Abstract
DNA rearrangements carried out by site-specific recombinases and transposases (Tpases) show striking similarities despite the wide spectrum of the catalytic mechanisms involved in the reactions. Here, we show that the bacterial insertion sequence (IS)30 element can act similarly to site-specific systems. We have developed an inversion system using IS30 Tpase and a viable λ phage, where the integration/excision system is replaced with IS30. Both models have been proved to operate analogously to their natural counterpart, confirming that a DDE family Tpase is able to fulfill the functions of site-specific recombinases. This work demonstrates that distinction between transposition and site-specific recombination becomes blurred, because both functions can be fulfilled by the same enzyme, and both types of rearrangements can be achieved by the same catalytic mechanisms.
It is now well documented that genetic information can be reshuffled by mobile elements that are usually distinguished as site-specific systems and transposons. Even though the output of their activity can be simplified to insertion, deletion, or inversion of DNA segments, transposition and site-specific recombination are generally regarded as distinct phenomena (1). Classification according to the protein-sequence motifs revealed that some transposases (Tpases) and site-specific recombinases (Recases) with entirely different functions fall within the same family (2-6). This finding supports that mechanistically dissimilar enzymes can perform similar biological functions and suggests that transposition and site-specific recombination may be closer to each other in some respect than was supposed earlier. Further support for this idea may emerge from insertion sequence (IS) elements that form an active junction composed of the inverted repeats (IRs) (7-10). This IR-IR junction shows similarities in its structure and function to the recombination sites of site-specific systems.
The Escherichia coli element, IS30, encoding a Tpase with the well conserved DDE signature (11), belongs to the increasing class of elements that transpose through an intermediate formed by abutted IRs (8). The IR-IR joint bracketing a 2-bp spacer is composed of the 26-bp left and right IR ends (IRL and IRR, respectively) that differ at three positions (12). IR-IR joints occur in both IS dimers and minicircles that can integrate into hot spot sequences or next to other IS30 ends (13-15). Whereas transposition into a hot spot is a conventional way of translocation, which accompanies the resolution of IR-IR junction, targeting an IR end seems more specific for IS30 and generates a new IR-IR joint. The IRs, together with the adjacent IS sequences, are important for the directionality of recombination, because, as always, IRL-IRR junction arises through dimerization or targeting an IR (8, 13, 16). The most frequent reaction has been observed when both donor and target DNA contain an IR-IR junction (13). In this case, the rearrangement is remarkably reversible and resembles site-specific recombination. Since joining the IRs and integrating the active junction next to an another IR seem to be highly site-specific activities, we decided to investigate whether IS30 Tpase is able to act similarly to the known Recases.
Materials and Methods
Development of the Inversion and Deletion Systems and the Recombination Assay. Both pJKI106 and pJKI179 carry the Tn5-derived KmR gene as a 1169-bp BglII-SalI fragment, and the Tn9-derived CmR gene as a 773-bp TaqI fragment, both lacking their own promoters (17). The oppositely oriented genes are delimited by two junction fragments both composed of the 459-bp left and 106-bp right IS30 ends separated by a GC spacer. Promoter activity of the right side junction was reduced by a TTG → GGT change at base pairs 1203-1205 of IS30 applying the Kunkel method with the mutagenic oligonucleotide 5′-TTCAATCTG-TaccTGCAACACCC-3′. The mutation destroys the -35 promoter box in IRR (18). To prevent any expression from outside promoters, the SmRΩ fragment from pHP45Ω (19) carrying transcriptional and translational stop signals was inserted adjacent to the mutated junction.
Both plasmids were introduced into recA- HB101 (20) cells harboring the Tpase producer pJKI133 (15), and the transformants were incubated overnight on solid LB+Tc+Ap medium at 37°C. For negative controls, HB101 lacking pJKI133 was used, and the transformants were selected on LB+Ap. Plasmid DNA was extracted by the alkaline lysis method (21) from single colonies and digested with XhoI, which eliminated the Tpase producer plasmid. Digested DNA was transformed into recA- TG2 (21) cells and the ApR transformants were replica plated on LB+Km and LB+Cm to detect rearrangements. Plasmid DNA extracted from randomly selected ApR colonies was tested by digestion with EcoRI and PvuII (pJKI106 ori and ori
and ori ) or HindIII and PvuII (pJKI179 and pJKI179Δ).
) or HindIII and PvuII (pJKI179 and pJKI179Δ).
Construction of λPC Phages. To construct λ red- ts cI Δ(xis, int, attP)::IS30 CmR phages, the regions flanking the xis, int, attP segment were PCR-amplified and cloned. The 1.7-kb region preceding attP (base pairs 24797-26536) was amplified with the following oligonucleotides: 5′-CCAGTTCGCCGGGCATTCA AC-3′ and 5′-tttgtcgacgcggccgCGCACGTGTGTTAAATGGTTTGC-3′ and cloned as a HindIII-SalI fragment including λ DNA from base pairs 25157-26536. The region upstream of the start codon of int (base pairs 28889-31941) was amplified with the following oligonucleotides: 5′-ataggatccctcgagTCCTCTTCAAAAGGCCACCTG-3′ and 5′-ACTTCCACACCCTGCTTGCTG-3′ and cloned as an EcoRI-BamHI fragment comprising base pairs 28889-31747 of λ DNA [lowercase indicates non-λ-specific sequences carrying restriction sites (italics) used for cloning]. Both fragments were cloned in the ApR vector, pEMBL19 (22), in the correct orientation, and the IS30-CmR cassettes were inserted in between as XhoI-NotI fragments. PC349 carries a single CmR gene; PC365 contains the CmR gene followed by IS30 from 63 to 1221 base pairs joined to the left end (1-169 base pairs) through a GC spacer. This construct comprises the full-length ORF of the Tpase from the first base of the start codon (ORF-A: base pairs 63-1211), whereas in PC395, the ORF begins from a second in-frame ATG codon (ORF-A*: base pairs 180-1211), otherwise PC395 is the same as PC365. Due to the cloning design, the start codon of the ORFs is at the same position as ATG of λ int (base pairs 28882). Consequently, in the segment spanning base pairs 25157-31747 of λ DNA, the region of base pairs 26537-28888, including attP, the whole int and 3′ part of xis (overlapping with int) is replaced with the IS30 and/or CmR gene cassettes.
The constructs were introduced into the λ genome by homologous recombination during lytic propagation of the λ red113 cI857 (a gift from H. Ikeda, Center for Basic Research, Kitasato Institute, Tokyo) in the recA+ TG1 strain (21) harboring one of the PC plasmids. This λ strain shows wild-type lysogenization at 30°C and is inducible at 37°C. The infected TG1 cultures were incubated at 37°C in Tryptone broth (TB) supplemented with 0.4% maltose and 10 mM MgCl2 until complete lysis. Lysates were sterilized with chloroform and used to infect TG2 host at a multiplicity of infection (moi) of ≈ 0.1. Cultures were incubated 24-30 h on solid LB+Cm at 30°C. Colonies were selected according to the following criteria: only (i) small (<1 mm after 30 h incubation) CmR colonies that were (ii) ApS, (iii) unable to grow at 37°C on LB+Cm, (iv) caused either no or a small lysis spot on TG1 indicator bacteria at 30°C, and (v) gave lysis at 37°C. Colonies fulfilling these requirements were considered as potential abortive lysogens carrying double recombinant phage. Phages were recovered as individual plaques grown on indicator bacteria at inducing temperature. Several plaques were tested again for the ability of CmR transduction, lysogen (abortive) formation, and induction. Suitable phage clones were propagated on TG1 strain cultured in TB medium at 37°C until complete lysis, and then phage DNA was purified and their structure was confirmed by extensive restriction and PCR analyses. The structural integrity of the IS30 components was proved also by sequencing. Restriction mapping and sequencing of λPC395 revealed a 5.5-kb deletion (base pairs 20301-25537) lying upstream from the replaced region and not affecting any essential λ genes.
Construction of attBIS30 Sites. For an attachment site (attBIS30) for λPC phages, a junction composed of the truncated left (base pairs 1-169) and right (base pairs 1115-1221) IS30 ends bracketing a GC spacer was inserted into the p15A-based plasmid pAW2015 (23) along with the SmRΩ fragment, resulting in the KmRSmR plasmid, pJKI373. TG2 strain harboring pJKI373 was the recipient host in the first lysogenization assay.
For developing a chromosomal attBIS30 site, the same junction was inserted into the Tn10-based KmR minitransposon located on the ApR-conjugative suicide plasmid, pLOFKm (24). This plasmid was transferred by conjugation into TG2 and KmRApS transconjugants carrying a single insertion of a mini-Tn10 copy, but lacking donor plasmid backbone, were selected by virtue of Southern analysis. Insertion sites of the minitransposon were sequenced in several clones. One of these clones, TG2i4, carrying the insertion at the 314-bp position of yhcL gene (GenBank accession no. AAC76259) was used for chromosomal integration assays.
Lysogenization Assay and Microbial Characterization of Lysogens. TG2 cells harboring pJKI373 or the TG2i4 strain were grown overnight in TB+Km supplemented with 0.4% maltose and 10 mM MgCl2 and infected with λPC395 at an moi of ≈ 0.01-0.05. Cells were spread onto LB+Cm plates and incubated 18-20 h at 30°C (only integrant lysogens form colonies during this time). For screening the potential lysogens, ≈50 normal-sized CmRKmR colonies were individually tested on LB+Cm and on indicator bacteria at 30 and 37°C. The integrant lysogen forms a colony on Cm at 30°C after 20 h incubation (Cm30+), whereas, at 37°C, no colony is formed due to the induction of the lytic cycle of the phage (Cm37-). The ability of phage production was tested on the phage-sensitive indicator strain, TG1. At the noninducing temperature, either no lysis spot, or a small lysis spot, is formed around the lysogen colony transferred to the lawn of indicator bacteria, indicating the correct repression of the phage (φ30-). Conversely, the same colony produces a large lysis spot at 37°C, indicating the production of viable phage particles (φ37+). The Cm30+-, Cm37--, φ30--, and φ37+-stable lysogen colonies were selected for further analyses by a microbial test where the proper clones were streaked for single colonies and at least 20 were individually tested for the original Cm30+, Cm37-, φ30-, and φ37+ phenotype.
Analysis of Integration and Excision of λPC395 in pJKI373. To verify the integrant lysogenic state, total DNA was isolated (15) from the colonies that provided positive signals for the attLIS30 and attRIS30 junctions in colony PCRs. Templates in colony PCRs were single colonies suspended in distilled water, the primers and cycling conditions were described as follows: The DNA was transformed into TG2 cells harboring a helper plasmid that expressed the λ repressor (ts cI857) to stabilize the lysogen status of the transformants. Total DNA was isolated from the KmR, Cm30+, Cm37-, φ30-, and φ37+ transformants grown overnight at 30°C in TB+Km+Cm and amplified with four primer combinations. The primers used were as follows: a-b for attBIS30, c-d for attPIS30, a-d for attLIS30, and c-b for attRIS30 (a, 5′-GAGCTGAATGAAGCCATACCAAACGAC-3′; b, 5′-GATGT TACCCGAGAGCT TGGC-3′; c, 5′-CAGCTCACCGTCTTTCATTGC-3′; and d, 5′-GACATCTAGAATTTACTGTCA-3′).
To examine excision, a single lysogen colony was grown overnight at 30°C in TB+Km+Cm, and the culture was then diluted to ≈108 cells per ml and incubated at 37°C under vigorous shaking. Samples were taken in every 10 min, phage titer was determined, and total DNA was isolated and analyzed by PCR as described above.
Integration and Excision Assays in the Genomic attBIS30 Site. Genomic DNA purified from the potential lysogens (phenotype: Cm30+, Cm37-, φ30-, and φ37+) was amplified with the following primer combinations for detecting junctions: e-f for attBIS30, c-d for attPIS30, e-d for attLIS30, and c-f for attRIS30 (e, 5′-TATCTTGTGCAATGTAACATCAGAGA-3′; and f, 5′-tgaattcATAATATACGGCGAGCGAATGAG-3′. Primers c and d were the same as in previous experiment). The phage induction assay with the TG2i4::λPC395 lysogens was carried out as described above. For detection of attPIS30 and attBIS30, the same primer combinations were used as in the test of lysogens.
Results
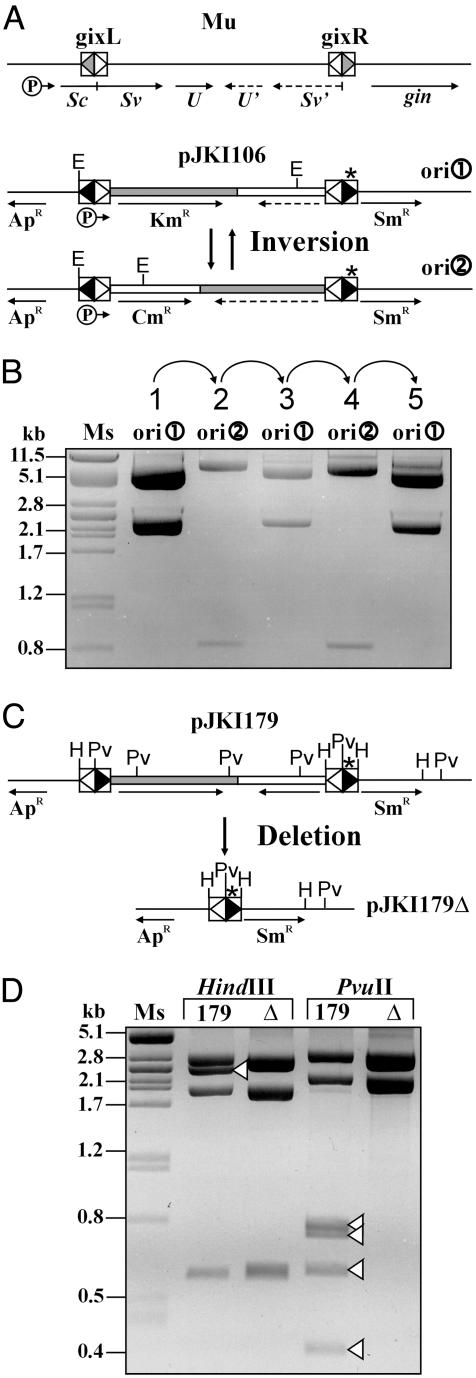
Site-Specific Inversion by IS30 Tpase. We have constructed an inversion model system that is structurally analogous to the invertible G segment of bacteriophage, Mu (Fig. 1A). In the model plasmid, the IR-IR joints having similar symmetry to those of gix sites (25) served as recombinogenic sequences, whereas IS30 Tpase was expressed from a compatible plasmid (see Materials and Methods). The resistance genes bracketed by the inversely oriented IR-IR joints could be expressed from the promoters located in the IRs (18). To avoid coexpression of both marker genes, one of these promoters has been destroyed by point mutations, so the expressed resistance marker refers to the actual orientation of the segment. The original construct providing high level of KmR (ori ) was introduced into recA- E. coli cells harboring the Tpase producer, then plasmid DNA was isolated and transformed again to sort out the rearranged species as individual clones. Among 30 randomly selected transformants, one showed a high level of CmR and turned out to carry an inversion (ori
) was introduced into recA- E. coli cells harboring the Tpase producer, then plasmid DNA was isolated and transformed again to sort out the rearranged species as individual clones. Among 30 randomly selected transformants, one showed a high level of CmR and turned out to carry an inversion (ori ). The inversion plasmid was reintroduced to the Tpase producer strain and analyzed again. In two parallel experiments, 28 of 75 and 19 of 30 colonies, respectively, contained the segment in the original direction (ori
). The inversion plasmid was reintroduced to the Tpase producer strain and analyzed again. In two parallel experiments, 28 of 75 and 19 of 30 colonies, respectively, contained the segment in the original direction (ori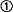 ). The next passage provided 8 of 30 and 2 of 30, whereas the last one yielded 1 of 20 and 4 of 20 inversions (Fig. 1B). Both ori
). The next passage provided 8 of 30 and 2 of 30, whereas the last one yielded 1 of 20 and 4 of 20 inversions (Fig. 1B). Both ori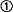 and ori
and ori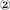 structures failed to undergo inversion (0 of 60) in similar experiments performed without Tpase producer. All constructs described carry a GC spacer in the IR-IR junction, which, together with the first two bases of the IRs, results in a PvuII restriction site. This result provides a simple and informative assay to test whether base changes occurred in the rearranged junctions during recombination (13). Analysis of the junctions by PvuII digestion confirmed that each inversion occurred without any base change in the recombinogenic sites.
structures failed to undergo inversion (0 of 60) in similar experiments performed without Tpase producer. All constructs described carry a GC spacer in the IR-IR junction, which, together with the first two bases of the IRs, results in a PvuII restriction site. This result provides a simple and informative assay to test whether base changes occurred in the rearranged junctions during recombination (13). Analysis of the junctions by PvuII digestion confirmed that each inversion occurred without any base change in the recombinogenic sites.
Fig. 1.
Inversion and deletion occurring between two IS30 IR-IR joints. (A) The invertible segment of pJKI106 and its structural and functional analogy to the G segment of bacteriophage Mu (25). Arrows and dashed arrows refer to expressed and nonexpressed genes, respectively, and “P” marks the promoters. Differently shaded triangles within boxes indicate the symmetric recombinogenic sites. The IS30-based inversion system contains the promoterless KmR (shaded box) and CmR (white box) genes bracketed by two inversely oriented IR-IR junctions that act as recombination sites. The white and black triangles refer to the IRL and IRR of IS30, respectively. *, the inactivated promoter in the right side junction (see Materials and Methods); E, EcoRI. (B) Consecutive inversions. Plasmid DNA exposed to Tpase was digested with EcoRI. Lane 1, original pJKI106; lanes 2-5, plasmid DNA having undergone inversion during consecutive passages. Each isolate derives from that shown in the preceding lane. Ms, λ DNA cleaved with PstI. (C) Deletion occurring between IR-IR joints arranged in direct orientation in pJKI179. H, HindIII; Pv, PvuII. (D) Comparison of pJKI179 and its site-specific deletion derivative, pJKI179Δ. Deleted fragments are indicated.
In addition to inversions (23% of the 235 analyzed events), two types of other reactions were also observed. One of them was the intermolecular recombination through two IR-IR junctions, resulting in dimer replicons (44%). This reaction showed the same conservative site-specific manner as did inversions. The apparently high frequency of dimerization may derive from the replication advantage of dimer replicons (26), which considerably distorts the estimation of the real contribution of dimerization in the rearrangements. The other was the intramolecular transposition leading to deletion formation (20%), where one of the IR-IR junctions targeted a non-IR sequence in the plasmid. In the remaining 13% of cases, alterations could not be detected.
To examine whether outcome of the reaction (i.e., inversion or deletion) depends on the relative direction of IR-IR joints, we constructed a plasmid that differed from the inversion model only in the direct orientation of the junctions (Fig. 1C). This plasmid exposed to Tpase frequently produced site-specific deletion removing the CmR and KmR marker genes along with one of the IR-IR junctions (90-95%), whereas the frequency of deletions was <0.1% in absence of Tpase (Fig. 1D). Other types of rearrangements were not observed and the remaining 5-10% of the analyzed clones were the same as the original structure. This site-specific behavior was strikingly similar to that of resolution systems, where the recombination occurs between two directly repeated resolution sites [e.g., two loxP or dif sites (27, 28)]. Conspicuous absence of inversions in case of directly oriented junctions (pJKI179) and, conversely, the absence of pJKI179Δ-type deletion products in the inversion model clearly showed that the orientation of the recombinogenic sites defined the product of recombination. Further analyses confirmed that the reaction between two IR-IR junctions was conservative, site-specific, and frequent enough to be applied in another recombination system that is closer to a natural situation.
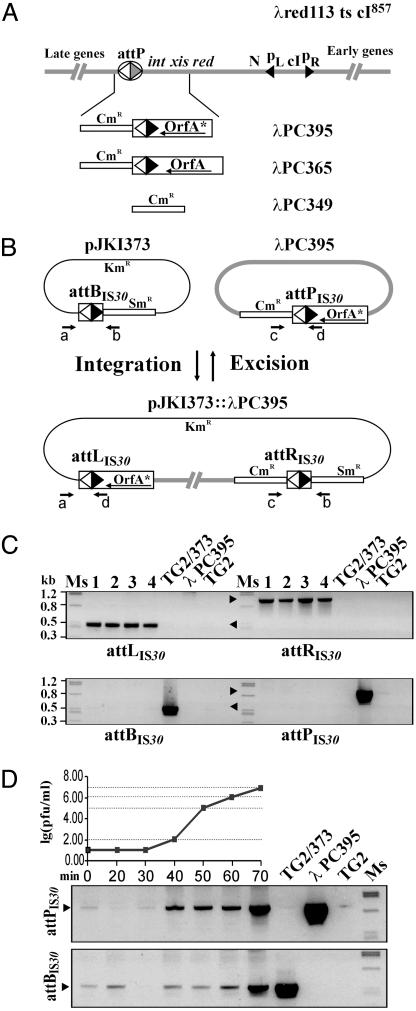
Replacement of the λ Integration/Excision System by Components of IS30. We assessed whether IS30 Tpase, along with the appropriately positioned junctions, was able to substitute for the site-specific recombination system of phage λ. A red- ts cI λ strain was used for replacing its int-attP region with the Tpase gene and an IR-IR junction (attPIS30). Thus, the recombinant phage did not contain any components of the original site-specific system. In addition, Tpase expression was placed under the control of λ cI repression. Three types of phages were used in the integration assays (see Materials and Methods), where an IR-IR junction located on a plasmid served as an attachment site (attBIS30). λPC349 carried a CmR gene alone, λPC365 and λPC395 further contained an IRL-IRR junction and a coding sequence of a full or an N-terminally truncated IS30 Tpase, respectively (Fig. 2A). A recA- E. coli strain harboring pJKI373 (Fig. 2B) was infected with λPC349, λPC365, or λPC395, and the potential integrant lysogens were selected for the CmR phenotype. Neither λPC349 nor λPC365 produced stable CmR lysogens, and integration into pJKI373 was never detected. The latter phage turned out to be quite unstable and produced various CmS deletion derivatives. However, this instability was not surprising as the wild-type Tpase has many known hot spots in λ DNA that may contribute to the decay of the phage (14). Conversely, λPC395, which expressed a shortened Tpase lacking the first 39 amino acids, which is defective in hot spot recognition (Z. Nagy, personal communication) yielded stable CmR lysogens (Fig. 2B). From three parallel infections, four of 40 CmR-stable lysogen colonies showed positive signal in colony PCR for attLIS30 and attRIS30 junctions. The pJKI373::λPC395 fusion products from these clones were transformed into a host expressing the cI repressor to stabilize lysogen transformants. Subsequent analyses of colonies obtained confirmed that attLIS30 and attRIS30 were present, whereas attPIS30 and attBIS30 were missing from the cointegrates (Fig. 2C). Sequencing of junction fragments also confirmed that λPC395 integrated into the attBIS30 site in a conservative and site-specific manner. The dynamics of the reverse process, the excision of the phage, was also investigated (Fig. 2D). During induction of a lysogen, the temporal change of both the phage titer and the relative intensity of attPIS30- and attBIS30-specific PCR fragments indicated the correct excision. Sequencing the PCR fragments also confirmed their identity to the original junctions.
Fig. 2.
Integration and excision of λPC395. (A) A schematic map of λ (35) and the IS30-based systems that substitute its site-specific system. λPC395, λPC365, and λPC349 carry the shortened IS30 ORF-A* (amino acids 40-383), the whole ORF-A, or the CmR gene alone, respectively. (B) Integration and excision of λPC395 in the target plasmid, pJKI373. Thin line, plasmid backbone; thick line, λ DNA; open boxes, resistance genes as indicated; black and white triangles, IR-IR junctions designated as att sites according to their location. Primers used for detecting att junctions are indicated as a-d (see Materials and Methods). (C) PCR detection of integrated λPC395. Samples 1-4 (independent isolates) gave no signal for attBIS30 and attPIS30 (arrowheads), but yielded PCR products indicative of attLIS30 (base pairs 500) and attRIS30 (base pairs 940). TG2/373 and λPC395, positive controls for attBIS30 (base pairs 534) and attPIS30 (base pairs 890); TG2, negative control. (D) Excision of λPC395 during induction of a lysogen strain. The excision was assayed by titration of phages and PCR detection of attBIS30 and attPIS30 every 10 min. Control PCRs for attBIS30 and attPIS30 were as in C.
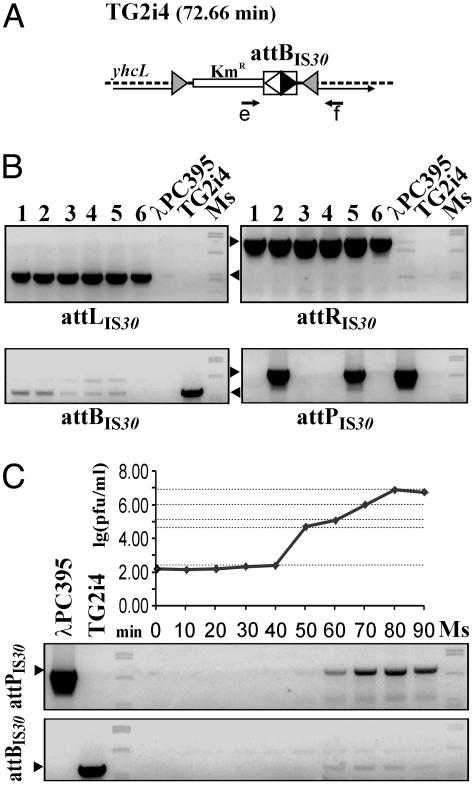
Genomic Integration and Excision of λPC395. Approaching this model system to the natural situation, the attBIS30 site was inserted into the host chromosome, yielding the TG2i4 strain (Fig. 3A), which was used to test the ability of λPC395 to integrate into a single attBIS30 site embedded in the 4.6-Mb E. coli genome. TG2i4 cells were infected with λPC395 and the lysogens were selected for the stable CmR phenotype. PCR analyses confirmed that 6 of 22 colonies selected from three parallel experiments according to phenotype tests (see Materials and Methods) carried the phage integrated into the attBIS30 site (Fig. 3B). The remaining 16 clones were also stable CmR lysogens, where the PCR test could not detect attPIS30 but could detect attBIS30, suggesting that the integration in these cases occurred elsewhere in the genome. The strong lysis and the very low final titer (<104 cfu/ml) produced by these clones under inductive circumstances also supports that the integrant phage could poorly excise from non-attB sites.
Fig. 3.
Integration and excision of λPC395 in the chromosomal attBIS30 site. (A) Location of the integrated attBIS30 in the E. coli genome. The shaded arrowheads bracketing the KmR gene with the IS30 IR-IR junction represent the ends of Tn10 minitransposon (see Materials and Methods). Thin arrow, yhcL gene; e and f, primers used in PCRs. (B) PCR analysis of six integrant lysogens. Lanes 1-6 show the amplification products obtained with the primer combinations of e-f for attBIS30 (base pairs 571), c-d for attPIS30 (base pairs 890; see Fig. 2B), e-d for attLIS30 (base pairs 512), and c-f for attRIS30 (base pairs 943). The weak attBIS30-specific signals suggest that these clones showed spontaneous excision similarly to a wild-type λ lysogen. Positive signals for attPIS30 in lanes 2 and 5 may also be derived from imperfect phage repression. (C) Excision of λPC395 during lytic phase of a lysogen. The excision was assayed by titration of phages and PCR detection of attBIS30 and attPIS30.
The lytic phase of the phage integrated into the attBIS30 site was examined as previously. In the course of phage induction assay, the temporal increase of phage titers from 102 to 107-108 pfu/ml, the gradual increase of the indicative PCR fragments (Fig. 3C), and the following sequence analysis of the restored attBIS30 and attPIS30 sites all supported that the phage correctly excised and propagated. Conversely, in a similar experiment where λPC395 had been integrated into a secondary (non-IR) site, the maximal titer did not exceed 103 pfu/ml during induction. This result clearly showed that effective excision required the recombination between intact attIS30 sites.
Discussion
In this work, we provide evidence that an enzyme belonging to the DDE family of Tpases can act as a site-specific Recase. The model systems described here fulfill the same functions as the well known site-specific inversion, resolution, or phage integration/excision systems. For such functions of IS30 Tpase, only three constraints are required: (i) the joined IRs are the reactive sites (8), (ii) the IRs (even more, the IR-IR junction) are preferred sites for integration (13), and (iii) the left and right ends of the element can be distinguished (8, 13). The last criterion is needed for determining the issue of intramolecular recombination. The relative orientation of the reactive sites prescribes that inversion or deletion will occur (Fig. 1). In the natural systems, the orientation of recombination sites has similar determinative function (29-31), although accessory functions have evolved in many cases to restrict the secondary activities [e.g., FIS enhances inversion by Gin-invertase (32)].
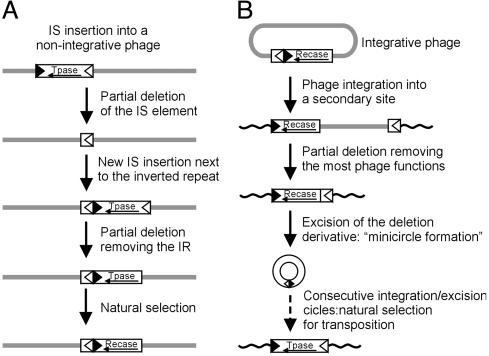
The known site-specific systems show many analogies with those based on IS30 (Table 1), indicating that certain transposons might have served as starting points for the evolution of site-specific systems. IS30 seems not exceptional in this respect. Accumulating data suggest that numerous IS elements may suit the above requirements (7, 9, 10, 33). Mutations such as deletions removing almost an entire element from the dimer, but leaving the IR-IR junction intact or eliminating the domain of Tpase involved in target selection, might easily establish the primitive form of a new site-specific system (Fig. 4A), which can improve further in the course of evolution. It is worth noting that the reverse scenario (i.e., establishment of a new transposable element based on a site-specific system) is also possible (Fig. 4B). Truncated IS30 Tpase carries out secondary reactions similarly to natural systems: Ser-invertases cause deletion or cointegration (30, 31, 34) and λ can integrate into secondary sites (35). More accurate functioning can be achieved by natural selection, as shown in the presented model (Fig. 4). λPC395 could not efficiently excise from secondary sites, therefore, those integrants produced orders of magnitude fewer phage particles during the lytic cycle. Thus, their extinction is more probable in the course of evolution.
Table 1. Comparison of the properties of site-specific, transpositional, and IS30 IR–IR-mediated recombination.
| Characteristics | Site-specific recombination | (IR–IR) × (IR–IR) | Transposition |
|---|---|---|---|
| Mechanism | Conservative | Conservative | Conservative or replicative |
| Frequency of recombination | High | High | Low |
| Specific sequence requirements on | |||
| Donor DNA | Yes (res, dix, att, lox, dif) | Yes (IR–IR) | Yes (IR) |
| Target DNA | Yes (res, dix, att, lox, dif) | Yes (IR–IR) | Variable specificity |
| Target site after recombination | Unaltered | Unaltered | Duplicated or unaltered |
| Host factors | None or IHF, HU, FIS, etc. | None | None or IHF, HU, FIS, etc. |
| Catalytic motif involved* | Active Ser/Tyr, DED/DDD, | DDE | DDE, active Tyr, DED/DDD |
Active Ser, Invertase/resolvase family; active Tyr, integrase family; DED/DDD, Piv/Moov/IS110 family; DDE, transposase/retroviral integrase family
Fig. 4.
Hypothetic possibilities for the evolution of a site-specific and a transposition system from each other. (A) A possible sequence of events potentially leading to the spontaneous development of a phage-encoded site-specific recombination system based on a transposable element. Most of these steps have already been described in different systems [IS insertion into a phage (36), partial deletion of an IS (37), and insertion adjacent to an IR (15)]. This series of events is only one of the numerous conceivable scenarios. Note that the third state is already able to fulfill the site-specific integration and excision by using an IR end as att site (13). (B) The reverse process resulting in a transposon is also possible. Note that the phage integrated into a secondary site is structurally analog to a transposon. Thick line, phage DNA; curved line, DNA of the host cell; black and white arrowheads, IRs; open box, coding sequence of the recombinase.
It is now clear that covalent protein-DNA intermediate formation is not an exclusive criterion of site-specific recombination. The same recombination function can be fulfilled by enzymes with divergent enzymatic mechanisms such as inversion by Ser-Recases, Tyr-Recases, or Piv, which is rather related to Tpases (3). Conversely, the same molecular mechanism can be applied to different functions (Table 2) such as inversion, resolution, integration, excision, or transposition, as is seen in the Int family (5). Here, we demonstrate the ability of a DDE Tpase to catalyze site-specific recombination. This finding may provide support for the theory that evolution of primitive recombinases can lead to the development of enzyme families, which can accomplish both transpositional and site-specific rearrangements independent of their catalytic mechanism.
Table 2. Functional classification of some well known recombinases.
| Functional classification of recombinases
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Family | Catalytic motif | Transposase | Invertase | Resolvase | Viral integrase | Integron integrase | V(D)J recombinase | Ref(s). |
| Integrase | Active Tyr | Tn916, Int, Tn554, Tnp A, B | Fim B, E, Flp | Cre, Xer C, D, Tn4430 TnpI | λ Int, P22 Int, ϕ80 Int | Tn21 Int 11, Tn7 Int 12 | — | 5 |
| Invertase/resolvase | Active Ser | — | Bin, Cin, Gin, Hin, Min, Pin | Tn3 TnpR, γδ TnpR, Tn501 TnpR | — | — | — | 1 |
| Piv/IS110 | DED/DDD | IS 110, IS492 | Piv | — | — | — | — | 3 |
| RAG | DDE?* | RAG-1 | — | — | — | — | RAG-1 RAG-2 | 4, 38, 39 |
| RAG-2 | ||||||||
| DNA Tpase/retroviral integrase | DDE | IS2, Tn10, IS911,*Tn3 TnpA | — | — | HIV-1 Int, RSV Int | — | — | 1 |
| IS30† | DDE | IS30 Tpase | IS30 Tpase | IS30 Tpase | IS30 Tpase | IS30 Tpase‡ | — | This work, 15 |
Spacing between D and D and E amino acids differs from that of classical DDE motif of Tpase/integrase family
IS30 family Tpases are members of the classical DDE Tpase/integrase family. This row represents which types of functions the IS30 Tpase could fulfill in different experimental situations
Integron-like functioning of an IS30-based system is described in ref. 15
Acknowledgments
We thank H. Ikeda for λ red113 cI857; I. Könczöl, E. Keresztúri, and M. Turai for skillful technical assistance; and K. Schlett for critical reading of the manuscript. This work was supported by Ministry of Education Grant NKFP4/006 and Hungarian National Scientific Research Foundation (Országos Tudományos Kutatási Alapprogramok) Grants T019365 and T025729 (to F.O.) and F038204 (to J.K.).
Abbreviations: IS, insertion sequence; IR, inverted repeat: IRL, left IR end; IRR, right IR end; Tpase, transposase; Recase, site-specific recombinase; TB, Tryptone broth.
References
- 1.Craig, N. L., Craigie, R., Gellert, M. & Lambrowitz, A. M., eds. (2002) Mobile DNA II (Am. Soc. Microbiol., Washington, DC).
- 2.Choi, S., Ohta, S. & Ohtsubo, E. (2003) J. Bacteriol. 185, 4891-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tobiason, M., Buchner, J. M., Thiel, W. H., Gernert, K. M. & Karls, A. C. (2001) Mol. Microbiol. 39, 641-651. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger, M. A. (1999) Curr. Opin. Cell Biol. 11, 325-329. [DOI] [PubMed] [Google Scholar]
- 5.Nunes-Düby, S. E., Kwon, R. S., Tirumalai, H. J., Ellenberger, T. & Landy, A. (1998) Nucleic Acids Res. 26, 391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenich, A. G. & Glasgow, A. C. (1994) J. Bacteriol. 176, 4160-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reimmann, C. & Haas, D. (1987) Genetics 115, 619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olasz, F., Stalder, R. & Arber, W. (1993) Mol. Gen. Genet. 239, 177-187. [DOI] [PubMed] [Google Scholar]
- 9.Turlan, C., Ton-Hoang, B. & Chandler, M. (2000) Mol. Microbiol. 35, 1312-1325. [DOI] [PubMed] [Google Scholar]
- 10.Szeverényi, I., Nagy, Z., Farkas, T., Olasz, F. & Kiss, J. (2003) Microbiology 149, 1297-1310. [DOI] [PubMed] [Google Scholar]
- 11.Mahillon, J. & Chandler, M. (1998) Microbiol. Mol. Biol. Rev. 62, 725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalrymple, B., Caspers, P. & Arber, W. (1984) EMBO J. 3, 2145-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olasz, F., Farkas, T., Kiss, J., Arini, A. & Arber, W. (1997) J. Bacteriol. 179, 7551-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olasz, F., Kiss, J., König, P., Buzás, Z., Stalder, R. & Arber, W. (1998) Mol. Microbiol. 28, 691-704. [DOI] [PubMed] [Google Scholar]
- 15.Kiss, J. & Olasz, F. (1999) Mol. Microbiol. 34, 37-52. [DOI] [PubMed] [Google Scholar]
- 16.Stalder, R. & Arber, W. (1989) Gene 76, 187-193. [DOI] [PubMed] [Google Scholar]
- 17.Szeverényi, I., Hodel, A., Arber, W. & Olasz, F. (1996) Gene 174, 103-110. [DOI] [PubMed] [Google Scholar]
- 18.Dalrymple, B. (1987) Mol. Gen. Genet. 207, 413-420. [DOI] [PubMed] [Google Scholar]
- 19.Prentki, P. & Krisch, H. M. (1984) Gene 29, 303-313. [DOI] [PubMed] [Google Scholar]
- 20.Boyer, H. W. & Roulland-Dussoix, D. (1969) J. Mol. Biol. 41, 459-472. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 22.Dente, L., Cesareni, G. & Cortese, R. (1983) Nucleic Acids Res. 11, 1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arini, A., Keller, M. P. & Arber, W. (1997) Biol. Chem. 378, 1421-1431. [DOI] [PubMed] [Google Scholar]
- 24.Herrero, M., Lorenzo, V. & Timmis, K. N. (1990) J. Bacteriol. 172, 6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, R. C. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambrowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 230-271.
- 26.Summers, D. K., Beton, C. W. H. & Withers, H. L. (1993) Mol. Microbiol. 8, 1031-1038. [DOI] [PubMed] [Google Scholar]
- 27.Hoess, R. H. & Abremski, K. (1984) Proc. Natl. Acad. Sci. USA 81, 1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuempel, P. L., Henson, J. M., Dircks, L., Tecklenburg, M. & Lim, D. F. (1991) New Biol. 3, 799-811. [PubMed] [Google Scholar]
- 29.Plasterk, R. H., Ilmer, T. A. & Van de Putte, P. (1983) Virology 127, 24-36. [DOI] [PubMed] [Google Scholar]
- 30.Haffter, P. & Bickle, T. A. (1988) EMBO J. 7, 3991-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandmeier, H., Iida, S., Meyer, J., Hiestand-Nauer, R. & Arber, W. (1990) Proc. Natl. Acad. Sci. USA 87, 1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch, C. & Kahmann, R. (1986) J. Biol. Chem. 261, 15673-15678. [PubMed] [Google Scholar]
- 33.Szeverényi, I., Bodoky, T. & Olasz, F. (1996) Mol. Gen. Genet. 251, 281-289. [DOI] [PubMed] [Google Scholar]
- 34.Iida, S., Huber, H., Hiestand-Nauer, R., Meyer, J., Bickle, T. A. & Arber, W. (1984) Cold Spring Harbor Symp. Quant. Biol. 49, 769-777. [DOI] [PubMed] [Google Scholar]
- 35.Weisberg, R. A. & Landy, A. (1983) in Lambda II, eds. Hendrix, R. W., Roberts, J. W., Stahl, F. W. & Weisberg, R. A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 211-250.
- 36.Caspers, P., Dalrymple, B., Iida, S. & Arber, W. (1984) Mol. Gen. Genet. 196, 68-73. [DOI] [PubMed] [Google Scholar]
- 37.Umeda, M. & Ohtsubo, E. (1990) Mol. Gen. Genet. 222, 317-322. [DOI] [PubMed] [Google Scholar]
- 38.Li, W., Chang, F. C. & Desiderio, S. (2001) Mol. Cell. Biol. 21, 3935-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Gent, D. C., Mizuuchi, K. & Gellert, M. (1996) Science 271, 1592-1594. [DOI] [PubMed] [Google Scholar]