Summary
HIV-1 assembly depends on its structural protein, Gag, which after synthesis on ribosomes, traffics to the late endosome/plasma membrane, associates with HIV Env glycoprotein, and forms infectious virions. While Env and Gag migrate to lipid microdomains, their stoichiometry and specificity of interaction are unknown. Pseudotyped viral particles can be made with one viral core surrounded by heterologous envelope proteins. Taking advantage of this property, we analyzed the association of HIV Env and Ebola glycoprotein (GP), with HIV-1 Gag coexpressed in the same cell. Though both viral glycoproteins were expressed, each associated independently with Gag, giving rise to distinct virion populations, each with a single glycoprotein type. Confocal imaging demonstrated that Env and GP localized to distinct lipid raft microdomains within the same cell where they associated with different virions. Thus, a single Gag particle associates “quantally” with one lipid raft, containing homogeneous trimeric viral envelope proteins, to assemble functional virions.
Keywords: MICROBIO, PROTEINS
Introduction
The assembly of HIV-1 is dependent on its major structural protein, Gag. The Gag precursor protein, Pr55 Gag, is synthesized on free cytosolic ribosomes and traffics to the late endosome/plasma membrane where it associates with the viral Env glycoprotein (Freed, 1998). While Gag has been detected in raft-like membrane microdomains (Lindwasser and Resh, 2001, Ono and Freed, 2001) and the matrix (MA) domain of HIV-1 Gag binds specifically to a plasma membrane-associated phospholipid, PI(4,5)P2 (Ono et al., 2004), the process by which Gag particles associate specifically with viral glycoproteins is not understood.
In HIV-infected T cells, newly synthesized Env transits from the ER to the Golgi where the newly added N-linked polysaccharides are modified. The subsequent trafficking of Env and its targeting to Gag is not completely understood. While a portion has been suggested to transit to CTLA-4-containing secretory granules (Miranda et al., 2002), 5%–15% of the newly synthesized gp160 traffics directly to the cell surface (Willey et al., 1988). Recent studies have shown that HIV-1 and Ebola virus, two unrelated viruses, both employ Tsg101, an endosomal protein sorting factor, for efficient budding (Martin-Serrano et al., 2001). Furthermore, budding of both HIV-1 and Ebola virus occurs through lipid rafts of the host cell plasma membrane (Bavari et al., 2002, Rousso et al., 2000). The Ebola glycoprotein (GP) is a typical trimeric viral spike protein with coiled coils and a fusion domain that mediates pH-dependent virus entry into host cells.
It is generally known that pseudotyped viral particles can be made with the core of one virus surrounded by a heterologous envelope protein. Lentiviral vector pseudotyping has been achieved using viral producer cells transfected with three expression plasmids: a lentiviral packaging construct encoding HIV Gag, an envelope-encoding plasmid, and a reporter vector expressing luciferase (Naldini et al., 1996). The use of this system in transiently transfected 293T cells allowed production of pseudotyped lentiviral vectors that could be used to study intracellular trafficking of HIV Gag and alternative envelope proteins, specifically HIV Env and Ebola GP. Here, we used this system to study the assortment of two distinct viral glycoproteins, synthesized from a single expression vector within the same cell, into viral particles. This analysis was performed by biochemical analysis and immunoprecipitation of virus preparations to determine the surface expression of each glycoprotein and by confocal imaging within virus producer cells and in HIV-1-infected primary T cells. Gaining a knowledge of this process advances a basic understanding of viral protein trafficking and HIV virion formation at the same time that it lends insight into potential targets of antiviral therapy and vaccine design.
Results
Physical and Biochemical Characterization of Pseudotyped Lentivirus Vectors
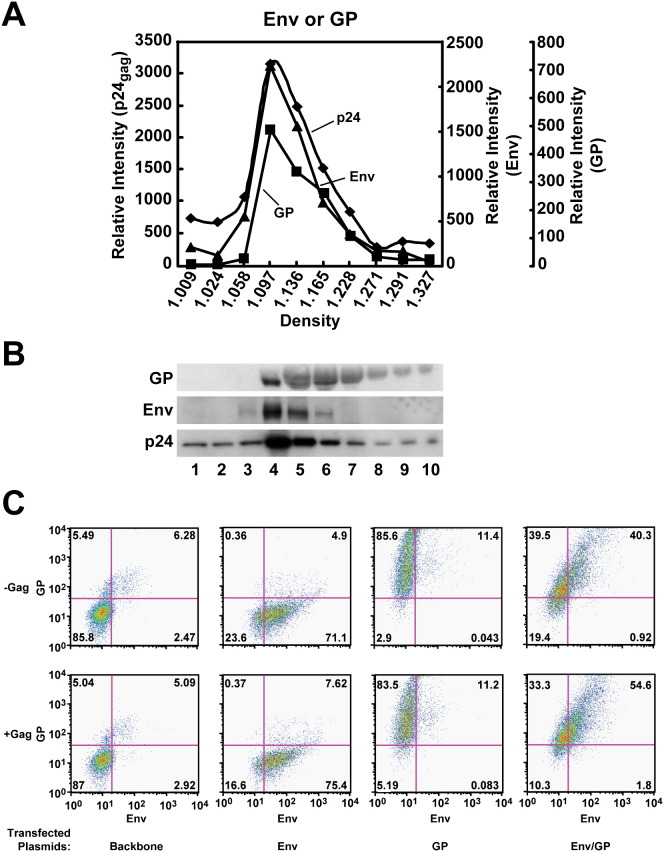
To examine the specificity and mechanism of HIV Gag and Env protein association, plasmid expression vectors encoding replication-defective lentiviral vectors were prepared that could be pseudotyped with both HIV-1 Env and Ebola GP viral glycoproteins. To ensure that both viral glycoproteins were synthesized in the same cell, a single eukaryotic expression vector was prepared that encoded Env and GP under independent promoters, and this plasmid was cotransfected along with Gag and a luciferase reporter gene linked to a packaging sequence that was used to quantitate infectivity. Expression of the envelope proteins was first confirmed biochemically: each viral glycoprotein was detected by western blotting in the same fractions after buoyant density sedimentation analysis ( Figures 1A and 1B), with Env, GP, and Gag expression observed within the range of densities expected for functional lentiviral particles (Yang et al., 2004). Cell surface expression of both Env and GP, the viral spikes, in the same cell was confirmed by flow cytometry (Figure 1C; right upper panel). Their expression was comparable to singly transfected viral spikes and was not significantly altered when expressed with Gag (Figure 1C; fourth versus second and third columns, and upper versus lower panels).
Figure 1.
Analysis of Buoyant Density Gradient Fractionated Env/GP Pseudotyped Lentiviral Vectors
293T cells were transfected with the packaging vector pCMVΔR8.2, pHR′-CMV-Luciferase, as well as the Env + GP expression vector.
(A) Virus supernatant was harvested 60 hr later for buoyant density gradient analysis.
(B) Rabbit anti-GP, anti-gp120, and anti-Gag were used for subsequent western blot detection of GP, Env, and Gag, respectively, in each fraction.
(C) Comparable expression of GP and Env was confirmed on viral producer cells transfected singly (GP or Env) or doubly (GP/Env) with the indicated spikes.
Functional and Immunological Analysis of Pseudotyped Lentivirus Vectors
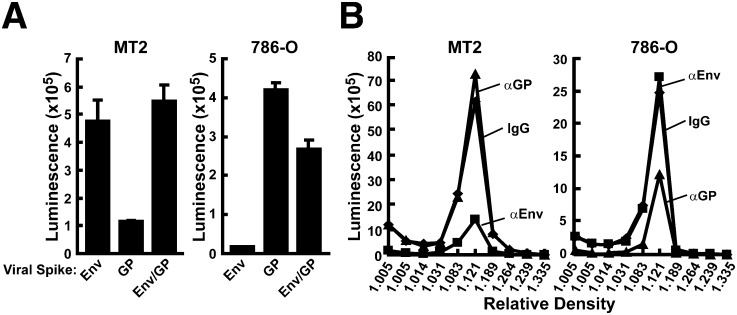
The functional activity of the pseudotyped lentiviral vector preparation was demonstrated by its ability to transduce both an HIV-1 target cell, the MT-2 T leukemia, as well as a cell infectable by Ebola GP, the 786-O cell ( Figure 2A). The specificity of each singly pseudotyped lentiviral vector was confirmed by incubation of buoyant density-purified virus with specific neutralizing antibodies known to interact with native, functional viral spikes. The 2F5 (Muster et al., 1993) and 2G12 (Trkola et al., 1996) monoclonal antibodies inhibited HIV lentiviral vector transduction while KZ52 (Maruyama et al., 1999) specifically reduced Ebola vector expression from viruses made in the doubly transfected (Env/GP) producer cells (Figure 2B, left, αEnv and right, αGP); however, neither inhibited entry of the pseudotyped vectors into the heterologous target cell (Figure 2B, left, αGP and right, αEnv), confirming their specificity.
Figure 2.
Specificity of Lentiviral Vectors Determined by Inhibition of Gene Transfer with Neutralizing Antibodies and Biochemical Evidence of Segregation of Viral Envelopes
(A) Pseudotyped lentiviral vectors produced from single (Env or GP) or doubly (GP/Env) transfected 293T cells infect corresponding HIV (MT2) and Ebola (786-O) target cells. Error bars indicate standard error of the mean of at least three independent transductions.
(B) Antibody neutralization of pseudotyped lentiviral vectors produced from doubly transfected 293T cells and fractionated in buoyant density gradient. For antibody neutralization, viruses were incubated at 37°C for 1 hr with 2F5 and 2G12 (5 μg/ml each), or for GP with KZ52 (20 μg/ml) before infecting the target cells.
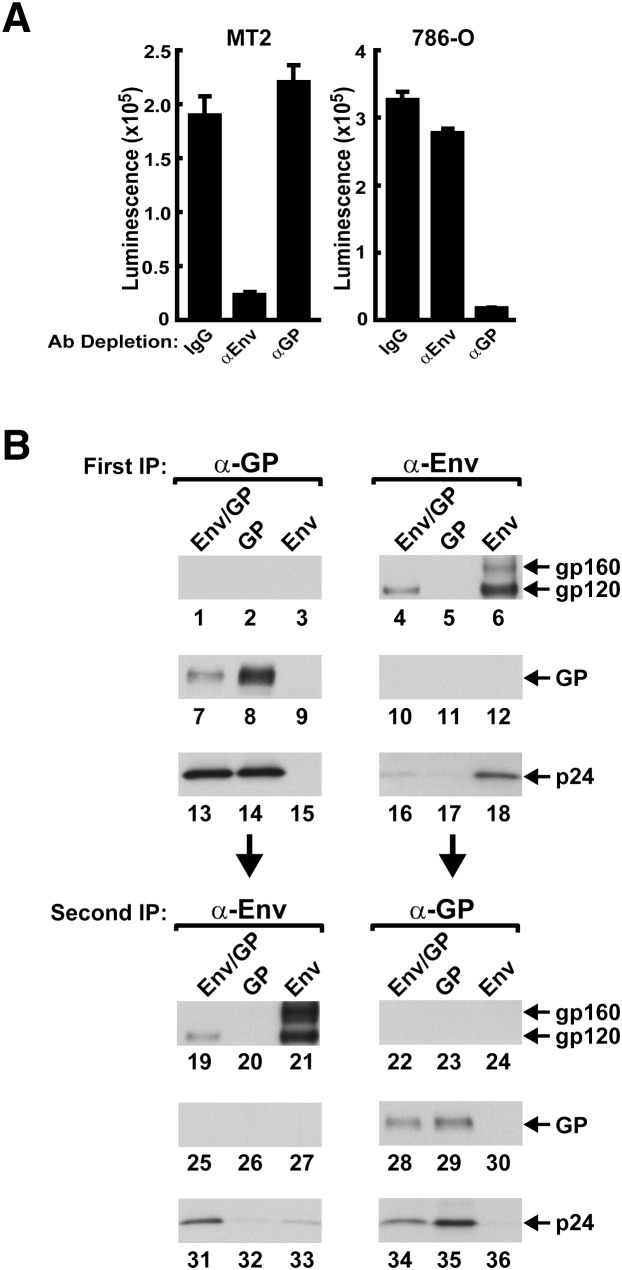
To determine whether entry in a population of virions pseudotyped with both viral glycoproteins in the same producer cell was mediated by single or mixed spike viruses, immunodepletion studies were performed. Neutralizing antibodies were used for immunodepletion and immunoprecipitation studies because they are the only ones known to react with native spike on virions. Unexpectedly, incubation followed by precipitation with Env- or GP-specific antibodies eliminated transduction of the relevant, but not the heterologous, target cell population compared to control IgG using comparable amounts of infecting vectors ( Figure 3A, left, αEnv versus αGP or IgG and right, αGP versus αEnv or IgG), implying that the viral envelopes segregated to distinct viral particles despite the fact that both were expressed in the same cells using the double expression vector. Immunodepletion studies were also performed on virions produced from singly (Env or GP) transfected 293T cells and mixed in different ratios. Similar patterns of infectivity and reduction after immunodepletion were observed with these mixed samples on the relevant target cells (Figure S1 available online). Similar to vectors from doubly transfected cells, a small amount of residual activity remained due to incomplete depletion. This analysis confirmed the specificity of the depletion and comparable behavior of mixed singly pseudotyped vectors.
Figure 3.
Evidence of Segregation in Virions Made from Producer Cells Expressing Two Alternative Viral Spikes by Immunodepletion and Biochemical Analyses
(A) Immunodepletion of Optiprep band-purified virions prepared from double expression plasmid (Env/GP) with anti-Env (2F5 and 2G12) or anti-GP (KZ52) as indicated. Error bars indicate standard error of the mean of at least three independent transductions.
(B) Primary (lanes 1–18) and secondary immunoprecipitation (lanes 19–36) of purified virions with heterologous antibody, anti-GP or anti-Env, respectively, followed by western blotting for the indicated gene products (right), providing biochemical evidence for the absence of chimeric virus. For immunodepletion, virus supernatants were incubated with 2F5 and 2G12 (5 μg/ml each) or for GP with KZ52 (20 μg/ml) at 4°C for 60 min before addition of biomagnetic particle-conjugated anti-human IgG for another 3 hr. Immune complexes were held in place with a strong magnet during the removal of the virus supernatant. Human IgG served as negative control. Immunoprecipitation of Env containing pseudotyped lentiviral vector was performed by incubating with human HIV Igb12 (1 μg/ml) and GP containing pseudotyped lentiviral vector with 13C6 (1 μg/ml) at 4°C for 60 min before adding protein A/G for another 3 hr. Immune complexes were separated from the virus supernatant by centrifugation at 4000 × g for 3 min. Between primary and secondary immunoprecipitation, the virus supernatants were precleared by incubating with protein G agarose for 3 hr before removal of the agarose beads. mAb 149, rabbit anti-gp120, and anti-Gag (Advanced Biotechnologies, Columbia, MD) were used for subsequent western blot detection of GP, Env, and Gag, respectively, in each immune complex.
The presence of the relevant viral glycoproteins on separate virions was demonstrated by biochemical analysis of lentiviral vectors made in cells expressing both Env and GP. Band-purified virus was analyzed by immunoprecipitation using specific antibodies against each protein followed by western blotting. No Env was detected on purified virions derived from GP/Env producer cells after immunoprecipitation with 13C6, a specific mouse antibody against GP, while it was readily detected after anti-Env precipitation (Figure 3B, lane 1 versus 4). Conversely, no GP was detected on purified virions from GP/Env producer cells after immunoprecipitation with Env antibody, while it was readily detected after anti-GP precipitation (Figure 3B, lane 10 versus 7). Viruses immunoprecipitated with Env antibodies showed western blot reactivity with Env antibody in the Env or doubly-transfected producer cells (Figure 3B, lanes 4 and 6 versus 5) and those from GP- and doubly-transfected producer cells reacted with anti-GP, but not anti-Env (Figure 3B, lanes 7, 8 versus 9), though the signals obtained from virions derived from singly expressing producer cells were stronger than those doubly expressing ones. When a secondary immunoprecipitation was performed with the heterologous spike antibody on the first immunodepleted supernatant, Env was detected in GP-depleted preparations of double expressors (Figure 3B; lane 19), and GP was found in Env-depleted supernatants (Figure 3B, lanes 28). These biochemical data document the segregation of the two viral glycoproteins to distinct virion populations, thus complementing and confirming the functional immune precipitation studies.
Segregated Localization of Envelope Proteins in MAGI-CCR5 Cells
The ability of different viral spikes to localize specifically with different virions was further analyzed by confocal imaging microscopy. Since the cell line 293T utilized as pseuodtyped virus producer was not readily adherent and therefore less optimal for confocal imaging, MAGI-CCR5 cells, a HeLa cell clone expressing human CD4 and CCR5, were analyzed by this technique. These cells exhibited Env and GP expression and distribution similar to that of the 293T cells (Figure S2) and are productively infected by HIV. They were, therefore, also amenable to analysis after infection with replication-competent virus.
Previous studies have shown that closely related HIV Env glycoproteins can form mixed heterotrimers that mediate functional viral entry (Yang et al., 2005b). To compare and contrast the behavior of such homologous, mixed heterotrimers to different viral glycoproteins that cannot form heterotrimers, two HIV Env expression vectors were tagged with distinct epitopes, HA and myc, in the V1 region and analyzed for their ability to colocalize in cotransfected cells. Three-dimensional visualization of transfected MAGI-CCR5 cells revealed colocalization of HA-Env and myc-Env ( Figure 4A, left panels; Pearson's correlation = 0.493), as would be expected from previous studies (Yang et al., 2005b). While this correlation is highly significant, the association of HA- and myc-Env trimers is not complete because of the statistical nature of trimer formation, where homotrimerization would be anticipated in approximately one-third of spikes. Thus, the degree of colocalization seen by confocal microscopy is within the expected range. In contrast, when cells were cotransfected with Env and Ebola GP, these two envelope proteins showed no colocalization and segregated to distinct sites within the same cell (Figure 4A, right panels; Pearson's correlation = −0.2633). Antibody staining of intact versus permeabilized cells had no effect on this pattern of distribution (data not shown), and cotransfection with Gag similarly did not alter the distribution or lack of colocalization (Figure 4B, upper- versus lower-right panels; Figure S3). Therefore, heterologous viral envelope proteins do not colocalize even when both are synthesized simultaneously in the same producer cells.
Figure 4.
Confocal Microscopy of Transfected MAGI-CCR5 Cells Revealed that GP Did Not Colocalize with Env, and This Pattern Did Not Change in the Presence or Absence of Gag
(A) MAGI-CCR5 cells were transfected with myc- or HA-Env expression vectors and tagged within the V1 region (see Experimental Procedures), which showed colocalization and served as a positive control (left panels). MAGI-CCR5 cells were transiently transfected with GP and HA-Env-expression vectors (right panels). Immunofluorescent staining was performed 40 hr later. Z stack sectioning of the cells was collected and analyzed by Imaris software to obtain 3D reconstruction and colocalization statistics.
(B) MAGI-CCR5 cells were transiently transfected with Env, GP, or double expression plasmid (Env/GP) in the presence or absence of packaging vector pCMVΔR8.2, which contains Gag. Immunofluorescent staining was performed 40 hr later. Z stack sectioning of the cells was collected and analyzed by Imaris software.
Segregated Colocalization of HIV-Gag and Envelope Proteins
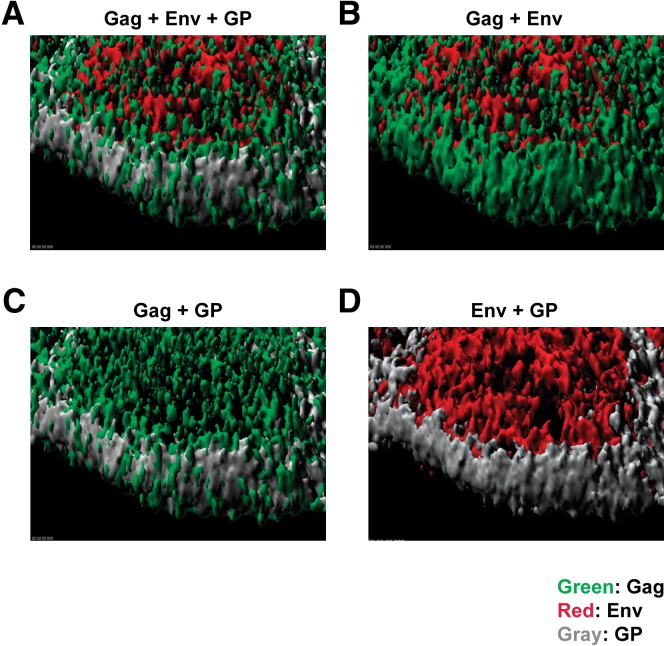
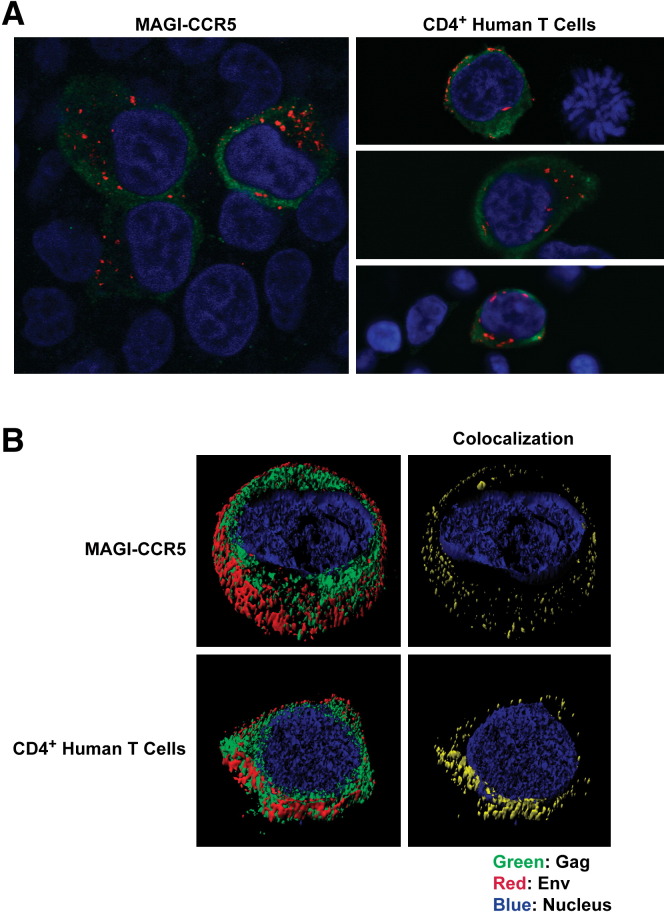
To quantify the interaction of Gag with Env or GP within the same cell, we examined transiently transfected MAGI-CCR5 cells for each component by immunofluorescence and confocal imaging. Z stack sections and the images were processed with Imaris software (version 5.3.3; Bitplane AG, Zurich, Switzerland). Using channel masking of contour surface objects and iso-surface modeling, colocalization of these respective gene products was defined. There was minimal colocalization of Gag and Env throughout the cell ( Figure 5A; Pearson's correlation = 0.13); however, when the analysis was limited to the plasma membrane region where virus assembly occurs by setting specific gating criteria (Figure S4), the correlation coefficient between Gag and Env increased to 0.26 (Figure 5B), suggesting that the association was greater at the site of budding on the plasma membrane. The cytoplasmic distribution of GP was too low to generate a meaningful correlation with Gag, but a positive correlation between GP and Gag was also evident at the plasma membrane (Figure 5C; 0.66). Interestingly, Env and GP showed no evidence of colocalization: the Pearson's correlation was close to zero both in the cytoplasm and on the membrane (Figure 5D), indicating these two viral spike proteins segregated to distinct sites in discrete lipid rafts. Therefore, it appeared that each viral glycoprotein associated independently with distinct Gag ribonucleoprotein complexes to give rise to distinct viral particles. Finally, to confirm the distribution of Gag and Env in HIV-infected cells, imaging was performed on MAGI-CCR5 cells and human CD4+ T cells stimulated with anti-CD3 and anti-CD28. Foci of Env staining similar to the cotransfected viral producer, MAGI-CCR5, were observed in both cell types ( Figure 6A), consistent with their localization to discrete lipid rafts. As in the viral producer cells, Gag was distributed throughout the cell cytoplasm. Colocalization was observed in HIV-infected primary T cells at discrete sites at the membrane similar to the viral producer cells, where 14.2% of Env was found in association with Gag (Figure 6B), also consistent with the findings in viral producer cells.
Figure 5.
Lack of Colocalization of Heterologous Envelope Proteins, Env and GP, in Producer Cells
Confocal microscopy revealed that GP did not colocalize with Env. MAGI-CCR5 cells were transiently transfected with packaging vector pCMVΔR8.2 and GP- and HIV-EnvADA-expressing vectors. Immunofluorescence staining was performed 40 hr later. Z stack sectioning of the cells was collected and analyzed by Imaris software to obtain iso-surface models. (A) Expanded view of a thick section of a single cell is presented here as an iso-surface model depicting Env, GP, and Gag distribution in the vicinity of the plasma membrane. (B)–(D) depict two of the three viral proteins presented in (A) for clear visual comparisons. Descriptions of the method used to identify membrane versus cytoplasmic proteins with Imaris software are described further in the Experimental Procedures and Figure S4.
Figure 6.
Distribution of Env and Gag in HIV-Infected Cells
(A) Confocal imaging confirms the expression of Env in discrete locations intracellularly, largely separate from Gag in MAGI-CCR5 and αCD3/CD28 stimulated human CD4 lymphocytes. MAGI-CCR5 or human CD4 cells were infected with HIV-1 89.6 and analyzed by immunofluorescent staining 48 hr later. Approximately one percent of cells were infected by this criteria.
(B) Z stack sectioning of the cells was collected and analyzed by Imaris software to obtain iso-surface models as in Figure 5. A thick section of a single cell is presented here as iso-surface model for each cell type depicting Env and Gag distribution within the cytoplasm and plasma membrane as well as areas of colocalization.
Discussion
In this study, the ability of HIV-1 Gag to associate with different viral glycoproteins within the same cell was examined. It was expected that a producer cell containing two viral spikes could give rise to virions containing both viral spike glycoproteins ( Figure 7). Though the producer cell was able to generate viral particles that incorporated each viral spike, these glycoproteins segregated to distinct lipid raft-like microdomains and ultimately separated pseudotyped lentiviral vectors (Figure 3, Figure 4, Figure 5). This finding suggests that a single Gag particle associates with one lipid raft that is homogeneous with respect to trimeric viral envelope. This observation was made for both Ebola and HIV glycoproteins, and we have observed it between HIV-1 Env and other viral glycoproteins as well, including HIV-1 Env and SARS, HIV-1 Env and FeLV, and HIV-1 and MuLV (data not shown). Previous studies have demonstrated that Envs derived from closely related lentiviruses, such as HIV-1 variants or HIV-2 and SIV, can form mixed heterotrimers (Boulay et al., 1988, Doms et al., 1990, Yang et al., 2005a, Yang et al., 2005b). Such heterotrimers would be expected to colocate to the same lipid raft as confirmed here (Figure 4A, left panels), but viral spikes from different glycoproteins did not colocalize to the same lipid rafts (Figure 4A, right panel). These observations suggest that this association is dependent on the coiled-coil region of the viral spike, which may also contribute to the formation of trimeric oligomers within the lipid raft. While each raft is homogeneous with respect to the viral spike, other host membrane glycoproteins are not likely excluded from the raft because many such cellular proteins are incorporated into mature virus particles (Arthur et al., 1992).
Figure 7.
Models of Segregation of Viral Spikes with Lentiviral Ribonucleoprotein Complexes
While it was expected that cells expressing two different trimeric viral spikes could give rise to virions containing both proteins, trafficking of such spikes to distinct lipid rafts that associate individually with Gag particles would give rise to virions containing one or the other viral glycoprotein, a model supported by the data in this study.
The assembly of virus particles requires packaging of viral proteins and RNA. For enveloped viruses like HIV and Ebola, assembly and budding occurs at the plasma membrane of infected cells (Cimarelli and Darlix, 2002, Maruyama et al., 1999, Hartlieb and Weissenhorn, 2006) and possibly through endosomal membranes (Martin-Serrano et al., 2001, Ono and Freed, 2004, Dong et al., 2005, Grigorov et al., 2006), though virus located at this latter site could arise by endocytosis of recently released virus (Jouvenet et al., 2006). In HIV-infected T cells, newly synthesized Env transits from the ER to the Golgi, where nascent N-linked polysaccharide side chains are modified, and the protein traffics to the cell surface. Previous studies have shown that HIV-1, and possibly Ebola virus, interacts with Tsg101, an endosomal protein sorting factor, as well as other members of the ESCRT pathway (reviewed in Goff et al., 2003, Martin-Serrano et al., 2001) to facilitate budding of the assembled virus from cells. Lipid rafts have been estimated at ∼100–200 nm in diameter (Pike, 2003), while an HIV virion is ∼125 nm (Briggs et al., 2006). These sizes, along with the finding that virions packaged in cells contain only one of the two expressed viral glycoproteins, are consistent with a model in which a virion arises from a single lipid raft domain that associates with an assembled Gag particle.
The replication-defective pseudotyped lentiviral vectors examined here facilitate the analysis of the molecular mechanisms of this process. It was generally known that pseudotyped viral particles could be made with the core of one virus surrounded by heterologous envelope proteins. But, the segregation of different spikes to distinct viral particles has not been previously appreciated, and this finding was unexpected. Confocal microscopy revealed distinct subcellular distribution of Env, GP, and Gag. Though at the plasma membrane, Env and GP colocalized separately with Gag at the presumed site of budding. Furthermore, Gag expression did not alter the subcellular distribution of envelope proteins by confocal imaging analysis (Figure 4B; Figure S2). A similar distribution of Env or GP was observed intracellularly in cells cotransfected with or without HIV-1 Gag. The association of a single viral spike with each particle suggests that the viral ribonucleoprotein complex associates with each lipid raft. This finding could arise if Gag oligomerization occurred prior to its association with an individual raft. Alternatively, the interaction of Gag with its relevant viral glycoproteins on a lipid raft may recruit additional Gag molecules to complete the formation of a single viral particle.
A recent paper by Brass et al. (Brass et al., 2008) employed small interfering RNA screening to identify more than 250 host proteins implicated in HIV replication. Among them, it is interesting to note that Caveolin-2 (Cav-2) is a lipid raft-associated host protein previously shown to inhibit HIV virion production more than 90% in 293T cells (Llano et al., 2002). This observation raises the possibility that this caveolin-related pathway may be involved in the segregation of viral spikes to distinct particles. Elucidation of such mechanisms will provide further understanding of this process and possible targets for antagonists of HIV replication.
Concurrent viral infections are well documented to occur in nature, and the ability of viral spike proteins to associate with their cognate viral ribonucleoprotein complex is critical to the generation of replication-competent viruses. The inclusion of the appropriate glycoproteins and exclusion of heterologous envelope proteins would, in fact, be crucial for the survival of a virus species. Here, the analysis of two envelopes expressed in a producer cell also served to define the mechanism of viral assembly further. The quantal nature of the spike association with Gag provides fundamental insight into the nature of HIV-1 assembly and suggests that the virus exploits normal cellular substructures and pathways of protein trafficking to allow its disparate components to interact selectively and to complete assembly. Knowledge of this mechanism also facilitates the development of antagonists of HIV-1 that could inhibit productive viral replication and lends insight into strategies for AIDS vaccine design.
Experimental Procedures
Plasmid Construction
Double expression vectors were developed to express both HIV gp16089.6P (Env) and SARS-CoV spike (S) protein (Huang et al., 2004), or Ebola GPZaireΔmuc (GP) (Yang et al., 2000). S or GP was expressed under the control of an mPGK promoter and bovine growth hormone polyadenylation expression cassette, whereas Env was regulated by the RSV promoter and SV40 polyadenylation sequence. Briefly, the mouse PGK promoter (McBurney et al., 1991) was amplified with the sense primer 5′-AAT TTG GCC AGG TAC CGA ATT CTA CCG GGT AGG GGA GGC GCT T-3′ and antisense primer 5′CCT TGA TAT CGG TCG AAA GGC CCG GAG ATG 3′. The MscI/EcoRV fragment of mPGK was then inserted into MscI/EcoRV-digested pVRC1012-gp145ΔCFIΔV12 (Yang et al., 2004), and gp145ΔCFIΔV12 was subsequently replaced by digestion with XbaI and treatment with Klenow, followed by BamHI digestion and insertion with S (isolated from CMV/R-S [Huang et al., 2004] by digestion with SalI followed by Klenow incubation, then BamHI digestion). Alternatively, gp145ΔCFIΔV12 was replaced by digestion with BamHI, treatment with Klenow incubation, and then XbaI digestion and insertion with GP (isolated from pGPΔMUC [Yang et al., 2000]) by incubation with KpnI, treatment with Klenow fragment, followed by XbaI digestion). The expression vector encoding Env was prepared by ligating an XbaI/Klenow/BamHI fragment of Env from plasmid pVRC1012-gp160(89.6P) (Yang et al., 2004) into the HindIII/Klenow/BglII-treated RSV-rabbit-β-globin plasmid (Gorman et al., 1983). The resulting plasmid RSV-Env was treated with NdeI/HpaI to excise the RSV/Env/SV40-polyA expression unit. This expression unit was then cloned into EcoRI/Klenow/NdeI-digested mPGK/S or mPGK/GP to make the double expression vectors. Protein expression was confirmed by western blot analysis.
The plasmids encoding HA or myc inserted into Env were prepared in the RSV-Env plasmid described above. The epitopes were inserted using the PCR-based QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions for tag insertion into the V1 region of Env. The resulting amino acid sequence for HA-Env plasmid at the V1 region was NLTSSYPYDVPDYAGIKNCSF. The resulting amino acid sequence for myc-Env plasmid at the V1 region was NLTSSEQKLISEEDLIKNCSF. The sequences of both plasmids were confirmed by double-strand DNA sequencing.
Antibodies
Anti-HIV type 1 (HIV-1) human monoclonal antibody 2F5, 2G12, and human HIV immunoglobulin G (HIV IgG) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Mouse anti-CD3 (12F6) was provided by Johnson Wong (Massachusetts General Hospital, Boston, MA). Rabbit anti-sGP/GP and anti-GP monoclonal antibody (MAb149) were provided by Anthony Sanchez (CDC). Human anti-GP antibody (KZ52) and human anti-SARS-S monoclonal antibody were provided by Dennis Burton and Antonio Lanzavecchia, respectively. 13C6 monoclonal antibody to Ebola GP was made available through USAMRIID (Frederick, MD). Rabbit anti-HIV-Env and Gag were obtained from Advanced Biotechnologies (Columbia, MD). Rabbit anti-HA tag and mouse anti-myc tag were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-SARS-S and rabbit anti-SARS-S antibody were generated in house and purchased from Imgenex (San Diego, CA), respectively.
Cell Lines
Human embryonic kidney 293T cells and human kidney adenocarcinoma 786-O were purchased from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, California) containing 10% fetal bovine serum (FBS) and 100 μg of penicillin-streptomycin/ml. The human T cell leukemia cell line MT-2 and the HeLa-derived cell line MAGI-CCR5 were obtained from the AIDS Research and Reference Reagent Program.
Western Blot Analysis
Virus supernatant was mixed with 4× sample loading buffer (100 mM Tris, 4% SDS, 20% glycerol, 5% 2-mercaptoethanol, 0.2% bromophenol blue) and boiled for 5 min. The sample was then resolved by 4–15% gradient SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad, Hercules, CA). The membrane was blocked twice with Tris-buffered saline containing 0.05% Triton X-100 (Sigma-Aldrich, St. Louis, MO) at room temperature for 5 min, followed by incubation with the appropriate antibody (human HIV Ig, 1:5000; KZ52, 1:10000; rabbit anti-Gag, 1:4000) in blocking buffer with 2% skim milk for 1.5 hr at room temperature. The membrane was washed twice with 50 ml of blocking buffer, followed by incubation with secondary antibody conjugated with horseradish peroxidase (Santa Cruz Biotech; 1:5,000) for 30 min at room temperature. After two washes with 50 ml of blocking buffer, the membrane was developed using ECL western blotting detection reagents (GE Healthcare, Piscataway, NJ) and was exposed on Biomax MR film (Kodak, Rochester, NY).
Generation of Replication-Defective Pseudotyped Lentiviral Vectors
Pseudotyped lentiviral vectors with Env and GP were prepared according to published methods (Yang et al., 2004). Briefly, the packaging vector pCMVΔR8.2, pHR′-CMV-Luciferase, and the envelope-expressing vector were transiently cotransfected into 293T cells by use of calcium phosphate reagent (Promega, Madison, WI). The amount of plasmid DNA used for making different pseudotyped vectors was as follows: for lentiviral vectors, 7 μg of pCMVΔR8.2 plus 7 μg of pHR′-CMV-Luc and 10 μg of VR1012/Ebola-GP/ HIV-Env89.6P. Cells were transfected overnight, and replenished with fresh medium. Supernatants were harvested 60 hr after transfection, filtered, and stored at −80°C. The same amount of virus, as determined by p24 ELISA assay (Beckman Coulter, Fullerton, CA), was added onto MT-2 (HIV target) and 786-O (Ebola-GP target) cells. The cells were harvested 48 hr after onset of incubation and lysed in cell culture lysis buffer (Promega). The luciferase assay was performed according to the manufacturer's recommendations (Promega).
Buoyant Density Gradient Analysis of Pseudotyped Lentiviral Vectors
Pseudotyped lentiviral vectors were prepared as mentioned in 293T cells. Sixty hours after transfection, the supernatants were harvested and filtered through a 0.45-μm syringe filter. The recombinant virus was first concentrated by layering a 10 ml sample in tissue culture media onto 2.5 ml of Optiprep (Iodixanol, 60% solution) (Accurate Biochemicals, Westbury, NY) and centrifuged at 53,000 × g for 1 hr in a Sorvall TH-641 rotor (Thermo Fisher Scientific, Waltham, MA). The top 7.5 ml of supernatant was removed, and the remaining solution was mixed uniformly to achieve a final concentration of 30% Optiprep in a 5 ml final volume. The gradient was formed by centrifugation at 363,000 × g for 3.5 hr with a Beckman NVT-100 rotor.
HIV Infection of MAGI-CCR5 and Activated Human CD4+ T Cells
Human peripheral blood CD4+ T cells were isolated from elutriated lymphocytes from a healthy adult donor at the Transfusion Medicine Department of Warren Grant Magnuson Clinical Center, National Institutes of Health, Bethesda, MD. CD4+ T cells were isolated from the lymphocyte fraction by depletion of non-CD4+ T cells with a CD4+ T cell isolation kit (Miltenyi Biotech, Auburn, CA). The enriched CD4+ T cells were stimulated with 12F6 (anti-CD3) and anti-CD28 (BD PharMingen) at 2 ng/106 cells/ml each in the RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. After 48 hr, replication competent HIV-1 89.6 at 100 ng/ml of p24 was added to the culture. MAGI-CCR5 cells were plated on the chamber slide (Lab-Tek) and HIV-1 89.6 at 50 ng/ml was added to the culture. Forty-eight hours later, the cells were stained for confocal microscopy.
Confocal Microscopy
After transfection or infection, cells were washed with PBS and then fixed and permeabilized for 30 min using Cytofix/Cytoperm solution (BD Biosciences). The cells were then stained with specific first antibody against the envelope and gag proteins (2G12 at 0.5 μg/ml, 13C6 at 0.1 μg/ml, rabbit-anti-HA at 1:100, and mouse-anti-myc at 1:500 and rabbit-anti-Gag at 1:4000) at room temperature. After 1 hr, the cells were washed twice with PBS, followed by incubation with secondary antibody conjugated with Alexa Fluor (Invitrogen) (1 μg/ml) for 30 min at room temperature. The cells were washed twice with PBS and then stained for the nuclei using DAPI in mounting medium (Santa Cruz Biotech). Enriched human CD4 T cells were deposited onto a glass slide using Cytospin (Thermo Fisher) before DAPI staining and mounting. Images of stained cells were obtained by confocal microscopy (Leica SP2, Leica Microsystems, Exton, PA) using a 63× oil immersion objective NA 1.4. All data were processed with Huygens Essential software for deconvolution (Version 3.0, Scientific Volume Imaging BV, Hilversum, The Netherlands). Sequential Z sections of stained cells were also recorded for 3D reconstruction and iso-surface modeling of representative cells with Imaris software (version 5.7, Bitplane AG, Zurich, Switzerland). To investigate colocalization in a cellular region, we used contour and channel masking techniques in the Imaris software package to create precise 3D regions of interest (ROI) for cytoplasm and membrane regions in 3D reconstructed confocal images of the cells (Figure S4). The channel masking technique was then used in conjunction with automatic thresholding to calculate colocalization statistics in the cytoplasm and membrane respectively. The fraction of one channel volume colocalized with another channel volume, and the Pearson channel correlation in colocalized volume was calculated for cytoplasm, plasma membrane, and whole cell using Imaris software colocalization package. Automatic thresholding performed on channels selected for colocalization analysis in Imaris is based on an algorithm developed by Costes and Lockett at NIH (Costes et al., 2002). This algorithm was based on the exclusion of intensity pairs that exhibit no correlation (Pearson's correlation below zero). The same thresholding was applied to each data set.
Acknowledgments
We dedicate this paper to Lakshmanan Ganesh M.D. Ph.D., friend and colleague, whose promising career was cut short tragically by illness.
We thank Ati Tislerics and Brenda Hartman for manuscript preparation; Zhi-yong Yang and Wataru Akahata for comments on the paper; Giulia Fabozzi and Devon Shedlock for helpful discussion and advice; Johnson Wong for providing 12F6; Anthony Sanchez for providing rabbit anti-sGP/GP and MAb149 to Ebola GP; Dennis Burton for providing KZ52; Antonio Lanzavecchia for providing human anti-SARS-S monoclonal antibody; and Mary Kate Hart, Shawn B. Guest, Ana I. Kuehne, Russell R. Bakken, and Michael Clayton for providing 13C6 monoclonal antibody. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID; 2F5, 2G12, HIV IgG, and MAGI-CCR5 cells were provided by Dr. Julie Overbaugh. This research was supported by the Intramural Research Program, Vaccine Research Center, NIAID, NIH.
Published: May 14, 2008
Footnotes
The Supplemental Data include four supplemental figures and two supplemental movies and can be found with this article online at http://www.cellhostandmicrobe.com/cgi/content/full/3/5/285/DC1/.
Supplemental Data
A 3D iso-surface model of two MAGI-CCR5 cells expressing HIV-1 Env (red), Ebola GP (grey), and area of colocalization (yellow). Using Imaris software, the model was generated from sequential Z stack sections of stained cells captured by confocal microscopy.
A 3D iso-surface model of two MAGI-CCR5 cells expressing HIV gp16089.6 with HA tag (red) and myc tag (green). Areas of colocalization of the two envelope spikes are shown in yellow. Using Imaris software, the model was generated from sequential Z stack sections of stained cells captured by confocal microscopy.
References
- Arthur L.O., Bess J.W., Jr., Sowder R.C., Benveniste R.E., Mann D.L., Chermann J.C., Henderson L.E. Cellular proteins bound to immunodeficiency viruses: Implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- Bavari S., Bosio C.M., Wiegand E., Ruthel G., Will A.B., Geisbert T.W., Hevey M., Schmaljohn C., Schmaljohn A., Aman M.J. Lipid raft microdomains: A gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay F., Doms R.W., Webster R.G., Helenius A. Posttranslational oligomerization and cooperative acid activation of mixed influenza hemagglutinin trimers. J. Cell Biol. 1988;106:629–639. doi: 10.1083/jcb.106.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Briggs J.A., Grunewald K., Glass B., Forster F., Krausslich H.G., Fuller S.D. The mechanism of HIV-1 core assembly: Insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Cimarelli A., Darlix J.L. Assembling the human immunodeficiency virus type 1. Cell. Mol. Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes S., Cho E., Catalfamo M., Karpova T., McNally J., Henkart P., Lockett S. Automatic 3D detection and quantification of co-localization. Microsc. Microanal. 2002;8:1040–1041. [Google Scholar]
- Doms R.W., Earl P.L., Chakrabarti S., Moss B. Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus env proteins possess a functionally conserved assembly domain. J. Virol. 1990;64:3537–3540. doi: 10.1128/jvi.64.7.3537-3540.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Li H., Derdowski A., Ding L., Burnett A., Chen X., Peters T.R., Dermody T.S., Woodruff E., Wang J.J. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120:663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Freed E.O. HIV-1 gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Goff A., Ehrlich L.S., Cohen S.N., Carter C.A. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 2003;77:9173–9182. doi: 10.1128/JVI.77.17.9173-9182.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C., Padmanabhan R., Howard B.H. High efficiency DNA-mediated transformation of primate cells. Science. 1983;221:551–553. doi: 10.1126/science.6306768. [DOI] [PubMed] [Google Scholar]
- Grigorov B., Arcanger F., Roingeard P., Darlix J.L., Muriaux D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 2006;359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Hartlieb B., Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang Z.-Y., Kong W.-P., Nabel G.J. Generation of synthetic Severe Acute Respiratory Syndrome coronavirus pseudoparticles: Implications for assembly and vaccine production. J. Virol. 2004;78:12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Neil S.J., Bess C., Johnson M.C., Virgen C.A., Simon S.M., Bieniasz P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser O.W., Resh M.D. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 2001;75:7913–7924. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M., Kelly T., Vanegas M., Peretz M., Peterson T.E., Simari R.D., Poeschla E.M. Blockade of human immunodeficiency virus type 1 expression by caveolin-1. J. Virol. 2002;76:9152–9164. doi: 10.1128/JVI.76.18.9152-9164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Rodriguez L.L., Jahrling P.B., Sanchez A., Khan A.S., Nichol S.T., Peters C.J., Parren P.W., Burton D.R. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 1999;73:6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney M.W., Sutherland L.C., Adra C.N., Leclair B., Rudnicki M.A., Jardine K. The mouse Pgk-1 gene promoter contains an upstream activator sequence. Nucleic Acids Res. 1991;19:5755–5761. doi: 10.1093/nar/19.20.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda L.R., Schaefer B.C., Kupfer A., Hu Z., Franzusoff A. Cell surface expression of the HIV-1 envelope glycoproteins is directed from intracellular CTLA-4-containing regulated secretory granules. Proc. Natl. Acad. Sci. USA. 2002;99:8031–8036. doi: 10.1073/pnas.122696599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T., Steindl F., Purtscher M., Trkola A., Klima A., Himmler G., Ruker F., Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Ono A., Ablan S.D., Lockett S.J., Nagashima K., Freed E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E.O. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike L.J. Lipid rafts: Bringing order to chaos. J. Lipid Res. 2003;44:655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- Rousso I., Mixon M.B., Chen B.K., Kim P.S. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA. 2000;97:13523–13525. doi: 10.1073/pnas.240459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J.P., Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey R.L., Bonifacino J.S., Potts B.J., Martin M.A., Klausner R.D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kurteva S., Lee S., Sodroski J. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 2005;79:3500–3508. doi: 10.1128/JVI.79.6.3500-3508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kurteva S., Ren X., Lee S., Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-Y., Duckers H.J., Sullivan N.J., Sanchez A., Nabel E.G., Nabel G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- Yang Z.-Y., Chakrabarti B.K., Xu L., Welcher B., Kong W.-P., Leung K., Panet A., Mascola J.R., Nabel G.J. Selective modification of variable loops alters tropism and enhances immunogenicity of Human Immunodeficiency Virus type 1 envelope. J. Virol. 2004;78:4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A 3D iso-surface model of two MAGI-CCR5 cells expressing HIV-1 Env (red), Ebola GP (grey), and area of colocalization (yellow). Using Imaris software, the model was generated from sequential Z stack sections of stained cells captured by confocal microscopy.
A 3D iso-surface model of two MAGI-CCR5 cells expressing HIV gp16089.6 with HA tag (red) and myc tag (green). Areas of colocalization of the two envelope spikes are shown in yellow. Using Imaris software, the model was generated from sequential Z stack sections of stained cells captured by confocal microscopy.