Abstract
The lethal genetic disease cystic fibrosis is caused predominantly by in-frame deletion of phenylalanine 508 in the cystic fibrosis transmembrane conductance regulator (CFTR). F508 is located in the first nucleotide-binding domain (NBD1) of CFTR, which functions as an ATP-gated chloride channel on the cell surface. The F508del mutation blocks CFTR export to the surface due to aberrant retention in the endoplasmic reticulum. While it was assumed that F508del interferes with NBD1 folding, biophysical studies of purified NBD1 have given conflicting results concerning the mutation's influence on domain folding and stability. We have conducted isothermal (this paper) and thermal (accompanying paper) denaturation studies of human NBD1 using a variety of biophysical techniques, including simultaneous circular dichroism, intrinsic fluorescence, and static light-scattering measurements. These studies show that, in the absence of ATP, NBD1 unfolds via two sequential conformational transitions. The first, which is strongly influenced by F508del, involves partial unfolding and leads to aggregation accompanied by an increase in tryptophan fluorescence. The second, which is not significantly influenced by F508del, involves full unfolding of NBD1. Mg-ATP binding delays the first transition, thereby offsetting the effect of F508del on domain stability. Evidence suggests that the initial partial unfolding transition is partially responsible for the poor in vitro solubility of human NBD1. Second-site mutations that increase the solubility of isolated F508del-NBD1 in vitro and suppress the trafficking defect of intact F508del-CFTR in vivo also stabilize the protein against this transition, supporting the hypothesize that it is responsible for the pathological trafficking of F508del-CFTR.
Keywords: cystic fibrosis, cystic fibrosis transmembrane conductance regulator (CFTR), protein thermodynamics, circular dichroism, fluorescence, static light-scattering
Introduction
Cystic fibrosis (CF) is a genetic disease caused by mutations in an ATP-gated chloride channel called the cystic fibrosis transmembrane conductance regulator (CFTR).1–6 CF is the most common fatal genetic disease among Caucasians and is prevalent in many other populations.7 CF causes pervasive defects in secretory processes, including most importantly water secretion in the epithelial tissues of the lung. This defect leads to insufficient hydration, which impairs bacterial clearance and leads to persistent cycles of infection/inflammation followed by eventual lung failure.8–11 While many advances have been made in treating CF during the past 20 years, most patients still die before age 30.8 There is intense interest in applying understanding of the molecular etiology of the disease to developing more efficacious pharmacological treatments.12–14
Population genetics shows that a single mutation, an in-frame deletion of phenylalanine 508 (F508del), accounts for ∼70% of the mutant CFTR alleles present in the human population.7 Therefore, ∼50% of CF patients have two copies and ∼90% have at least one copy of this specific mutation. Therefore, a substantial proportion of CF drug-discovery efforts have focused on correction of the molecular defect caused by the F508del mutation.12,13
F508del is located in the first nucleotide-binding domain (NBD1) of CFTR,15–19 which is homologous to proteins in the ABC Transporter superfamily.20–22 This name derives from the stereotyped nucleotide-binding domains (NBDs), or ATP-binding cassettes (ABCs), that are conserved among superfamily members. While CFTR is the only member known to function as an ATP-gated ion channel rather than an ATP-fueled transmembrane pump, its overall domain organization and ATP-dependent mechanochemistry are equivalent to that of ABC Transporters.4,6,18 These all contain a pair of transmembrane domains (TMDs) that interact with a pair of cytoplasmic ABC domains, which control protein conformation by binding ATP at their mutual interface.23–25 In CFTR, these domains are encoded in a single polypeptide along with a regulatory (R) domain, in the order TMD1, NBD1, R, TMD2, NBD2.
F508del-CFTR displays a severe temperature-dependent defect in protein biogenesis.26–29 While ∼10% of it is properly exported to the plasma membrane in cells growing at 25°C, less than 1% is properly exported at 37°C. The remainder is retained in the endoplasmic reticulum (ER) and eventually degraded via retrograde transport to the cytoplasmic proteasome complex. Furthermore, F508del-CFTR channels exported to the plasma membrane at 25°C are relatively stable at that temperature but are destabilized and degraded much more rapidly at 37°C.28,30 The obvious temperature-dependent defects in the biogenesis and stability of F508del-CFTR have led to a widespread assumption that the mutation interferes with protein folding.31 However, structural and thermodynamic studies have led to conflicting conclusions regarding the exact molecular defect caused by the F508del mutation.15,18,19,32–34
Previously published isothermal denaturation studies have failed to reveal a perturbation in the major chemical unfolding transition of human NBD1 (hNBD1).15,19 Moreover, extensive crystallographic analyses demonstrated that F508del only perturbs the equilibrium structure of Mg-ATP-bound hNBD1 locally at the site of the mutation, which is the region likely to mediate interaction with the transmembrane domains of CFTR.18 High-resolution hydrogen/deuterium-exchange mass spectrometry analyses of Mg-ATP-bound hNBD1 have measured the influence of the mutation on protein backbone dynamics, which are modestly enhanced near the site of the mutation but not elsewhere in the domain.18 These results have led to the hypothesis that F508del may interfere with biogenesis by blocking proper interdomain interaction in nascent CFTR15,35 or by promoting adventitious chaperone interactions near the site of the mutation,18 rather than by destabilizing hNBD1 itself. On the other hand, other results provide evidence that F508del does perturb hNBD1 stability. First, the mutation lowers the thermal melting temperature of hNBD1 by ∼6 to 7°C (see Ref. 36). Moreover, three second-site mutations, selected by Teem and coworkers to reverse the in vivo trafficking defect caused by F508del,37 appeared to stabilize hNBD1 against chemical unfolding.15 Because this “Teem suppressor triplet” (G550E, R553Q, and R555K) does not significantly alter the structure of F508del-hNBD1,18 its effect increasing the thermodynamic stability of hNBD1 seems likely to be responsible for reversing the deleterious effects of the F508del mutation.
While the F508del mutation has complex effects on the structure and stability of hNBD1, this mutation very clearly impairs the in vitro solubility of the domain compared to matched protein constructs containing F508.15 Even wild-type hNBD1 has such low solubility in aqueous buffers that various “solubilizing” mutations were needed to obtain well-behaved protein preparations for biophysical studies.15 These mutations included the Teem suppressor triplet as well as substitutions of surface-exposed hydrophobic residues with more hydrophilic residues found at equivalent positions in other vertebrate CFTR sequences. Surprisingly, several of these solubilizing surface mutations in hNBD1, identified in a screen focused exclusively on the in vitro solubility of hNBD1, were shown to suppress the in vivo trafficking defect of F508del-CFTR more strongly than the best existing pharmacological agents.32,38 Notably, the mutated residues (e.g., F429S, F494N, and Q637R) are not in direct contact with F508 and do not appear to be allosterically coupled.18 A similar hydrophobic-to-hydrophilic substitution in the immediate vicinity of F508, the V510D mutation, also strongly suppresses the in vivo trafficking defect of F508del-CFTR.39,40 It was proposed that these substitutions could block adventitious chaperone interactions that prevent proper ER export.18 However, there is as yet no concrete evidence explaining the tight correlation between the effects of mutations on the in vitro solubility properties of hNBD1 and the in vivo trafficking properties of human CFTR.
To clarify the relationship between the folding, stability, and solubility properties of hNBD1 and the in vivo trafficking of CFTR, we have undertaken coordinated isothermal (this paper) and thermal (see Ref. 36) denaturation studies of human NBD1 (hNBD1), using a variety of biophysical techniques. These studies produce a thermodynamic and structural model for hNBD1 unfolding and importantly identify a partial unfolding transition that is destabilized by F508del and stabilized by second-site mutations suppressing the in vivo trafficking defect of F508del-CFTR. This transition, which precedes the major chemical unfolding transition and seems likely to include unfolding of the α-helical (ABCα) subdomain, leads to aggressive self-association in vitro. The observed biophysical properties of hNBD1 variants harboring different mutations suggest that this initial unfolding transition is responsible for both the poor in vitro solubility of F508del-hNBD1 and the pathological ER retention of F508del-CFTR in vivo. This hypothesis, along with our model for hNBD1 unfolding, should help guide development of more efficacious drugs to treat the ∼90% of CF patients carrying the F508del mutation.
Results
Overview of Design of Isothermal Denaturation Experiments
Figure 1 shows isothermal urea denaturation profiles at 25°C for F508 and F508del-hNBD1-Δ(RI,RE). This protein construct has the wild-type human sequence except for deletion of most of the regulatory insertion (RI) and regulatory extension (RE).16,17,41 These protein segments, 37 and 39 residues in length, respectively, are disordered in hNBD1 in solution in vitro18,42 and dispensable for normal gating by intact CFTR in vivo.43 Their removal substantially improves both F508del CFTR biogenesis in vivo41 and hNBD1 solubility in vitro,17 enabling the F508del domain to be purified in sufficient yield for biophysical characterization in the absence of additional solubility-enhancing point-mutations. The F508del mutation in the full-length domain reduces its solubility to the point where such solubility-enhancing mutations are required to obtain a practically useful yield of purified protein.16 While these mutations represent natural sequence variations found in some vertebrate CFTR orthologues, their influence on the biophysical properties of hNBD1 are controversial.18,32 Therefore, we exploited the improved solubility of hNBD1-Δ(RI,RE) to explore the folding properties of the domain in the absence of solubility-enhancing mutations and compare its behavior to the full-length domain containing solubilizing mutations. Furthermore, we conducted experiments in a Standard Stabilizing Buffer, containing 10% (v/v) glycerol and 10% (v/v) ethylene glycol, that was developed to inhibit hNBD1 aggregation.15
Figure 1.
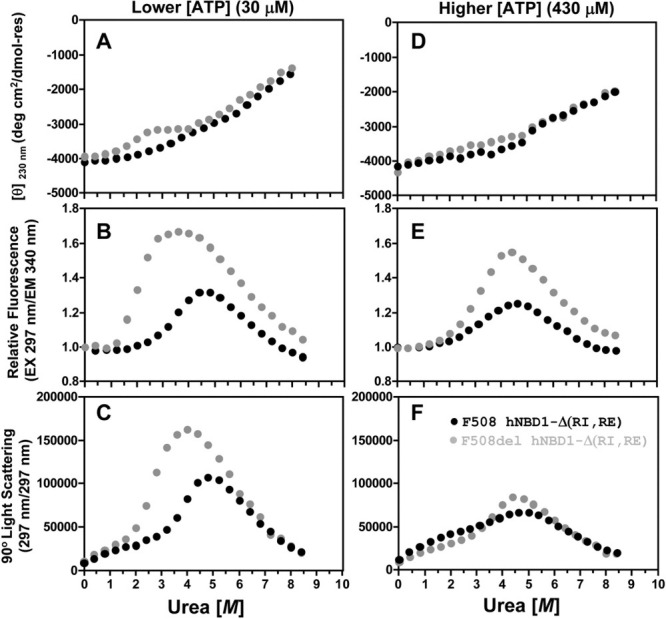
Simultaneous monitoring of CD, trp fluorescence, and SLS during isothermal urea denaturation at 25°C of hNBD1-Δ(RI,RE) ± F508del. Protein samples (2 μM) in the absence or presence of 8 M urea were progressively mixed every 800 seconds to achieve the indicated concentration of urea, using an autotitrator for CD measurements or an equivalent kinetic protocol for fluorescence and SLS measurements. The buffer contained 150 mM NaCl, 0.5 mM MgCl2 10% glycerol, 10% ethylene glycol, 0.8 mM TCEP, 20 mM Na-HEPES, pH 7.5. Data for F508 and F508del constructs are shown in black and gray, respectively. The buffer used to purify and store the protein, which is catalytically inactive,15–19 introduces 30 μM Mg-ATP. Additional Mg-ATP was added to the indicated final concentration. CD (top) was monitored at 230 nm instead of 222 nm to enable use of a higher concentration of Mg-ATP without interference from nucleotide absorbance. Intrinsic trp fluorescence (middle) was excited at 297 nm and monitored at 340 nm. SLS at 297 nm (bottom) was measured at a 90° angle simultaneously with trp fluorescence in a T-format fluorimeter (see Methods). SLS counts are background-subtracted for scattering from the protein-free 0 M urea buffer for measurements in the fluorimeter, so the ratio of the SLS signal at a given urea concentration to that at the start of the titration gives a rough estimate of weight-averaged aggregation state. Note, however, that this calculation underestimates the aggregation state at higher urea concentrations because it ignores the effect of urea in reducing protein contrast.
Denaturation of hNBD1-Δ(RI,RE) was performed using an autotitrator protocol and followed simultaneously by circular dichroism (CD) spectroscopy to monitor protein secondary structure (Fig. 1A,D), fluorescence spectroscopy to monitor the environment of the two tryptophan (trp) residues in hNBD1 (Fig. 1B,E), and static light-scattering (SLS)44 to monitor its aggregation state (Fig. 1C,F). The low solubility of hNBD1 has raised questions concerning the relationship between unfolding and self-association processes.18,32 This relationship can be addressed explicitly by monitoring protein aggregation state using SLS44 simultaneously with monitoring of protein conformation using the traditional spectroscopic methods. Supporting Information Figure S1 for this paper presents data validating our SLS methods, by showing minimal changes in SLS during chemical denaturation of a well behaved soluble protein that undergoes a two-state unfolding transition without alteration of its aggregation state.
The hNBD1-Δ(RI,RE) variants without or with F508del both display significantly different isothermal denaturation properties at a nominally saturating Mg-ATP concentration (430 μM) compared to a lower concentration (30 μM) giving only partial occupancy of the binding site (Fig. 1 and Supporting Information Fig. S2). The latter concentration is the lowest that is readily achievable because a high concentration of Mg-ATP (∼2 mM) must be maintained during purification to obtain high protein yield.15 Surface plasmon resonance studies have demonstrated that hNBD1-Δ(RI,RE) binds Mg-ATP with 0.8 μM affinity at 4°C.17 Calorimetric studies of this construct presented in Ref. 36 indicate that the enthalpy of Mg-ATP binding is 20–25 kcal/mole, so binding-affinity will be reduced ∼10-fold at 25°C versus 4°C. Therefore, the affinity for Mg-ATP should be ∼8 μM at the temperature of the chemical denaturation experiments shown in Figure 1, producing ∼80% occupancy of the binding site at 30 μM Mg-ATP compared to ∼98% at 430 μM. Most importantly, irrespective of the exact binding affinity, the ratio of Mg-ATP-free versus Mg-ATP-bound protein should decrease ∼14-fold (30/430) between the experiments in Figure 1 conducted at a high concentration of Mg-ATP compared to those conducted at a low concentration (because [hNBD1free]/[hNBD1bound] = Kd/[ATPfree]). The qualitatively different behavior in isothermal denaturation at different Mg-ATP concentrations indicates that hNBD1 has substantially different conformational behavior with versus without bound Mg-ATP (see also Ref. 36).
F508del Facilitates the First of Two Sequential Isothermal Unfolding Transitions in hNBD1
At a subsaturating 30 μM concentration of Mg-ATP, hNBD1-Δ(RI,RE) denatures in two successive steps at 25°C, the first resulting in ∼25% loss of secondary structure and the second resulting in ∼75% loss of secondary structure as monitored by CD (Fig. 1A). Note that the second transition is not completed at the highest urea concentration attainable in these experiments (8M) conducted in Standard Stabilizing Buffer, so there is no post-denaturation baseline visible in the CD spectra in Figure 1A or D. However, experiments conducted in Simple Buffer with urea (Supporting Information Fig. S3) or in Standard Stabilizing buffer with Gdn-HCl (data not shown), which is more denaturing than urea, demonstrate that the later phase of the denaturation profile does represent a discrete unfolding transition. The biphasic nature of the unfolding process is also clearly visible in the fluorescence traces in Figure 1B,E and the SLS traces in Figure 1C,F.
The F508del mutation moves the midpoint of the initial unfolding transition to substantially lower urea concentration (∼1.8 M vs. ∼3.6 M), while the second unfolding transition (with midpoint at ∼6.5 M) is unaffected. This pattern suggests that the ABCα subdomain45 of hNBD1,16,18 which contains F508, unfolds during the initial unfolding transition to produce a partially folded intermediate. The observation that F508del does not influence the second transition15 indicates that this transition does not produce an energetically significant conformational change in the vicinity of F508 in the ABCα subdomain and suggests that it is likely to involve predominantly conformational changes in hNBD1's nucleotide-binding core (which comprises an F1 ATPase-like core subdomain and the ABCβ subdomain16,18,45). It is difficult to obtain a reliable estimate of the free energy of the initial unfolding transition (ΔG0unfolding) in F508-hNBD1 due to the lack of a clear baseline between the two successive transitions (although a crude estimate is presented in Supporting Information Fig. S9 later). For F508del-hNBD1, it is ∼+3.3 kcal/mole at 25°C in the presence of 30 μM Mg-ATP, based on extrapolation of the equilibrium constant to zero urea concentration (Supporting Information Fig. S9 later).46 This estimate is only a rough approximation due to partial occupancy of the Mg-ATP binding site and the incomplete reversibility of the transition in these experiments (see below). However, it is generally consistent with the value of +1.8 kcal/mole for Mg-ATP-free hNBD1 at 25°C determined via detailed mathematical modeling of DSC data in Ref. 36.
The initial unfolding transition observed by CD spectroscopy is followed by an increase in trp fluorescence (Fig. 1B) and subsequently a strong increase in SLS (Fig. 1C). The fluorescence transition occurs at a slightly higher urea concentration than the CD transition, and the SLS transition occurs at a significantly higher urea concentration than the fluorescence transition. These offsets indicate that, rather than a simple two-state transition, the protein is undergoing sequential changes in its physical state resulting in the formation of protein aggregates. These aggregates probably correspond to the “N*” species previous identified by Thomas and coworkers.47 Their elevated fluorescence seems likely to be attributable to burial of the one or both of the trp residues in hNBD1 in intersubunit interfaces in the aggregates. The inference that aggregation follows an initial unfolding transition is strongly supported by a lag between the CD and fluorescence/SLS changes in kinetic studies of the denaturation process (Fig. 2 later). Moreover, although the forward titration behavior is reproducible, reverse titrations show significant hysteresis in the region of the initial transition (data not shown), and the magnitude of the increase in trp fluorescence varies somewhat in experimental replicates even though the qualitative features of the curve are reproducible (data not shown). These observations are consistent with irreversible aggregation influencing behavior in this range of urea concentration. Such aggregation is consistent with the kinetic trapping of partially unfolded intermediates inferred in Ref. 36 to occur during thermal denaturation. Both the trp fluorescence and SLS changes that follow the initial unfolding transition are reversed by the second unfolding transition that results in the loss of most protein secondary structure as monitored by CD. Therefore, full denaturation of hNBD1 reverses aggregation of the partially unfolded intermediate produced by the initial unfolding transition.
Figure 2.
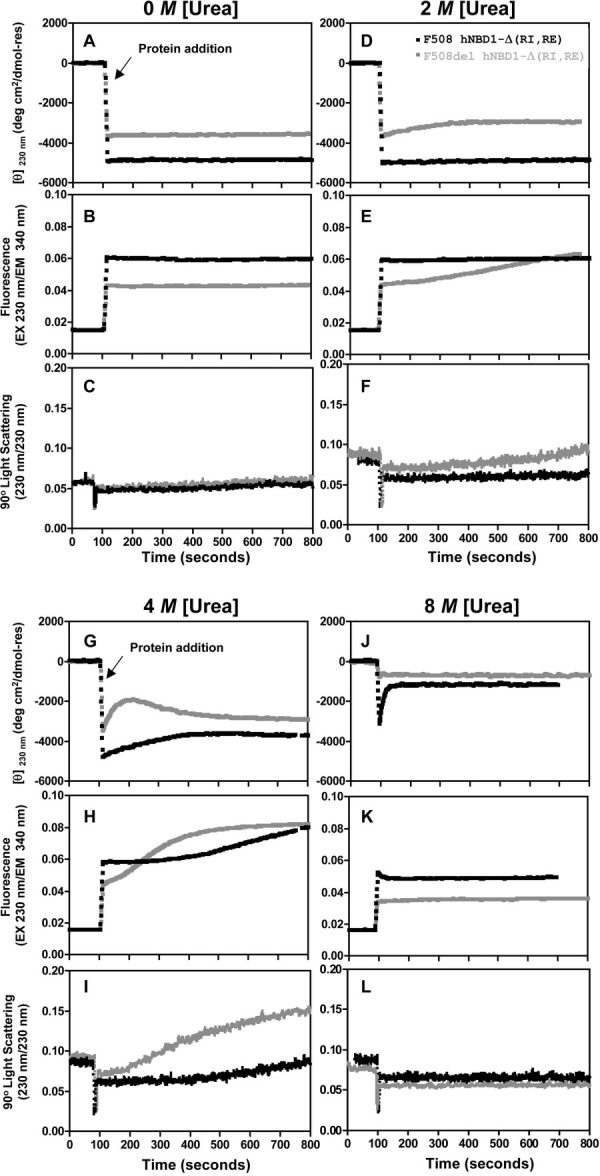
Kinetics of isothermal urea denaturation at 25°C of hNBD1-Δ(RI,RE) ± F508del. CD (top), trp fluorescence (middle), and SLS (bottom) signals were monitored using identical methods to Fig. 1 except that fluorescence emission at 340 nm and 90° SLS were measured in the Jasco spectropolarimeter using 230 nm incident light. At ∼100 seconds, 2 μM hNBD1-Δ(RI,RE) with (black) or without (gray) F508del was added from urea-free storage buffer to the Standard Stabilizing Buffer containing 30 μM Mg-ATP and the indicated concentration of urea.
Increasing Mg-ATP to a more highly saturating concentration of 430 μM shifts the first unfolding transition to higher urea concentration, and it suppresses the differences in the fluorescence and SLS signals observed during urea denaturation of the F508 versus F508del domains at 30 μM Mg-ATP (Fig. 1D–F compared to Fig. 1A–C; see Supporting Information Fig. S2 for direct overlay of the experiments in Fig. 1 conducted at high versus low Mg-ATP concentrations). The SLS changes during urea denaturation of hNBD1-Δ(RI,RE) are reduced in magnitude at the higher Mg-ATP concentration, especially in the F508del construct (Fig. 1F and Supporting Information Fig. S2C,F), indicating that aggregation is suppressed by greater saturation of the Mg-ATP binding site. This effect is presumably due to inhibition of the initial unfolding transition, which limits the concentration of the aggregation-prone, partially unfolded intermediate until the urea concentration is high enough to inhibit aggregation. The observed effect of Mg-ATP in suppressing aggregation is likely to explain the beneficial influence of super-saturating Mg-ATP concentrations on the yield of the domain during purification.15
Characterization of the Second Isothermal Chemical Unfolding Transition
Rigorous characterization of the second unfolding transition in urea is not possible in Standard Stabilizing Buffer because the domain is not fully denatured at the highest accessible denaturant concentration. However, this transition is completed at accessible urea concentrations in buffers that do not contain stabilizing osmolytes, for example, the Simple Buffer used in earlier chemical denaturation experiments.15 Supporting Information Figure S3 shows the results of autotitrator denaturation experiments conducted on hNBD1-Δ(RI,RE) constructs at 25°C in this buffer. All qualitative features of the unfolding behavior are the same as in Standard Stabilizing Buffer, although the lower stability of the domain in Simple Buffer shifts all conformational transitions 1–2 M lower in urea concentration. (The significant differences in these results compared to those reported earlier in the same buffer15 are likely to be attributable to the faster denaturation/observation protocol used here, as explained in the Discussion section below.) Similar glycerol concentrations to those in Standard Stabilizing Buffer have been observed to suppress the protein trafficking defect caused by the F508del mutation in full-length CFTR in vivo in tissue culture cells,48,49 consistent with the hypothesis that unfolding of the domain contributes to aberrant ER retention of F508del-CFTR. Increasing Mg-ATP concentration has at most a minor effect on the progression of the second unfolding transition in Simple Buffer (Supporting Information Fig. S3), in contrast to the obvious inhibition of the first unfolding transition observed at the higher Mg-ATP concentration in either buffer (Supporting Information Figs. S2–S3). Additional investigations of the influence of adenine nucleotides on the progression of the second unfolding transition in several buffers have failed to detect a reproducible effect (data not shown).
These results indicate that there is a substantial reduction in Mg-ATP binding affinity during the initial urea unfolding transition but not during the ensuing transition that yields fully unfolded hNBD1-Δ(RI,RE). Therefore, the partially unfolded intermediate produced by the initial unfolding transition is unlikely to have significant affinity for Mg-ATP. While aggregation of the intermediate or the elevated urea concentrations (>4M) needed to drive the second unfolding transition could potentially interfere with Mg-ATP binding, it is also possible that the intermediate does not retain a functional Mg-ATP binding site, even though it retains 75% of the native secondary structure. Such characteristics would be consistent with formation of a “molten globule” conformation,50,51 which has significant secondary structure but lacks stable tertiary structure. The accompanying paper36 demonstrates that thermal denaturation produces an aggregation-prone intermediate with generally similar biophysical properties that seems likely to have molten globule properties based on its minimal enthalpy of unfolding. Therefore, thermal denaturation and isothermal chemical denaturation of hNBD1-Δ(RI,RE) may proceed via similar aggregation-prone molten globule intermediates. (See Discussion section.)
Evidence for Multiple Self-Association Pathways
It is noteworthy that, as opposed to the order of events observed during urea denaturation in the presence of 30 μM Mg-ATP (Fig. 1A–C), the fluorescence and SLS changes precede the CD changes during urea denaturation of F508-hNBD1-Δ(RI,RE) in the presence of 430 μM Mg-ATP (Fig. 1D–F). These observations raise the possibility that self-association of the domain leading to an increase in trp fluorescence can also occur via a distinct pathway involving a native-like conformation that has not lost significant secondary structure. Kinetic studies (Fig. 2G later) show that aggregation of the partially unfolded intermediate leads to a small increase in secondary structure content as monitored by CD, meaning that aggregation tightly coupled to unfolding could potentially obscure a secondary structural change. However, equivalent denaturation experiments conducted on constructs harboring suppressor and solubilizing mutations (Fig. 4 later) strongly support the inference that hNBD1-Δ(RI,RE) can aggregate in a native-like conformational state to produce a species with elevated trp fluorescence. In addition, the SLS data demonstrate that the domain has a tendency to self-associate in the native conformational state to form aggregates with unaltered trp fluorescence (i.e., at the points below 1.5 M urea in Fig. 1C,F). Therefore, hNBD1-Δ(RI,RE) displays three distinct aggregation pathways in vitro. However, the biggest aggregates (i.e., those giving the largest SLS signals in Fig. 1), which also have the most strongly elevated trp fluorescence, are formed subsequent to the initial unfolding transition, and this pathway clearly dominates the aggregation behavior of the domain harboring the F508del mutation.
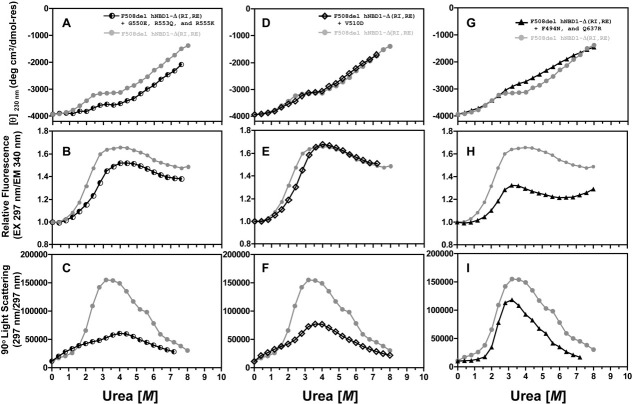
Figure 4.
Trafficking suppressor mutations stabilize hNBD1-Δ(RI,RE)-F508del against the initial unfolding transition. Urea denaturation at 25°C in the presence of 30 μM Mg-ATP was conducted under conditions identical to Fig. 1 and monitored using CD spectroscopy (top), trp fluorescence spectroscopy (middle, with 297 nm excitation and 340 nm emission), and SLS (bottom, at 297 nm). The denaturation of hNBD1-Δ(RI,RE)-F508del is compared to that of the same construct containing in addition the Teem suppressor triplet37 (left), V510D39 (center), or F494N/Q637R15 (right).
Similar Chemical Unfolding Behavior of hNBD1-Δ(RI,RE) at Higher Temperature
Isothermal urea denaturation of hNBD1-Δ(RI,RE) using an autotitrator protocol at 35°C (top of Supporting Information Fig. S4) yields qualitatively similar results to those obtained at 25°C. Most importantly, the F508del mutation clearly promotes unfolding (lowering the midpoint of the transition by ∼1.5 M in urea concentration), while raising the Mg-ATP concentration clearly inhibits unfolding (raising the midpoint of the transition by ∼1.5 M in urea concentration). Therefore, F508del and Mg-ATP have equivalently strong and opposing influences on domain stability at 35° and 25°C. Furthermore, there is evidence of self-association of the F508del domain in a native-like conformational state at both temperatures in the presence of 430 μM Mg-ATP, because the increase in trp fluorescence precedes the secondary structure transition monitored by CD spectroscopy. However, there is one noteworthy difference in the biophysical behavior of the domain at the higher temperature. Only a single CD transition is observed at 35° for either the F508 or F508del domain in the presence of 30 μM Mg-ATP (Supporting Information Fig. S4A,B), compared to two successive transitions under the same conditions at 25°C (Fig. 1 and Supporting Information Fig. S4C,D). While additional analyses would be required for rigorous mechanistic dissection of this behavior, the thermal denaturation studies presented in Ref. 36 indicate a two-step unfolding pathway for the same protein constructs in the absence of urea in this temperature range, with the relative secondary structure content of the thermal unfolding intermediate closely matching that of the chemical unfolding intermediate observed at 25°C. Therefore, the simplest explanation for the observed behavior is that unfolding of hNBD1-Δ(RI,RE) still proceeds through a partially unfolded intermediate but that this intermediate is less stable to urea denaturation at 35°C and therefore does not accumulate at this temperature, at least on the kinetic timescale of isothermal chemical denaturation experiments (∼5 minutes per point). Increasing urea concentration above that required for full denaturation of the domain at 35°C continues to reduce trp fluorescence, likely reflecting dissociation of unfolded protein aggregates by urea (Supporting Information Fig. S4B,F).
Notably, chemical denaturation of the domain occurs at substantially lower urea concentrations at 35 versus 25°C (by ∼1.5 M), consistent with the calorimetric studies in Ref. 36 showing substantially reduced stability at the higher temperature. Consistent with the results of these studies, the data in Supporting Information Fig. S4A suggest that the unfolding transition in the F508del domain is already underway at 35°C in the absence of urea, while the initial unfolding transition in this construct is delayed at 25° to urea concentrations in excess of 1 M. The lower stability of hNBD1-Δ(RI,RE) at 35° versus, especially in the presence of the F508del mutation, parallels the dramatic exacerbation of the in vivo trafficking defect produced by this mutation during cell growth at 37° versus 25°C.26–29 These results are consistent with the initial unfolding transition contributing to the pathological ER retention of F508del-CFTR.
Equivalent Two-Step Chemical Unfolding of Full-Length hNBD1
Isothermal urea denaturation of full-length hNBD1 (i.e., containing both the RI and RE) using an autotitrator protocol at 25°C yields essentially equivalent results to those obtained with the Δ(RI,RE) construct under the same solution conditions (Supporting Information Fig. S5). Solubilizing surface mutations need to be introduced into full-length hNBD1 to obtain sufficient material for biophysical studies.15 Supporting Information Figure S5 compares the behavior of matched full-length and Δ(RI,RE) constructs containing F429S, F494N, and Q637R mutations15 in the absence or presence of the F508del mutation. Both constructs show two sequential unfolding transitions equivalent to those observed for the hNBD1-Δ(RI,RE) construct without any solubilizing mutations; the first of these transitions is inhibited by a higher concentration of Mg-ATP and facilitated by the F508del mutation.
Supporting Information Figure S6 replots the same data to compare directly the equivalent constructs with and without the RI and RE segments. The presence of the RI and RE destabilize the domain, moving the initial unfolding transition to lower urea concentration (by 1.5–2.5 M). This observation suggests that one or both of these disordered protein segments interact with and stabilize the partially unfolded conformation of hNBD1, thereby reducing the magnitude of the free energy change during the initial unfolding transition.17 The thermal denaturation studies in Ref. 36 which examine a ΔRI construct in addition to Δ(RI,RE) and full-length constructs, demonstrate that the increase in domain stability in the Δ(RI,RE) construct compared to the full-length construct is attributable primarily to deletion of the RI rather than the RE. Recently, it has been demonstrated that deletion of the RI strongly suppresses the trafficking defect in F508del-CFTR in vivo in tissue culture cells.41 These observations provide further evidence that the initial unfolding transition of hNBD1 contributes to aberrant trafficking of F508del-CFTR in vivo. Supporting Information Figure S6 shows that the trp fluorescence changes, which likely reflect domain aggregation, track the initial unfolding transition in all experiments conducted on full-length hNBD1 constructs. Therefore, aggregation subsequent to the initial unfolding transition also contributes to the low solubility of the domain in vitro, suggesting that the improved solubility of the Δ(RI,RE) domain17 is also attributable at least in part to the enhanced stability of the truncated domain.
Kinetic Studies Demonstrate that the Initial Unfolding Transition Induces Aggregation
Figure 2 shows CD, intrinsic trp fluorescence, and SLS signals as a function of time after introducing hNBD1-Δ(RI,RE) into solutions containing a low concentration of Mg-ATP (30 μM) and 0, 2, 4, or 8 M urea. Without urea (Fig. 2A–C), no significant changes are observed during the 13 minutes of the experiment except for a very small increase in SLS at the latest time points, likely reflecting a minor degree of protein self-association. Similarly, under denaturing conditions at 8 M urea (Fig. 2J–L), the signals are all constant after rapid completion of the unfolding transition (which occurs more slowly in the F508 domain, with a half-time of ∼20 seconds).
In contrast, multiphase kinetic processes are observed for the F508 and F508del domains at urea concentrations leading to accumulation of partially unfolded intermediates. At 2 M urea, the initial unfolding transition is mostly completed for F508del but only beginning for F508 hNBD1-Δ(RI,RE) (Fig. 1A). The F508 domain remains stable under these conditions but the F508del domain shows an initial loss of secondary structure as monitored by CD followed ∼3 minutes later by a gradual increase in trp fluorescence and SLS (Fig. 2D–F). The similarity in the kinetics of the fluorescence and SLS changes suggests the alteration in trp environment leading to increased fluorescence quantum yield occurs during aggregation of the domain (presumably due to solvent-shielding of at least one of its two trps in interprotein interfaces in the aggregates, as explained above). In any event, the different kinetics observed for the SLS versus CD changes demonstrate that aggregation of the partially unfolded intermediate occurs on the time-scale of minutes only after occurrence of the initial unfolding transition.
In 4 M urea (Fig. 2G–I), the F508 domain shows CD, trp fluorescence, and SLS changes with very similar magnitudes and kinetics to those observed for the F508del domain in 2 M urea. These observations suggest that F508-hNBD1 adopts a partially unfolded conformation in 4M urea similar to that adopted by F508del-hNBD1 in 2 M urea, consistent with the data in Figure 1. Note that the F508del domain unfolds more rapidly than the F508 domain in 4 M urea (as monitored by CD – Fig. 2G) and then aggregates more rapidly and aggressively (as monitored by trp fluorescence and SLS – Fig. 2H–I). Under these conditions, aggregation of F508del-hNBD1 (Fig. 2I) is accompanied by a small increase in its secondary structural content (Fig. 2G,I). When aggregation is very rapid, this increase might potentially obscure the loss of CD signal occurring during the initial unfolding transition. Therefore, caution must be exercised in interpreting results from individual isothermal chemical denaturation experiments.
Distinct Effects of Mg-ADP Versus Mg-ATP on the Initial Unfolding Transition
Figure 3 examines the influence of Mg-ADP versus Mg-ATP on the isothermal urea denaturation of hNBD1-Δ(RI,RE) at 25°C in the absence (Fig. 3A,B) or presence (Fig. 3C,D) of the F508del mutation. In all cases, unfolding occurs in two successive steps, as observed in Figure 1 above. The addition of 400 μM Mg-ADP (in addition to 30 μM Mg-ATP) stabilizes the domain in the native conformational state and increases the midpoint of the initial unfolding transition in urea, although to a lesser extent than addition of the same concentration of Mg-ATP. Surface plasmon resonance measurements indicate that both nucleotides have similar binding and release rates and affinities for hNBD1-Δ(RI,RE).17 To verify that they exert somewhat different effects on the unfolding of the domain, we monitored CD and trp fluorescence as a function of time after dilution of F508del-hNBD1-Δ(RI,RE) into buffers containing 0, 2, or 4 M urea (Supporting Information Fig. S7). These kinetic studies verify the qualitative features of the data in Figure 3. Notably, Mg-ATP retards the unfolding and subsequent self-association of the domain substantially more than the equivalent concentration of Mg-ADP (Supporting Information Fig. S7C–F). While ADP interacts exclusively with the nucleotide-binding core of hNBD1, the γ-phosphate of ATP makes a bridging contact between the ABCα subdomain and the nucleotide-binding core via the γ-phosphate switch (or Q-loop),18,23,52 a short loop at the N-terminus of the ABCα subdomain.18,45 This subdomain packs tightly against the core in a defined orientation when the γ-phosphate of Mg-ATP is bound,18,23,52 but it is otherwise flexibly attached,45,53 including when Mg-ADP is bound. Mg-ATP thus stabilizes the interaction of the core of hNBD1 with the ABCα subdomain, an interaction that would be disrupted by unfolding of this subdomain. The more rapid unfolding and aggregation of Mg-ADP-bound versus Mg-ATP-bound F508del-hNBD1-Δ(RI,RE) suggests that the ABCα subdomain plays a critical role in controlling the kinetics of unfolding and provides additional evidence that its conformation is altered significantly during the initial unfolding transition.
Figure 3.
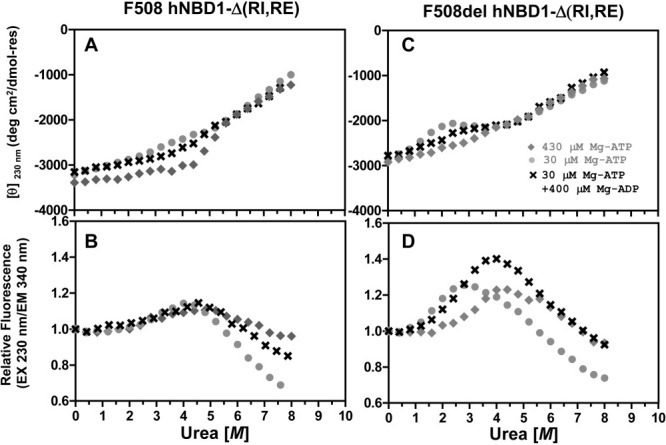
Effects of Mg-ADP versus Mg-ATP on isothermal urea denaturation of hNBD1-Δ(RI,RE). Denaturation of the F508 (left) or F508del (right) domain was conducted at 25°C under conditions identical to Fig. 1 and monitored using CD (top) and trp fluorescence (bottom, with 230 nm excitation and 340 nm emission). Proteins were diluted into Standard Stabilizing Buffer containing 400 μM Mg-ADP (crosses), 400 μM Mg-ATP (gray diamonds), or no additional nucleotide (gray closed circles). Because purified proteins are stored in Standard Stabilizing Buffer containing Mg-ATP, they introduce 30 μM Mg-ATP into every experimental sample in addition to any nucleotide present in the measurement buffer. The final Mg-ATP concentration in each experiment is indicated in the legends.
Second-Site Solubilizing/Suppressor Mutations Delay the Initial Unfolding Transition
Figure 4 shows the isothermal denaturation behavior of F508del-hNBD1-Δ(RI,RE) constructs harboring additional point mutations known to suppress the trafficking defect caused by the F508del mutation.32,37,39,40 The G550E, R553Q, and R555K mutations in the Teem suppressor triplet were isolated in vivo using a selection for mutations restoring the export of a yeast CFTR homolog bearing the equivalent of the F508del mutation. They were subsequently introduced into intact F508del-CFTR and demonstrated to suppress its aberrant trafficking in vivo in tissue culture cells.37 Introducing all three mutations simultaneously into full-length hNBD1 led to improved yield and stability of the purified soluble domain after overexpression in E. coli.15 The V510D mutation was made to explore the molecular pathology caused by the F508del mutation after crystallographic studies demonstrated that F508del produces a large change in the conformation of val-510.15 The hydrophobic sidechain of val-510 is mostly buried on the surface of hNBD1 in the absence of the F508del mutation,18 at a site where it is likely to make direct packing interactions to the transmembrane domains of CFTR.35 However, its backbone conformation is altered so that it projects from the surface of hNBD1 and is completely solvent-exposed in the presence of the F508del mutation.18 Surprisingly, the V510D mutation strongly suppresses the trafficking defect caused by the F508del mutation in vivo in tissue culture cells.39,40 Finally, the F494N/Q637R mutations were identified in a screen for surface substitutions that improve the solubility of purified hNBD1 in vitro.15 This screen focused on replacement of hydrophobic residues with more hydrophilic residues present at equivalent sites in other vertebrate CFTR sequences. These mutations also show significant efficacy in suppressing the trafficking defect caused by the F508del mutation in vivo in tissue culture cells,32 although they are less effective that the Teem suppressors,37 the V510D mutation,39,40 or deletion of the RI.41
The Teem suppressor triplet (Fig. 4A–C), the V510D mutation (Fig. 4D–F), and the F494N/Q637R mutations (Fig. 4G–I) all stabilize hNBD1 against the initial unfolding transition, shifting its midpoint ∼0.25–0.50 M higher in urea concentration. The magnitude of the SLS increase following the initial unfolding transition is greatly reduced by the Teem suppressor triplet and the V510D mutation and significantly reduced by the F494N/Q637R mutations. Therefore, the suppressor and solubilizing mutations all reduce the extent of aggregation of the partially unfolded intermediate, possibly by delaying its formation until a higher concentration of urea is present.
These second-site mutations have generally similar effects improving the stability and reducing the aggregation of hNBD1-Δ(RI,RE) bearing the native phe residue at position 508 (Supporting Information Fig. S8). In the presence of the Teem suppressor triplet or the V510D mutation, which significantly stabilize the domain, the urea-induced increase in trp fluorescence (Supporting Information Fig. S8B,E) precedes the initial unfolding transition as monitored by CD spectroscopy (Supporting Information Fig. S8A,D). This observation provides further evidence that the domain can also self-associate in a native-like conformation before formation of the partially unfolded intermediate, as suggested by the analysis of Figure 1D–F above.
While the different suppressor and solubilizing mutations studied here all have similar biophysical effects, some minor differences in behavior are worth noting. First, the SLS data in Figure 4I demonstrate that the F494N/Q637R mutation pair in the F508del domain is unique in suppressing the self-association that occurs without an increase in trp fluorescence before the onset of the initial unfolding transition. Therefore, this mutation pair may influence the solubility of hNBD1 via more than one mechanism. Second, the F494N/Q637R mutation pair appears to decrease the secondary structure content in the partially unfolded intermediate formed by both the F508del (Fig. 4G) and F508 (Supporting Information Fig. S8G) domains, while the Teem suppressor triplet may increase the secondary structure content in this state just in the F508del domain (Fig. 4A). Therefore, the second-site suppressor mutations may alter the structure of the partially unfolded intermediate produced by the initial unfolding transition, as predicted in computational studies of the folding pathway of hNBD1,54 although aggregation of the intermediate could also influence its secondary structure content (as shown in Fig. 2G). Conformational changes in the partially unfolded intermediate could produce complex behavior in some experiments.
Discussion
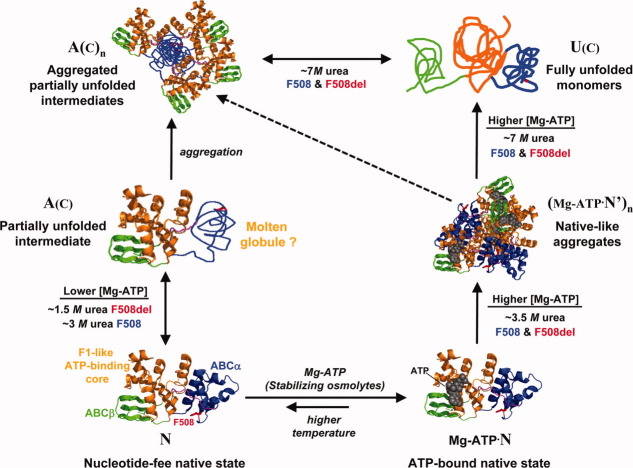
The isothermal denaturation studies in this paper combined with the thermal denaturation studies in Ref. 36 provide a consistent model for the unfolding of NBD1 from human CFTR (Fig. 5) and its perturbation by F508del, the predominant mutation causing cystic fibrosis. This model and the data used to develop it lead to a coherent hypothesis concerning the thermodynamic and structural effects of F508del that cause pathogenic retention of CFTR in the ER. Specifically, we hypothesize that F508del promotes the initial step in the unfolding of hNBD1, which produces an aggregation-prone, partially unfolded intermediate (species A(c) in Fig. 5) that is targeted for degradation rather than exported to the cell surface. Several lines of evidence support this hypothesis, including the observations that that the initial unfolding transition in hNBD1 is facilitated by F508del and inhibited by the second-site mutations that suppress the trafficking defect caused by F508del in vivo32,37,39 (Supporting Information Fig. S9).
Figure 5.
Schematic model of isothermal unfolding pathway of hNBD1. The crystal structure of hNBD1 (2PZE) is shown at the bottom either with (right) or without (left) bound Mg-ATP. The F1- like core subdomain is shown in orange, the ABCβ subdomain in green, and the ABCα subdomain in blue.39,45 A(C) represents the aggregation-prone intermediate produced by the initial chemical unfolding transition, while U(C) represents the fully denatured state produced by urea. The conformation and biophysical properties of the chemical unfolding intermediate A(C) are likely to be very similar to those of the thermal unfolding intermediate A(T) described in Ref. 36. Unfolding at high urea concentration may still proceed through A(C) as a kinetic intermediate, but it does not accumulate to appreciable concentration under these conditions. See Lewis et al.39 for a detailed description of the subdomain organization of hNBD1 and the stereochemical effects of the F508del mutation, the Teem suppressor mutation triplet,37 and the F494N/Q637R solubilizing mutations.15
Beyond agreement with a wide variety of biophysical and physiological studies, our model linking partial unfolding of hNBD1 with the molecular etiology of CF explains several enigmatic observations. Most importantly, this model explains why mutations improving the in vitro solubility of hNBD1 have consistently shown efficacy in suppressing the in vivo trafficking defect caused by the F508del mutation in intact CFTR.32,39 To obtain constructs of full-length hNBD1-F508del sufficiently soluble for in vitro purification and characterization, an extensive screening program was carried out to identify second-site mutations that improve solubility.15 Many candidate mutations were considered, which generally involved substitution of surface-exposed hydrophobic residues with more polar residues naturally occurring at the equivalent position in other vertebrate CFTR sequences. This research program led to the identification of several point mutations that improve the solubility and yield of purified hNBD1, including the Teem suppressor triplet (G550E, R553Q, and R555K) isolated as suppressors of the in vivo trafficking defect of F508del-CFTR.37 However, surprisingly, the other efficacious solubilizing mutations chosen exclusively on the basis of polarity and consistency with the CFTR sequence profile also generally suppress the defective in vivo trafficking of F508del-CFTR.32,39 This correlation is readily explained if the initial unfolding transition in hNBD1 (left side of Fig. 5) controls both aggregation of the purified domain in vitro and aberrant ER retention and degradation of intact CFTR in vivo.
The SLS studies reported in this paper demonstrate that the initial unfolding transition in hNBD1 leads to aggregation in vitro, and the totality of the available data supports the inference that this transition is at least partially responsible for the poor solubility properties of the domain, especially in the presence of the F508del mutation. For example, improvements in hNBD1 solubility were obtained by increasing Mg-ATP concentration further over levels already expected to produce >99.9% saturation of its binding site.15 Our experiments (Fig. 1) demonstrate that lowering the fractional population of nucleotide-free hNBD1 strongly inhibits the initial unfolding transition and greatly reduces the degree of protein self-association, especially in the presence of the F508del mutation. As noted above, the hypothesis that the aggregation-prone intermediate produced by the initial unfolding transition in hNBD1 also causes aberrant ER retention of CFTR in vivo rationalizes the observation that second-site mutations isolated solely based on their influence on in vitro solubility are generally efficacious is suppressing the F508del-induced in vivo trafficking defect.15,32,39 Moreover, second-site in vivo suppressor mutations isolated using completely orthogonal approaches (i.e., the Teem suppressor triplet37 vs. F949N/Q637R15 vs. V510D39) all suppress the initial unfolding transition in isolated hNBD1 and its resulting aggregation (Fig. 4).
The isothermal chemical denaturation studies reported in this paper (summarized in Fig. 5) and the thermal denaturation studies reported in Ref. 36 reveals that hNBD1 unfolds via two sequential transitions which have similar spectroscopic and aggregation properties whether unfolding is driven by urea or by heat. The first transition, which results in loss of 20–25% of the overall secondary structure content (as assessed by CD spectroscopy), is facilitated by the F508del mutation and inhibited by second-site suppressors of the in vivo and in vitro pathologies caused by this mutation. The second transition, which results in loss of the remaining secondary structure, is not strongly influenced by these sequence variations. The F508del mutation destabilizes the native conformational state of the domain compared to the partially unfolded intermediate produced by the initial unfolding transition, while the suppressor mutations tested so far stabilize it. Mg-ATP binding inhibits the initial unfolding transition in isothermal urea denaturation experiments (Fig. 1 and Supporting Information Fig. S1), consistent with the conclusion from thermal unfolding studies that the initial unfolding transition occurs from the nucleotide-free state (based on nonlinear least squares fitting of differential scanning calorimetry (DSC) data in Ref. 36). This transition leads to an aggregation-prone intermediate, as revealed explicitly by the SLS data collected during our chemical denaturation experiments and inferred indirectly from the observation that thermal denaturation is kinetically controlled and largely irreversible (see Ref. 36).
The initial unfolding transition may produce a “molten globule”50,51 conformation that retains significant secondary structure but lacks stable tertiary structure (Fig. 5). This possibility is supported by a series of different observations including the minimal enthalpy of unfolding of the resulting intermediate (see Ref. 36), its enhanced binding of the hydrophobic reporter dye bis-anilinonaphthalene-disulfonic acid (data not shown), and its minimal near-UV circular dichroism from the two trp residues in the domain (data not shown and Ref. 36). The minimal influence of Mg-ATP on the second unfolding transition (Supporting Information Fig. S3 and Ref. 36) is also consistent with formation of a molten globule during the initial unfolding transition, because this observation indicates that the partially unfolded intermediate does not possess a high-affinity nucleotide binding site.
While further investigation will be required for definitive characterization of the global conformational properties of the partially unfolded intermediate, it clearly involves significant conformational changes in the ABCα subdomain (Fig. 5) compared to the native-state conformation of hNBD1. This inference is supported by the location in the ABCα subdomain of the F508del mutation that strongly facilitates the initial unfolding transition and all but Q637R among the second-site suppressor mutations demonstrated in Figures 4 and Supporting Information Figure S8 to inhibit this transition. The Teem suppressor triplet and the F494N mutation are located on the opposite face of the ABCα subdomain from the V510D mutation, which is adjacent to the F508del site, and crystallographic studies indicate that the direct stereochemical effects of these different mutations are likely to be unrelated.18 The influence of all of these mutations on the initial unfolding transition suggests that it involves a global change in ABCα subdomain conformation. This inference is further supported by the inhibition of the transition by Mg-ATP (Figs. 1 and Supporting Information S1), as well as by the greater stability and slower unfolding kinetics of the domain in the presence of Mg-ATP compared to Mg-ADP (Fig. 3), because the γ-phosphate of ATP specifically bridges the interface between the nucleotide-binding core of hNBD1 and the ABCα subdomain at a site on that subdomain that is distal to the location of the F508del mutation.18
The multimode spectroscopic studies reported in this paper demonstrate that hNBD1 self-associates via three distinct pathways. The first precedes unfolding of the domain as monitored by CD and occurs without an increase in intrinsic trp fluorescence. The second also precedes the unfolding of the domain but produces a substantial increase in trp fluorescence, presumably due to burial of at least one of the two trp residues in the domain in interprotein interfaces in the aggregates. The third pathway, which produces the largest aggregates (as assessed by SLS) and dominates the behavior of the F508del domain, involves aggregation of the partially folded intermediate concomitant with a substantial increase in trp fluorescence. Note that the influences of protein mutations, nucleotide-binding, stabilizing osmolytes, and temperature on this last aggregation pathway correlate consistently with their influence on CFTR biogenesis in vivo. The other two aggregation pathways, which seem likely to involve distinct conformational phenomena, do not have any clear relationship to the physiological properties of CFTR. However, these pathways could readily be confused with the physiologically relevant unfolding/aggregation pathway in studies employing only a single spectroscopic probe rather than the multimode spectroscopic methods employed here (e.g., in Fig. 1).
An earlier isothermal chemical denaturation study15 showed minimal differences in the behavior of full-length hNBD1 in the presence or absence of the F508del mutation. There are several technical differences between the current study and the previously reported study that could potentially account for the different observations. Supporting Information Figure S3 shows that the equivalent denaturation behavior is obtained in either the Standard Stabilizing Buffer developed to optimize hNBD1 solubility or in the Simple Buffer not containing glycerol or ethylene glycol used in the earlier study.15 Therefore, the divergent results cannot be due to differences in buffer composition and are likely to be attributable to the different protocols used to perform isothermal chemical denaturation in the two studies. In contrast to the autotitrator protocol used here, the earlier study used a standard titration protocol involving lengthy pre-incubation of parallel samples at each denaturant concentration.46 Therefore, except at fully denaturing urea concentrations, all samples in the earlier study were likely to be aggregated at the time of measurement (i.e., kinetically trapped in an aggregated state after undergoing the initial unfolding transition). Even though that study employed a widely used protocol for chemical denaturation experiments,46 aggregation went undetected because parallel SLS measurements were not performed, and protein aggregation likely masked the influence of the F508del mutation on domain stability. Furthermore, the stabilizing influence of the Teem suppressor triplet reported in the earlier study differs in detail from that reported in Figure 4 above, so even those results seem likely to have been distorted by protein aggregation. The results presented here highlight the greater power of chemical denaturation studies monitored simultaneously by SLS.
Materials and Methods
Buffers
Standard Stabilizing Buffer, developed to optimize yield of purified hNBD1,15,16 contains 150 mM NaCl, 0.5 mM MgCl2, 10% (v/v) glycerol, 10% (v/v) ethylene glycol, 0.8 mM TCEP, 20 mM Na-HEPES, pH 7.5. The Simple Buffer used in previously published chemical denaturation experiments15 contains 0.8 mM TCEP, 20 mM sodium phosphate, pH 8.0.
Protein Expression and Purification
The hNBD1-Δ(RI,RE) construct comprises residues 387–646 of human CFTR with residues 405–436 deleted. All hNBD1 constructs were expressed in E. coli at 18°C and purified using previously published methods.15–17 In brief, domains with an N-terminal His6-Smt3 fusion55 were purified by Ni-NTA chromatography, cleaved by Ulp1 protease, purified by S200 gel filtration chromatography, recovered from the flow-through of a second Ni-NTA column (to remove residual His6-Smt3 tag), and concentrated to 2–10 mg in Standard Stabilizing Buffer containing 3 mM MgCl2 and 2 mM ATP.
Chemical Denaturation Protocol
Titrations were monitored in a 1 cm path-length Hellma quartz fluorescence cuvette (Thermo, Waltham, MA) filled initially with 1.6 mL of protein in urea-free buffer and continuously mixed with a magnetic stir-bar. The reservoir solution contained 8–10 mL of an identical concentration of protein in the same buffer with 8.5–9.0 M spectroscopic grade urea (Fluka, St. Louis, MO). The urea-containing protein solution in the reservoir was progressively mixed with the protein sample in the cuvette to achieve 0.4 M steps in urea concentration, with 800 second equilibration time between successive concentration steps and spectroscopic measurements, using a computer-controlled autotitrator on the Jasco CD instrument or by hand on the PTI fluorimeter. Urea concentration was determined using http://sosnick.uchicago.edu/gdmcl.html ([urea] = 118•Δη + 29.8•Δη2 + 186•Δη3, with Δη being the difference in refractive index relative to urea-free buffer). Refractive index was measured at room temperature using a refractometer (Baush & Lomb, catalogue no. 33.46.10). Control titrations of protein-free buffers with or without 400 μM ATP showed no significant changes in the spectroscopic signals monitored in experiments reported in this paper (data not shown).
CD, Fluorescence, and SLS Measurements on the Jasco Spectropolarimeter
Measurements were conducted using a J-815 spectropolarimeter (Jasco, Easton, MD) equipped with a PFD-425 Peltier temperature-controlled cell, an FMO-427 fluorescence detector, and an ATS-429 autotitrator. Wavelength scans were conducted from 220 to 300 or 340 nm at 100 nm/second. The monochromator of the FMO-427 detector was set to 340 nm for fluorescence and 230 or 340 nm for SLS44 with sensitivity of 600–650 Volts. Either fluorescence or SLS were acquired simultaneously with CD data, which were collected at 230 nm rather than 222 nm to allow use of a higher Mg-ATP concentration without interference from nucleotide absorbance. At lower nucleotide concentrations, equivalent CD results were observed at either wavelength (data not shown).
Fluorescence and SLS Measurements on the PTI Fluorimeter
Measurements were alternatively conducted in a T-format PTI QuantaMaster C-61 spectrofluorimeter (Monmouth Junction, NJ), using 5 nm slits and photomultipliers in digital photon-counting mode. The temperature of the jacketed cell-holder was maintained by a circulating water bath and monitored by a Digi-Sense type T thermocouple thermometer. Unpolarized fluorescence and 90° SLS44 signals were acquired simultaneously with emission monochromators set to 340 nm and 297 nm, respectively, using 297 nm excitation light passing through a vertical Glan-Thompson polarizer.
Data Analysis
Prism 5 (GraphPad, San Diego, CA) was used for plotting and least-squares curve fitting. Background-subtraction used protein-free buffer at 0 M urea for CD and 90° SLS measurements in the fluorimeter or protein-free buffer containing the same urea concentration for fluorescence measurements. CD values were normalized to mean residue ellipticity using protein concentration measured in a Bradford assay;56 for the constructs with suppressor mutations in Figure 4 and Supporting Information Figure S8 and for the full-length F508 construct in Supporting Information Figures S5 and S6, an additional linear normalization factor was applied to produce equivalent CD signals at 0 M urea, to correct for likely inaccuracy in protein concentration determination. Fluorescence was normalized to the value in 0 M urea. While only two of the three possible measurements could be conducted simultaneously, combined CD-fluorescence-SLS datasets were assembled from pairs of experiments showing equivalent results for one redundant measurement.
Acknowledgments
The authors thank the Cystic Fibrosis Foundation for their long-term commitment to basic research. They also thank Scott Banta and members of his lab for access to their Jasco spectrometer. They acknowledge an anonymous referee for a critical review that led to significant improvements in this manuscript.
References
- 1.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MP, Rich DP, Gregory RJ, Smith AE, Welsh MJ. Generation of cAMP-activated chloride currents by expression of CFTR. Science. 1991;251:679–682. doi: 10.1126/science.1704151. [DOI] [PubMed] [Google Scholar]
- 4.Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang TC, Sheppard DN. Gating of the CFTR Cl- channel by ATP-driven nucleotide-binding domain dimerisation. J Physiol. 2009;587:2151–2161. doi: 10.1113/jphysiol.2009.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riordan JR. CFTR function and prospects for therapy. Annu Rev Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- 7.Bobadilla JL, Macek M, Jr, Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations—correlation with incidence data and application to screening. Hum Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 8.Cystic Fibrosis Foundation Patient Registry 2008 Annual Report. Bethesda, MD: Cystic Fibrosis Foundation; 2008. [Google Scholar]
- 9.Zemanick ET, Harris JK, Conway S, Konstan MW, Marshall B, Quittner AL, Retsch-Bogart G, Saiman L, Accurso FJ. Measuring and improving respiratory outcomes in cystic fibrosis lung disease: opportunities and challenges to therapy. J Cyst Fibros. 2010;9:1–16. doi: 10.1016/j.jcf.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkby S, Novak K, McCoy K. Update on antibiotics for infection control in cystic fibrosis. Expert Rev Anti Infect Ther. 2009;7:967–980. doi: 10.1586/eri.09.82. [DOI] [PubMed] [Google Scholar]
- 11.Cleveland RH, Zurakowski D, Slattery D, Colin AA. Cystic fibrosis genotype and assessing rates of decline in pulmonary status. Radiology. 2009;253:813–821. doi: 10.1148/radiol.2533090418. [DOI] [PubMed] [Google Scholar]
- 12.Amaral MD, Kunzelmann K. Molecular targeting of CFTR as a therapeutic approach to cystic fibrosis. Trends Pharmacol Sci. 2007;28:334–341. doi: 10.1016/j.tips.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Ashlock MA, Beall RJ, Hamblett NM, Konstan MW, Penland CM, Ramsey BW, Van Dalfsen JM, Wetmore DR, Campbell PW., 3rd A pipeline of therapies for cystic fibrosis. Semin Respir Crit Care Med. 2009;30:611–626. doi: 10.1055/s-0029-1238919. [DOI] [PubMed] [Google Scholar]
- 14.Frerichs C, Smyth A. Treatment strategies for cystic fibrosis: what's in the pipeline? Expert Opin Pharmacother. 2009;10:1191–1202. doi: 10.1517/14656560902928896. [DOI] [PubMed] [Google Scholar]
- 15.Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, et al. Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 16.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atwell S, Brouillette CG, Conners K, Emtage S, Gheyi T, Guggino WB, Hendle J, Hunt JF, Lewis HA, Lu F, et al. Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel. 2010 doi: 10.1093/protein/gzq004. [DOI] [PubMed] [Google Scholar]
- 18.Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, Lu F, Sauder JM, Molnar KS, Coales SJ, et al. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2010;396:406–430. doi: 10.1016/j.jmb.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 19.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins CF. ABC transporters: from microorganisms to man. Ann Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 21.Linton KJ, Higgins CF. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 22.Dassa E, Bouige P. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 23.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mense M, Vergani P, White DM, Altberg G, Nairn AC, Gadsby DC. In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J. 2006;25:4728–4739. doi: 10.1038/sj.emboj.7601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 27.Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 28.Sharma M, Benharouga M, Hu W, Lukacs GL. Conformational and temperature-sensitive stability defects of the delta F508 cystic fibrosis transmembrane conductance regulator in post- endoplasmic reticulum compartments. J Biol Chem. 2001;276:8942–8950. doi: 10.1074/jbc.M009172200. [DOI] [PubMed] [Google Scholar]
- 29.Lukacs GL, Chang XB, Kartner N, Rotstein OD, Riordan JR, Grinstein S. The cystic fibrosis transmembrane regulator is present and functional in endosomes. Role as a determinant of endosomal pH. J Biol Chem. 1992;267:14568–14572. [PubMed] [Google Scholar]
- 30.Cholon DM, O'Neal WK, Randell SH, Riordan JR, Gentzsch M. Modulation of endocytic trafficking and apical stability of CFTR in primary human airway epithelial cultures. Am J Physiol Lung Cell Mol Physiol. 2010;298:L304–314. doi: 10.1152/ajplung.00016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu BH, Thomas PJ. Alteration of the cystic fibrosis transmembrane conductance regulator folding pathway. J Biol Chem. 1996;271:7261–7264. doi: 10.1074/jbc.271.13.7261. [DOI] [PubMed] [Google Scholar]
- 32.Pissarra LS, Farinha CM, Xu Z, Schmidt A, Thibodeau PH, Cai Z, Thomas PJ, Sheppard DN, Amaral MD. Solubilizing mutations used to crystallize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem Biol. 2008;15:62–69. doi: 10.1016/j.chembiol.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 33.Brouillette C, Protasevich I, Yang Z, Atwell S, Zhao X, Emtage S. (conference abstract) Pediatric Pulmonology. 2008;43:227. [Google Scholar]
- 34.Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, Wetmore D, Hunt J, Brouillette C. (conference abstract) Pediatric Pulmonology. 2009;44:227. [Google Scholar]
- 35.Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci U S A. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Protasevich I, Yang Z, Wang C, Zhao X, Atwell S, Emtage JS, Wetmore DR, Hunt JF, Brouillette CG. Thermal unfolding studies show the disease causing F508 deletion mutation in cystic fibrosis transmembrane conductance regulator (CFTR) thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010;19:1917–1931. doi: 10.1002/pro.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeCarvalho AC, Gansheroff LJ, Teem JL. Mutations in the nucleotide binding domain 1 signature motif region rescue processing and functional defects of cystic fibrosis transmembrane conductance regulator delta f508. J Biol Chem. 2002;277:35896–35905. doi: 10.1074/jbc.M205644200. [DOI] [PubMed] [Google Scholar]
- 38.He L, Aleksandrov LA, Cui L, Jensen TJ, Nesbitt KL, Riordan JR. Restoration of domain folding and interdomain assembly by second-site suppressors of the {Delta}F508 mutation in CFTR. FASEB J. 2010 doi: 10.1096/fj.09-141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Loo TW, Bartlett MC, Clarke DM. Correctors promote maturation of cystic fibrosis transmembrane conductance regulator (CFTR)-processing mutants by binding to the protein. J Biol Chem. 2007;282:33247–33251. doi: 10.1074/jbc.C700175200. [DOI] [PubMed] [Google Scholar]
- 40.Loo TW, Bartlett MC, Clarke DM. The V510D Suppressor Mutation Stabilizes DeltaF508-CFTR at the Cell Surface. Biochemistry. 2010 doi: 10.1021/bi100807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aleksandrov AA, Kota P, Aleksandrov LA, He L, Jensen T, Cui L, Gentzsch M, Dokholyan NV, Riordan JR. Regulatory Insertion Removal Restores Maturation, Stability and Function of DeltaF508 CFTR. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Csanady L, Chan KW, Nairn AC, Gadsby DC. Functional roles of nonconserved structural segments in CFTR's NH2-terminal nucleotide binding domain. J Gen Physiol. 2005;125:43–55. doi: 10.1085/jgp.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi Y, Matsui H, Takagi T. Membrane protein molecular weight determined by low-angle laser light-scattering photometry coupled with high-performance gel chromatography. Methods Enzymol. 1989;172:514–528. doi: 10.1016/s0076-6879(89)72031-0. [DOI] [PubMed] [Google Scholar]
- 45.Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure. 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 46.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 47.Richardson JM, Thibodeau PH, Watson J, Thomas PJ. Identificatin of a non-native state of NBD1 that is affected by F508del (conference abstract) Pediatric Pulmonology 42:203. 2007 [Google Scholar]
- 48.Brown CR, Hong-Brown LQ, Biwersi J, Verkman AS, Welch WJ. Chemical chaperones correct the mutant phenotype of the delta F508 cystic fibrosis transmembrane conductance regulator protein. Cell Stress Chaperones. 1996;1:117–125. doi: 10.1379/1466-1268(1996)001<0117:ccctmp>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato S, Ward CL, Krouse ME, Wine JJ, Kopito RR. Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J Biol Chem. 1996;271:635–638. doi: 10.1074/jbc.271.2.635. [DOI] [PubMed] [Google Scholar]
- 50.Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Kelly SM, Price NC. The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci. 2000;1:349–384. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- 52.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 53.Diederichs K, Diez J, Greller G, Muller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon thermococcus litoralis. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serohijos AW, Hegedus T, Riordan JR, Dokholyan NV. Diminished self-chaperoning activity of the DeltaF508 mutant of CFTR results in protein misfolding. PLoS Comput Biol. 2008;4:e1000008. doi: 10.1371/journal.pcbi.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 56.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]