Summary
Background
Clathrin-actin-mediated endocytosis in yeast involves the progressive assembly of at least 60 different proteins at cortical sites. More than half of these proteins are involved in the assembly of a branched network of actin filaments to provide the forces required for plasma membrane invagination.
Results
To gain insights into the regulation of endocytic actin patch dynamics, we developed an in vitro actin assembly assay using microbeads functionalized with the nucleation promoting factor (NPF) Las17 (yeast WASP). When incubated in a yeast extract, these beads assembled actin networks and a significant fraction became motile. Multi dimensional Protein Identification Technology (MudPIT) showed that the recruitment of actin binding proteins to these Las17-derived actin networks is selective. None of the proteins known to exclusively regulate the in vivo formation of actin cables or the actin contractile ring were identified. Intriguingly, our analysis also identified components of three other cortical structures, eisosomes, PIK patches and the TORC2 complex, establishing intriguing biochemical connections between four different yeast cortical complexes. Finally, we identified Aim3 as a regulator of actin dynamics at endocytic sites.
Conclusions
WASP is sufficient to trigger assembly of actin networks composed selectively of actin-patch proteins. These experiments establish that the protein composition of different F-actin structures is determined by the protein factor that initiates the network. The identification of binding partners revealed new biochemical connections between WASP derived networks and other cortical complexes and identified Aim3 as a novel regulator of the endocytic actin patch.
Introduction
Clathrin-actin-mediated endocytosis is a fundamental process in living cells, important for capturing macromolecules from the extracellular environment and regulating plasma membrane composition. Many aspects of the molecular mechanism of endocytosis are conserved among eukaryotes [1, 2]. In budding yeast at least 60 proteins implicated in this process have been identified so far based on genetic analysis and localization with fluorescent markers [3–5]. In a highly invariant sequence of events, recruitment of these proteins at the cell cortex is coordinated to 1) initiate the endocytic site and recruit cargo, 2) internalize the cell membrane and 3) release the endocytic vesicle into the cytoplasm [4, 5].
More than half of the endocytic patch proteins are responsible for the regulated turnover of a network of actin filaments, nucleated and crosslinked by the Arp2/3 complex around the clathrin-coated pit [1, 5]. The actin network provides the force necessary for invagination of the membrane and later for scission [3]. The precise patch protein recruitment and disassembly time sequence at endocytic sites has been quantitatively determined using powerful live cell imaging techniques [4, 6–8], and the peak numbers of molecules that accumulate in patches has been measured in fission yeast [9]. An ongoing crucial challenge is to further determine how the endocytic proteins act in concert. The contributions of nearly every identified endocytic patch protein to these dynamics have been tested in vivo by analyzing the defects caused by mutations [4, 8, 10]. However, these in vivo approaches provide limited mechanistic information and likely fail to identify a complete inventory of proteins. An alternative approach has been to analyze in vitro the mechanism of action of one or a few interacting proteins. This approach has been especially effective for studies of proteins controlling nucleation of the actin network by four NPFs, Abp1 (Actin Binding Protein 1), Pan1, Las17 and the type I myosins [8, 11–14]. However, such studies are also limited because they only allow analysis of the activities of a few proteins at a time and therefore fail to reveal synergistic properties that emerge in the presence of a multitude of factors.
This study was motivated by the fact that comprehension of diverse cellular processes has been realized through use of reconstituted systems. Of particular relevance to our study, microbeads functionalized with NPFs have been shown previously to stimulate actin assembly in cellular extracts, which often results in bead motility [15]. Developing such a system for yeast would provide new tools for analysis of the collective behavior of the actin cytoskeleton proteins involved in endocytosis, an opportunity to increase the inventory of proteins involved in actin assembly, and possibilities for synergy with genetic studies. Here we report the successful development of such a system for yeast with the identification of the proteins involved, establishing that Las17 is sufficient to recruit an actin network composed specifically of endocytic patch proteins, and increasing knowledge of the complex interactions that regulate patch function.
Results
Assembly of Las17-Derived Actin Structures in a Yeast Extract
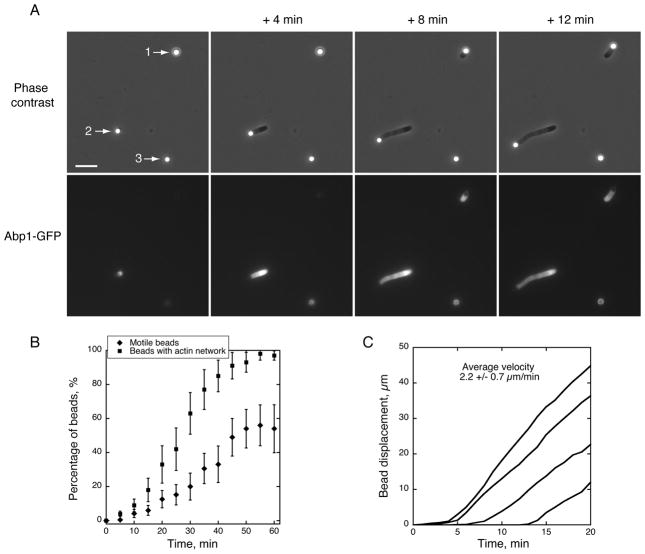
In this study, we utilized the fact that Las17 is among the first actin regulators recruited to endocytic sites where it plays a key role initiating actin assembly. We used purified full length Las17, which is fully active in vitro in stimulating Arp2/3-mediated actin filament nucleation [8, 11]. We prepared extracts from yeast cells grown in log-phase, and their protein concentration was 21.1 +/− 1.6 mg. ml−1. When 2 μm polystyrene microbeads functionalized with Las17 were incubated in the extract, we observed under phase contrast optics progressive assembly of dense clouds around the beads (Figure 1A beads 1 and 2 and Movie S1). We used an extract prepared from a strain in which Abp1 was fused to green fluorescent protein (GFP) in order to determine whether the clouds consisted of actin and associated proteins. Abp1 is a faithful reporter of actin assembly at endocytic sites in vivo, and was detected in association with the clouds (Figure 1A and Movie S1).
Figure 1. Reconstitution of Endocytic Actin Patches around Polystyrene Microbeads in Yeast Cytoplasmic Extracts.
Conditions: Las17-functionalized microbeads were added to a yeast cytoplasmic extract generated from a yeast strain expressing Abp1-GFP. Images were taken at the indicated time intervals.
(A) Timecourse of actin network assembly from the beads, which either became motile (beads 1 and 2) or formed a symmetric shell (bead 3). Upper panels: phase contrast microscopy images. Lower panels: GFP fluorescence images. See also Movie S1. Scale bar: 10 μm. (B) Time course of actin assembly on beads. Squares: Percentage of beads that assembled an actin network as a function of time (motile or non motile). Diamonds: Percentage of motile beads as a function of time. (C) Bead centroid displacement over time for several motile beads. The slopes of the curves indicate a rate of actin assembly of 2.2 +/− 0.7 μm.min−1 (n = 30).
We observed by time-lapse microscopy that beads existed without any detectable fluorescent signal until a sudden flash of fluorescence corresponding to initiation of actin assembly appeared (Figure 1A – bead 1 and Movie S1). This peculiar behavior might be explained by the need for a drifting actin filament to contact the bead to initiate autocatalytic actin assembly [16]. Approximately 98% of the beads were able to trigger actin assembly by 1 h (Figure 1B). When actin network assembly occurred on one side of a bead, the resultant force induced its motility (Figure 1A – beads 1 and 2 and Movie S1), similar to the motility of ActA-coated beads in Xenopus laevis egg extracts [15], demonstrating that even though yeast are non-motile organisms, their actin machinery can generate forces comparable to more complex eukaryotes. Approximately 20% of the beads became motile by 30 min and a maximum of 55% motile beads was reached by 50 min (Figure 1B). The remaining beads assembled robust actin filament shells but did not become motile, reflecting an inability to break the actin network symmetry (Figure 1A – bead 3 and Movie S1).
Bead velocity measurements indicate that actin assembly occurs at a rate of 2.2 +/− 0.7 μm. min−1 (Figure 1C). This rate is similar to what has been reported in vivo for actin assembly at yeast actin patches (ie. 2.7 +/0.3 μm. min−1 [3]), and is also in the same order of magnitude to actin assembly rates in a Xenopus laevis egg extracts and with purified proteins [15, 17], and in vivo at the leading edge of motile cells [18].
MudPIT Analysis Reveals the Protein Composition of the Las17-Derived Actin Networks
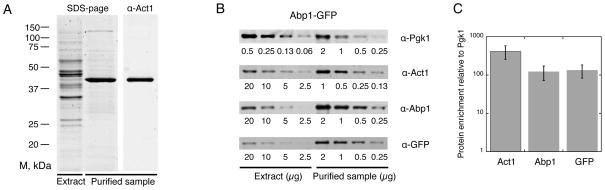
The design of this assay offered a unique opportunity to purify the Las17-derived actin networks assembled in the extract, in order to further identify their components. In this procedure, we deliberately avoided the use of an actin filament stabilizing drug such as phalloidin in order to maintain the structural integrity of the filament network and to avoid potential exclusion of actin binding partners due to competition for binding sites [19, 20]. We estimated by western-blot that no more than 15% of the actin filaments are lost during our purification procedure (data not shown). SDS-PAGE and western blot analysis of the purified samples confirmed that actin is the main protein associated with the purified beads (Figure 2A). This protocol specifically enriched actin cytoskeleton proteins such as actin and Abp1 up to several hundred fold relative to Pgk1, a 3-phosphoglycerate kinase with no known affinity for actin (Figure 2B and 2C).
Figure 2. Purification of Reconstituted Endocytic Actin Patches Assembled in Yeast Cytoplasmic Extracts.
(A) Coomassie-stained SDS-page gel of 2.5 μg of yeast cytoplasmic extract (left lane) and of 7.5 μg of purified actin structures (central lane). Immunoblot of 7.5 μg of purified actin structures probed with antibody to yeast actin (right lane). (B) Immuno-blots showing the enrichment of actin (Act1) and Abp1-GFP compared to the reference protein Pgk1. Numbers below each lane represent the amount of extract or purified protein loaded by serial dilutions (unit: μg). (C) Quantification of (B): Histogram plots showing the fold enrichment of actin and Abp1-GFP (detected independently using Abp1 or GFP antibodies) compared to Pgk1.
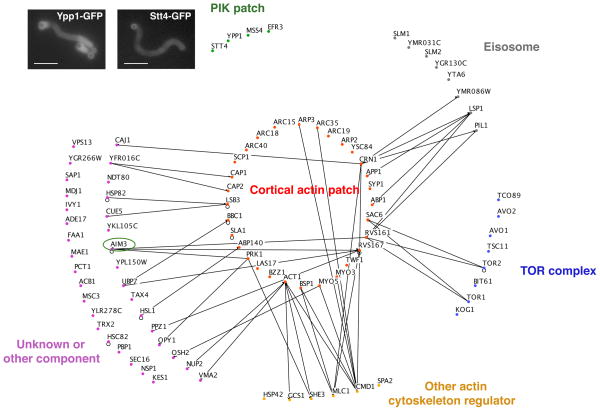
We analyzed the samples by MudPIT to determine the protein composition of these WASP-derived actin networks. MudPIT identifies the peptides generated from very complex protein mixtures, because peptides are first sorted by multidimensional liquid chromatography before identification by tandem mass spectrometry [21]. The MudPIT analysis of three independent experiments identified a total of 32,695 peptides corresponding to 1,113 yeast proteins (Figure S1 and S2, Table S1 and S2). Because MudPIT analysis is so sensitive, we developed a statistical method to distinguish enriched proteins from background, based on a measure of the relative peptide enrichment for each protein. This strategy, detailed in the Supplemental Experimental Procedures, enabled us to identify 90 of these proteins as the most highly enriched components (Figure 3 and S2 and Table S3).
Figure 3. MudPIT Analysis of Actin Patch Protein Composition.
Functional classification of the most probable actin tail components based on their GO annotations. Lines represent an Osprey-generated network showing published physical interactions between the identified actin patch components and other identified components. Inset micrographs show validation of MudPIT analysis by tail fluorescence in bead assembly experiments using extracts from cells expressing GFP-tagged PIK patch proteins Stt4 and Ypp1. The circle indicates Aim3, a focus of further analysis (see Figures 5 and 6). Scale bars: 10 μm.
Las17 is Sufficient to Form Actin Patch-Like Structures of Appropriate Protein Composition
Actin patch proteins are recruited to the plasma membrane in a precisely timed and ordered sequence, allowing their classification into one of several protein modules involved in different steps of endocytosis [4, 5]. Of the 90 proteins identified by MudPIT, 31 have been shown previously to colocalize with endocytic actin patches in vivo [4, 5, 22] (Figure 3 and Table S3). Thirty of these 31 actin patch proteins are recruited either at the same time as Las17 or later in vivo, reinforcing the notion that Las17 plays a central role in formation of these actin structures, and establishing that it can by itself trigger recruitment of most of the proteins that form branched actin filament networks in actin patches. Syp1, which was reported to be a negative regulator of Las17-Arp2/3 complex activity [23], is the only patch protein detected in our analysis that is recruited to cortical patches before Las17.
Even though positions of many endocytic proteins at the base, tubule or tip of invaginations have been deduced from positions of fluorescence centroids [3, 4], the size of endocytic sites is comparable to the diffraction limit for light microscopy, making it difficult to precisely determine the position of each protein relative to the plasma membrane and the actin cytoskeleton during internalization. Moreover, electron microscopy analysis of protein locations, though powerful, has so far only been applied to a handful of proteins [24, 25]. The spatial scale of our bead system provided a new perspective by allowing us to determine whether proteins were localized at the bead with Las17, or in the actin tails, as detailed below.
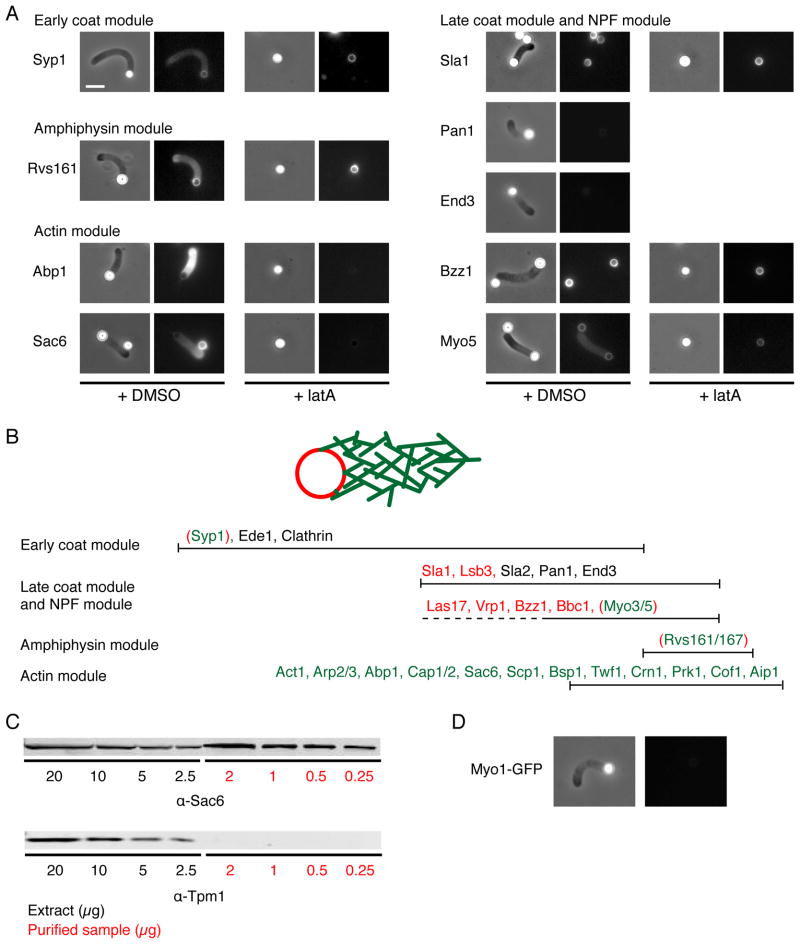
“Coat” and NPF-Module Proteins
Coat- and NPF- module proteins arrive around the same time, shape the membrane and/or regulate actin assembly [3, 5, 10, 11, 13]. The localization of these components to endocytic sites in vivo is not F-actin dependant, reinforcing the idea that these proteins function upstream of actin assembly [4, 8]. Localizing with Las17 to the bead surface were Sla1, Vrp1 (yeast WIP), Bbc1 and Bzz1 (yeast syndapin), while the type I myosins Myo3 and Myo5 could also be detected along the actin networks (Figure 4A and 4B). We used the actin monomer sequestering drug latrunculinA (latA) to inhibit actin network assembly, and found that none of these proteins requires actin assembly to localize to the bead surface.
Figure 4. Actin Patch Protein Localization on Reconstituted Structures and Specificity of Protein Recruitment.
(A) Protein localization on motile Las17-functionalized microbeads in the absence or presence of latA for several representative proteins from each module. Extracts were prepared from strains expressing the indicated proteins tagged with GFP. MudPIT identified proteins were detected only around the beads (Sla1, Bzz1) or also in association with the F-actin structures (Syp1, Rvs161, Sac6, Abp1, Myo5) based on their fluorescence signal. Proteins not identified by MudPIT (Pan1, End3) could not be detected. LatA (180 μM) prevents the recruitment of only a subset of these proteins (Abp1, Sac6). Left: Phase contrast images; Right: Fluorescence images. Scale bar: 5 μm. (B) In vivo time line for endocytic actin patch protein recruitment, based on [5]. Red proteins localized only around the Las17 functionalized microbeads, and do not require F-actin for their recruitment. Green proteins localized along the lengths of the F-actin structures. Green proteins require F-actin for their recruitment, except that the ones indicated within red brackets were recruited to the bead surface in the absence of actin assembly. Black proteins were not detected. (C) Immuno-blots showing the presence of Sac6 in the purified samples and the absence of any detectable Tpm1. (D) Absence of type II myosin Myo1 on motile Las17-functionalized microbeads.
“Actin module” proteins
Twenty proteins identified in the purified samples have been previously classified as members of the “actin module” directly responsible for organization and dynamics of the actin network (Figure 4A and 4B). In contrast to proteins of the coat module, localization of these proteins in vivo depends on F-actin [4]. Consistent with this observation, these proteins decorate the actin network formed from the beads in our system (Figure 4A and 4B), but could not be detected on the beads in the presence of latA.
Membrane shaping proteins
Several proteins conserved from yeast to mammals cooperate with actin to deform the plasma membrane in order to help its invagination and scission during endocytosis (Kishimoto et al., in preparation and [26–28]). In budding yeast, these proteins include the Bin-Amphiphysin-Rvs (BAR) domain heterodimer Rvs161/167 and the FER-CIP4 homology-BAR (F-BAR) domain protein Bzz1. We found that while Bzz1 is localized with Las17 around the beads, Rvs161 also decorates the actin filament networks, and that neither protein requires actin assembly for recruitment to the bead surface (Figure 4A and 4B). These results suggest that while Bzz1 provides a biochemical link between the membrane and some coat components and/or NPFs in order to regulate actin nucleation, the Rvs complex can also be recruited to the membrane by the actin cytoskeleton itself. Thus, we suggest that the Rvs complex provides a mechanical link between the actin network and the plasma membrane.
Among the proteins known to interact with branched actin networks in vivo, only Aip1 and Gmf1 were missing from the MudPIT data set, and Cof1 and Srv2 were not highly enriched relative to other actin-associated proteins (Table S2) [22, 29]. However, these proteins could be detected along the branched actin networks as well (Figure 4B and data not shown). Since Aip1, Gmf1, Srv2 and Cof1 are all involved in actin filament disassembly, it is likely that the actin filaments decorated by these proteins were primarily disassembled during the preparation of the MudPIT samples.
Strikingly, none of the proteins known to exclusively bind and regulate actin cables or the actomyosin ring, each composed of parallel networks of actin filaments, was detected by the MudPIT analysis. These missing candidates include the two isoforms of tropomyosin, Tpm1 and Tpm2, the yeast IQGAP Iqg1, the type II myosin Myo1 and the 2 type V myosins Myo2 and Myo4, even though each of these purified proteins has been shown to interact with actin filaments in vitro [22]. We verified that the actin binding protein Sac6 (yeast fimbrin), a component of actin patches, decorates the actin tails (Figure 4A) and can be detected readily by western-blot (Figure 4C), while the actin binding protein tropomyosin (Tpm1) could not be detected in the same samples (Figure 4C). We also verified by fluorescence microscopy that the type I myosin Myo5 could be detected along the actin tails (Figure 4A), but not the type II myosin Myo1 (Figure 4D).
In total, our results show that a pathway initiated by an NPF triggers recruitment of a specific subset of actin binding proteins to the filaments in order to generate an actin network of a specific composition and properties, and establish that the structures induced on Las17-functionalized beads are actin patch-like structures.
Identification of Novel Biochemical Connections to Other Cortical Patch Structures
Some unexpected proteins were identified by MudPIT analysis in addition to the 31 known endocytic and actin patch components. To gain an overview of the biological structures and processes represented by the remaining interacting proteins, we used their respective gene annotations (GO, The Gene Ontology Consortium, 2000) (Figure 3 and Table S3). GO annotations draw on many types of data from the literature to organize genes into cellular processes, components or functions. Interestingly, we found that 20 of these proteins could be grouped into 3 distinctive protein complexes, namely eisosomes, TOR (Target of Rapamycin) complexes (TORC1 and/or TORC2) and phosphoinositide kinase (PIK) patches (Figure 3). These complexes have each previously been implicated in in vivo organization of the actin cytoskeleton and/or in endocytosis, however the underlying mechanisms remain unclear [30–34]. Co-purification of these protein complexes with F-actin networks assembled on Las17-functionalized beads was surprising because none had been reported to colocalize with actin patches in vivo, but rather appear in distinct neighboring domains on the cell cortex (except the TORC1 complex, which shows a vacuolar localization) [35–37]. However, Osprey-generated networks [38] indicate that several eisosome and TOR complex components interact physically with certain components of the reconstituted actin patch structures (Figure 3). Only components of the PIK patches did not have previously described interactions with components of the actin patches. For this reason, we decided to focus on this group of proteins, and showed by fluorescence microscopy that they interact with actin tails in our system (Figure 3 - inset pictures). Together, these results suggest physical communication between these various structures in vivo.
Identification of Aim3 as a New Endocytic Actin Patch Component of the Actin Module
Our analysis identified other proteins with unknown function or localization. We noticed that many of these uncharacterized proteins are expressed at lower levels in cells. Thus, we hypothesized that some of these proteins might be cortical patch components that had not been discovered previously due to their low abundance. We decided to focus on Aim3 (Altered Inheritance rate of Mitochondria, also called Ybr108w) because of reported interactions with several patch proteins, including the BAR proteins Rvs161 and Rvs167 (Figure 3) [39].
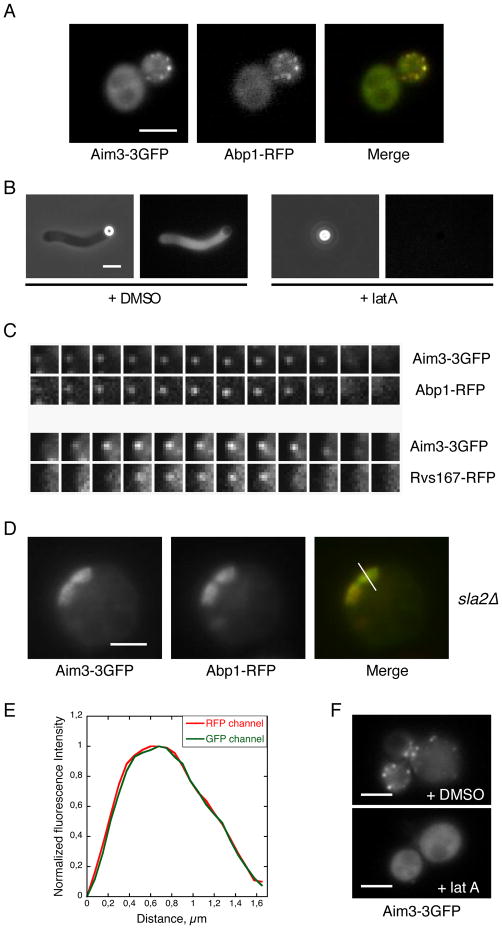
We investigated the in vivo localization of Aim3 by two-color fluorescence microscopy. Aim3-3GFP was detected as puncta at the cell cortex (Figure 5A), and simultaneous imaging with Abp1-RFP showed that more than 96% (n = 125 cortical patches) of the green foci simultaneously colocalized with red foci (Figure 5A), confirming that Aim3 is an actin patch protein. Aim3-3GFP localized with the actin networks in the bead assay and this recruitment was F-actin dependant (Figure 5B). Supporting this conclusion, Aim3 was recruited to and disassembled from cortical patches concomitantly with actin module proteins such as Abp1 (Figure 5C). Although a biochemical link had been previously detected with proteins of the Rvs/amphiphysin module [39], Aim3 was recruited before and disassembled after Rvs167 (Figure 5C).
Figure 5. Aim3 is a Novel Patch Protein in the Actin Module.
Scale bars: 3 μm.
(A) Colocalization of Aim3-3GFP with the actin patch component Abp1-RFP. (B) Aim3-3GFP associates with the F-actin structures in vitro. LatA (180 μM) prevents this recruitment. (C) Simultaneous two-color imaging reveals synchronous appearance and disappearance of Aim3-3GFP with Abp1-RFP, and subsequent appearance of Rvs167-RFP. Images captured at a 1.3 s time interval. (D) Colocalization of Aim3-3GFP with the actin network in sla2Δ cells expressing Abp1-RFP. (E) Normalized fluorescence intensity profiles along the length of the actin tails in (D), from the cell surface toward the cell interior. (F) Aim3-3GFP does not associate with endocytic actin patches in the presence of Lat A (180 μM).
To further investigate Aim3’s spatio-temporal localization to actin structures in vivo, we generated an sla2 strain which expressed both Aim3-3GFP and Abp1-RFP. In this mutant background, Arp2/3-dependent, elongated actin networks accumulate at the cell cortex. These structures allow for the spatial distinction between protein complexes binding actin versus proteins localized to endocytic components [3]. Consistent with an interaction between Aim3 and filamentous actin at the membrane, we observed that Aim3 and Abp1 colocalize along the elongated actin structures (Figure 5D and 5E). Lastly, we used latA to disassemble the actin patches, finding that Aim3-3GFP could no longer be detected as foci at the cell cortex, but showed cytoplasmic staining instead, demonstrating that Aim3 recruitment to the plasma membrane requires F-actin (Figure 5F).
A Role for Aim3 in Actin Patch Dynamics
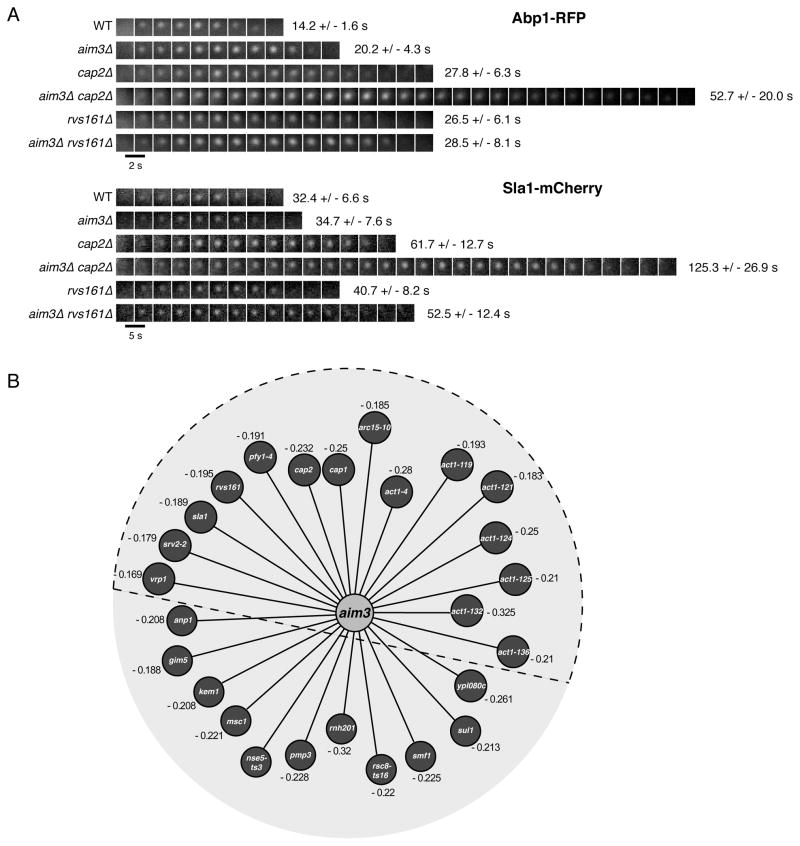
We next sought to investigate the contribution of Aim3 to actin patch dynamics. A sensitive diagnostic is to measure in a deletion mutant strain the lifetimes of the endocytic patch components Abp1 and Sla1 [4]. Absence of Aim3 increased Abp1 lifetime at the plasma membrane from 14.2 +/− 1.6 s to 20.2 +/− 4.3 s, while having hardly any effect on Sla1 dynamics (Figure 6A). Next, we considered that functions of numerous proteins are only fully revealed in the absence of a second protein, which may have a redundant function. To test this possibility for Aim3, we performed a genome-wide Synthetic Genetic Array (SGA) screen with an aim3Δ deletion allele as a query to uncover AIM3 genetic interactions [40]. Quantitative assessment of aim3Δ double mutant fitness phenotypes identified 26 mutants showing pronounced growth defects [40]. This subset was significantly enriched for genes encoding an actin cytoskeleton regulating or interacting protein (15/26), including genes encoding capping protein (Cap1 and Cap2), amphiphysin (Rvs161), and numerous different actin alleles (Figure 6B and Table S4). We measured the impact on Abp1 and Sla1 lifetimes of double knock-outs of Aim3 and proteins of the actin and amphiphysin modules. While absence of Rvs161 induced only moderate additional effect on Abp1 and Sla1 lifetimes, absence of capping protein had a dramatic effect on Abp1 and Sla1 dynamics in cells lacking Aim3, wherein these markers had much longer and more variable lifetimes (Figure 6A; 52.7 +/− 20.0 s and 125.3 +/− 26.9 s for Abp1 and Sla1, respectively) suggesting an important role for Aim3 in regulating actin patch dynamics when another actin regulating function is compromised.
Figure 6. Deletion of Aim3 and its Effects on Actin Patch Dynamics.
(A) Timecourse of representative Abp1-RFP and Sla1-Cherry patches in various strain backgrounds as indicated. Images were captured at the indicated time intervals. Lifetimes are indicated for n = 50 patches for all strains. (B) Genome-wide Synthetic Genetic Array (SGA) screen with an aim3Δ defined confidence threshold (ε < −0.15, p < 0.01). Numbers represent the SGA genetic interaction score ε. Proteins contained within the dashed semi-circle correspond to actin cytoskeleton regulating or interacting proteins. See also Table S4.
Discussion
In this article, we described our reconstitution of actin patches in yeast cell extracts. This work provided new insights into molecular pathways for endocytic vesicle formation, clues about mechanisms for assembly of a complex network of actin filaments of defined protein composition, identified a new actin patch protein, Aim3, and interactions with additional cortical patch structures.
Patch Reconstitution Identifies Distinct Pathways Mediating Endocytosis
Our experiments demonstrated that Las17 plays a central role in the formation of endocytic actin patches, since it can by itself recruit the proteins from the “actin module” that appear at endocytic sites at the same time as Las17 or later. Abp1, which shows a modest NPF activity, was recruited to the actin tails in the bead assay. We suggest therefore that a role of Abp1 might be to interact with the Arp2/3 complex and actin filaments in order to compete with additional interactors such as GMF in order to stabilize the actin network against debranching [29]. In addition, proteins from the coat and NFP modules were recruited directly to the beads, even in the presence of latA, indicating that Las17 is sufficient to assemble a scaffold of these proteins without actin. A subset of these proteins (Sla1, Bbc1, Syp1) negatively regulates Las17 activity, while others (Bzz1, Vrp1 and the type I myosins) release this inhibition or provide additional NPF activity [8, 11, 23]. Type I myosins were also identified along the actin tails, reflecting a probable ability of their motor domains to interact with actin filaments organized in branched networks.
In contrast, several other coat module proteins (Sla2p, Pan1, End3) were not identified in our studies, and these proteins are not known to regulate Las17 activity. End3 has been identified as a stoichiometric component of the Pan1 complex [13], and Sla2 has been shown to down-regulate the NPF activity of Pan1 [13] but not any other NPF. We therefore propose that the NPF Pan1 and its cofactors are recruited by and function in a distinct biochemical pathway from Las17 and the type I myosins. In agreement with our results, each NPF exhibits a unique dynamic behavior at endocytic sites [8, 10]. Together, these observations indicate that cells initiate assembly of independent actin filament networks at the same endocytic sites and at the same time. Evidence for independent pathways raises the interesting question of how they are coordinated in time and space.
The MudPIT analysis also revealed the presence of the amphiphysin/Rvs complex, which interacts with the plasma membrane via its BAR domains. In vivo data show that the actin cytoskeleton is important for the recruitment of the Rvs complex at the membrane [4], and a recent model suggests that the initial plasma membrane deformation induced by the actin cytoskeleton could subsequently drive BAR domain accumulation [41]. Our data now reveal that the Rvs complex can be recruited to the plasma membrane by the actin cytoskeleton itself, independant of membrane curvature. Therefore the Rvs complex may represent a physical link between the plasma membrane and the actin cytoskeleton. It will be crucial in the future to determine whether the actin cytoskeleton uses the Rvs proteins as a scaffold to exert an invaginating or pinching force on the endocytic membrane.
Understanding How Actin Structures of Distinct Organization and Protein Composition Assemble in a Common Cytoplasm
Eukaryotic cells are able to form higher order actin filament-based structures of distinct organization and protein composition from identical actin monomers. Our work shows that these structures are able to auto-regulate themselves in order to recruit the appropriate actin binding partners required to optimally perform their particular cellular function. It is, for example, interesting that tropomyosin, which binds to actin filaments in vitro with a very high affinity, but is not a component of cortical patches in vivo, did not decorate our reconstituted actin patches. Thus, our work demonstrates the existence of a mechanism for controlling actin network composition while filaments continue to assemble, and to segregate proteins to different filament populations. This observation suggests that at the time they assemble, actin filaments acquire a specific identity characteristic of the network to which they belong, and that populations of actin filaments with distinct identities modulate their affinity for various actin-binding proteins.
Our results raise an important question concerning how the same actin subunit can in the same cytoplasm assemble into structurally and biochemically distinct filament assemblies. One possible explanation is that different actin binding proteins might compete for binding to actin filaments due to a steric hindrance [42, 43]. However, this explanation does not explain how different filament populations inside a cell can be decorated differently at the same time since the binding of these proteins to actin is the result of a chemical equilibrium. This feature suggests that actin filaments acquire their specific identity while they assemble, and maintain this identity until they age or disassemble. Thus, another reasonable explanation derives from the fact that several proteins including ADF/cofilin [44, 45], tropomyosin [42] and barbed end cappers such as gelsolin [42] and formins [46] can alter the filament conformation. Hence, an hypothesis to consider is that Arp2/3-mediated actin assembly (such as actin patch assembly in yeast) prevents tropomyosin binding to actin filaments [47] but allows the binding of other factors such as fimbrin or calponin, while formin mediated actin assembly (into actin cables and the acto-myosin ring in yeast) leads to formation of linear actin filaments decorated mainly by tropomyosin. Actin filaments decorated by one set of proteins can provide a substrate for the optimal binding of other proteins. This model is consistent with several recent papers showing that tropomyosins dramatically regulate the biochemical properties of several F-actin interacting proteins such as formins, fimbrins or myosins [48–50].
Identification of New Biochemical Connections at the Cell Cortex
Purifying the reconstituted actin patches and determining their composition by MudPIT also gave us a unique opportunity to identify new biochemical connections at the cell cortex.
First, our results suggest that some patch proteins may not have been discovered previously because they are expressed at low levels. Here, we demonstrated that Aim3 colocalizes with actin patches in vivo, and is recruited in vivo with the proteins of the actin module. In agreement with this observation, we revealed that Aim3’s function is important for maintaining rapid and efficient actin patch dynamics, especially when another actin module protein such as Cap2 is simultaneously not expressed.
We also identified among proteins associated with reconstituted patches multiple components of three different cortical patch complexes, according to their GO annotations. First, 8 proteins were identified as components of the eisosomes, static cortical patch-like structures postulated to be involved in endocytosis [30]. At least two components from the eisosomes, Pil1 and Lsp1, have been shown to interact with multiple components of actin patches (Figure 3).
We also identified 4 protein components of the recently described PIK patches [35] as interacting with reconstituted actin patches. PIK patches are reported as static sites at the plasma membrane, and are implicated in PI(4,5)P2 synthesis, which is important for actin cytoskeleton organization and endocytosis.
Finally, 8 other interacting proteins are all components of the TORC1 and/or the TORC2 complexes. Both complexes have been shown to be important for organization of the actin cytoskeleton and endocytosis in different eukaryotes [31–33]. In particular, Tor2 signals to components required for proper actin organization, such as the GTPase Rho1, and to two eisosome proteins, Slm1 and Slm2 (reviewed in [33]). Aronova et al. showed that many TORC1 and TORC2 components fractionate in vitro with detergent-resistant membranes, and a proteomic analysis of these TOR-associated membranes revealed the presence of regulators of endocytosis and the actin cytoskeleton [32]. These results may explain the presence of these TOR components in our system, even though we were unable to detect the presence of lipids around the beads or along the actin tails using FM4–64 dye staining (our unpublished observations).
Collectively, our work and work reported by others suggest functional interplay between at least four different cortical structures whose components show different localization patterns and different dynamics at the plasma membrane. Future studies are required to elucidate the underlying basis and biological consequences of these interactions.
Experimental procedures
Strains
Fluorescent tags were integrated chromosomally to generate C-terminal fusions of each protein as described in [51]. Yeast strains used for this study are listed in Table S5.
Protein Purification and Microbead Functionalization
Las17 was overexpressed and purified as described in [11]. Polystyrene microbeads were functionalized according to pre-existing protocols [15]. Briefly, 2 μm polystyrene microbeads (Polybead® Microspheres, Polysciences, Inc.) were incubated on ice at 0.2% solid with 100 nM Las17 in 50 μl of HK buffer (10 mM Hepes buffer pH 7.8, 0.1 M KCl, 1 mM MgCl2, 1 mM ATP, 0.1 mM CaCl2, 0.05% Triton X100). After 45 min, 1% BSA was added for 15 min. Beads were washed and stored in 50 μl of HK buffer containing 0.1% BSA, and were used within 12 h without any significant loss of activity.
Yeast Extract Preparation and Actin Patch-Like Structure Formation
Yeast strains were cultured in standard rich media (YPD) at 30°C to an OD600 of 1 – 1.5. Cells were harvested by centrifugation, resuspended in cold water, and centrifuged again. Pellets were flash-frozen in liquid N2, and ground by mechanical shearing in a Waring blender. To each gram of yeast powder were added 100 μl of 10X HK buffer and protease inhibitors (Protease Inhibitor Cocktail Set IV, Calbiochem, Merck4Biosciences). Yeast powder was gently mixed on ice with the buffer, progressively thawed, incubated on ice for 20 min, and centrifuged for 20 min at 350,000 g. The cleared supernatant was collected, kept on ice and used within 3 h. For all of our experiments, functionalized microbeads were added to the extract to induce formation of actin patch-like structures at a ratio of 1 μl of the bead stock solution for 19 μl of extract.
Actin Patch-Like Structure Purification and Sample Preparation for Multi dimensional Protein Identification Technology (MudPIT)
Actin patch-like structures were assembled around the polystyrene microbeads for 40 min at room temperature in 500 μl of extract. Beads were pelleted by low-speed centrifugation (1 min at 1,000 × g), the supernatant was removed, and the pellet was rapidly and carefully washed twice with 1 ml of ice-cold HK buffer. For western-blot analysis, samples were run on a polyacrylamide gel and transferred to a nitrocellulose membrane. Proteins were immuno-blotted with primary antibodies, and then with secondary antibodies conjugated with a fluorescent moiety. Western-blots were imaged using an Odyssey Imaging System (LI-COR BioSciences). Sample preparation for MudPIT analysis is described in Document S1.
Multi dimensional Protein Identification Technology (MudPIT) and Analysis of Tandem Mass Spectra
MudPIT experiments were processed and tandem mass spectra were analyzed according to pre-existing protocols and are detailed in Document S1.
Microscopy and Image analysis
In vitro bead assay: After addition of the functionalized beads to the extract, a 3.3 μl sample was placed between a slide and a coverslip, and sealed with VALAP (vaseline, lanolin, paraffin 1:1:1). Images were acquired using an Olympus IX71 microscope equipped with a 60X PlanApo objective and a camera (Orca II; Hamamatsu Photonics). In vivo yeast cell observations: Cells were immobilized and observed as described in [4]. Images were acquired using an Olympus IX81 microscope equipped with a 100X PlanApo objective, a two-channel simultaneous imaging system (DV2, Photometrics) and a camera (Orca R2; Hamamatsu Photonics). Images were processed using MetaMorph 7.1.7.0 or ImageJ 1.39t (http://rsb.info.nih.gov/ij/).
Deletion mutants genome-scale genetic interactions analysis
This analysis was performed as described in [40].
Highlights.
Reconstitution of endocytic actin patches in a yeast extract
Yeast WASP is sufficient to nucleate networks of biologically relevant composition
Novel biochemical connections with other cortical complexes detected
Identification of Aim3 as a novel regulator of actin patch dynamics
Supplementary Material
Acknowledgments
We thank the members of the Drubin/Barnes lab for helpful discussions, including Voytek Okreglak, Yutian Peng and Yidi Sun for their critical reading of the manuscript, and Eric Li for his assistance. This work was supported by Fondation pour la Recherche Médicale (SPE20070709969) and Human Frontier Science Program (LT00565/2008-L) fellowships to A.M. and by NIH grant R37 GM42759 to D.G.D and P41 RR011823 to J.R.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 2.Conibear E. Converging views of endocytosis in yeast and mammals. Curr Opin Cell Biol. 2010;22:1–6. doi: 10.1016/j.ceb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 4.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 6.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 7.Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative Analysis of the Mechanism of Endocytic Actin Patch Assembly and Disassembly in Fission Yeast. Mol Biol Cell. 2010;21:2894–904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- 12.Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toshima J, Toshima JY, Duncan MC, Cope MJ, Sun Y, Martin AC, Anderson S, Yates JR, 3rd, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geli MI, Lombardi R, Schmelzl B, Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. Embo J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4908–4913. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Achard V, Martiel JL, Michelot A, Guerin C, Reymann AC, Blanchoin L, Boujemaa-Paterski R. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 2010;20:423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 17.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 18.Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- 19.Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 20.Mahaffy RE, Pollard TD. Influence of phalloidin on the formation of actin filament branches by Arp2/3 complex. Biochemistry. 2008;47:6460–6467. doi: 10.1021/bi702484h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boettner DR, D’Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idrissi FZ, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson SM, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, EOT, De Camilli P. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, Lappalainen P, Goode BL. GMF Is a Cofilin Homolog that Binds Arp2/3 Complex to Stimulate Filament Debranching and Inhibit Actin Nucleation. Curr Biol. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 31.Araki T, Uesono Y, Oguchi T, Toh EA. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet Syst. 2005;80:325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]
- 32.Aronova S, Wedaman K, Anderson S, Yates J, 3rd, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Flower TR, Clark-Dixon C, Metoyer C, Yang H, Shi R, Zhang Z, Witt SN. YGR198w (YPP1) targets A30P alpha-synuclein to the vacuole for degradation. J Cell Biol. 2007;177:1091–1104. doi: 10.1083/jcb.200610071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, Opekarova M, Tanner W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol. 2008;183:1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009;20:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Germann M, Swain E, Bergman L, Nickels JT., Jr Characterizing the sphingolipid signaling pathway that remediates defects associated with loss of the yeast amphiphysin-like orthologs, Rvs161p and Rvs167p. J Biol Chem. 2005;280:4270–4278. doi: 10.1074/jbc.M412454200. [DOI] [PubMed] [Google Scholar]
- 40.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindberg U, Schutt CE, Goldman RD, Nyakern-Meazza M, Hillberg L, Rathje LS, Grenklo S. Tropomyosins regulate the impact of actin binding proteins on actin filaments. Adv Exp Med Biol. 2008;644:223–231. doi: 10.1007/978-0-387-85766-4_17. [DOI] [PubMed] [Google Scholar]
- 43.Pruyne D. Tropomyosin function in yeast. Adv Exp Med Biol. 2008;644:168–186. doi: 10.1007/978-0-387-85766-4_14. [DOI] [PubMed] [Google Scholar]
- 44.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bugyi B, Papp G, Hild G, Lorinczy D, Nevalainen EM, Lappalainen P, Somogyi B, Nyitrai M. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem. 2006;281:10727–10736. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 48.Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 20:1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 49.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.