, a new model of spontaneous hypertension. Clin Sci (Lond) 1999;97:91–8. [PubMed] [Google Scholar]
- 38.Newton-Cheh C, et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nature Genetics. 2009 doi: 10.1038/ng.328. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacMahon S, Peto R, Cutler J, Collins R, Sorlie P, Neaton J, Abbott R, Godwin J, Dyer A, Stamler J. Blood pressure, stroke, and coronary heart disease. Part 1, Prolonged differences in blood pressure: prospective observational studies corrected for the regression dilution bias. Lancet. 1990;335:765–74. doi: 10.1016/0140-6736(90)90878-9. [DOI] [PubMed] [Google Scholar]
- 40.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 41.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ NIDDK IBD Genetics Consortium Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Ding J, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 44.Servin B, Stephens M. Imputation-Based Analysis of Association Studies: Candidate Regions and Quantitative Traits. PLoS Genet. 2007;3(7):e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui JS, Hopper JL, Harrap SB. Antihypertensive Treatments Obscure Familial Contributions to Blood Pressure Variation. Hypertension. 2003;41(2):207–210. doi: 10.1161/01.hyp.0000044938.94050.e3. [DOI] [PubMed] [Google Scholar]
- 46.Senn S. Trying to be precise about vagueness. Stat Med. 2007;26:1417–30. doi: 10.1002/sim.2639. [DOI] [PubMed] [Google Scholar]
- 47.Gordon A, Glazko G, Qiu X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Annals of Applied Statistics. 2007;1:179–190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.