Abstract
Hydrogen sulfide (H2S), a messenger molecule generated by cystathionine γ-lyase, acts as a physiologic vasorelaxant. Mechanisms whereby H2S signals have been elusive. We now show that H2S physiologically modifies cysteines in a large number of proteins by S-sulfhydration. About 10 to 25% of many liver proteins, including actin, tubulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), are sulfhydrated under physiological conditions. Sulfhydration augments GAPDH activity and enhances actin polymerization. Sulfhydration thus appears to be a physiologic posttranslational modification for proteins.
INTRODUCTION
The gases nitric oxide (NO) and carbon monoxide (CO) are physiologic messenger molecules that influence various physiological processes, including blood vessel relaxation and neurotransmission (1). Recent evidence indicates that hydrogen sulfide (H2S) also acts as a messenger to elicit hibernation states (2), inhibit insulin signaling (3), and regulate inflammation (4) and blood vessel caliber (5). H2S is physiologically generated from L-cysteine by cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE). H2S formation by blood vessels and the heart is abolished in CSE−/− mice, which display hypertension and loss of endothelium-dependent vasorelaxation (6). Molecular mechanisms whereby H2S influences its targets have been elusive. NO physiologically S-nitrosylates diverse proteins (7). We now show that endogenous H2S physiologically S-sulfhydrates (SHY) proteins (that is, converts cysteine -SH groups to -SSH) to regulate their function. Whereas nitrosylation appears to diminish cysteine reactivity, sulfhydration seems to enhance it. Similar to nitrosylation, however, the covalent modification in sulfhydration is reversed by reducing agents, such as dithiothreitol (DTT). Given the high abundance of protein S-sulfhydration, we propose that this posttranslational modification may affect a variety of biological pathways.
RESULTS
H2S covalently modifies cysteine residues through S-sulfhydration
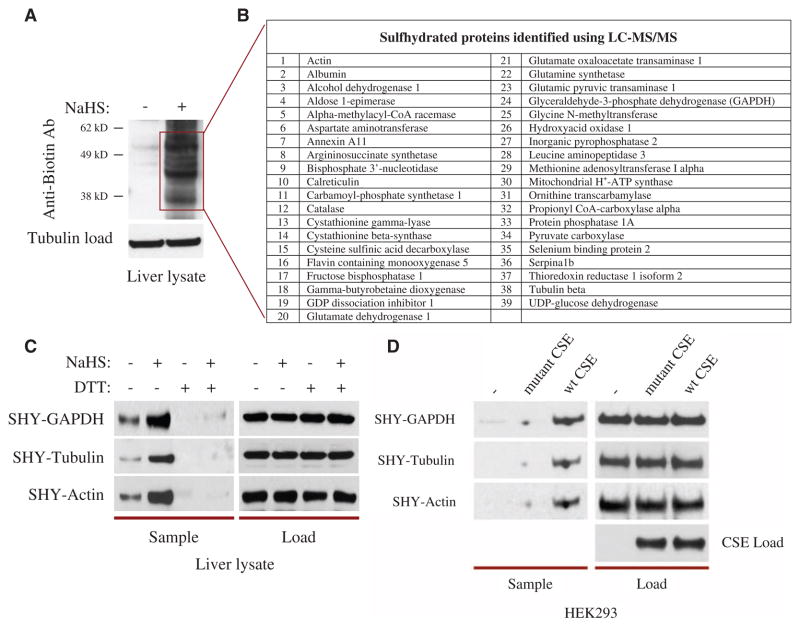
S-Nitrosylation can be detected by the biotin switch assay, in which free thiols are blocked by a highly specific free sulfhydryl-reactive compound, methyl methanethiosulfonate (MMTS), which does not interact with nitrosylated thiols or any other forms of oxidized thiols (8, 9). The nitrosylated thiols are then selectively exposed by treatment with ascorbate and labeled with N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP), a compound that interacts with sulfhydryl groups. Even in the absence of ascorbate treatment, there is some biotin labeling of proteins in the brains of both wild-type and neuronal nitric oxide synthase (NOS) knockout (nNOS−/−) animals, perhaps indicating that the biotin switch assay is also detecting protein S-sulfhydration (9). We elected to study sulfhydration in the liver because it generates high amounts of H2S (4). In mouse liver lysates in the absence of ascorbate, we detected basal labeling of multiple proteins using an antibody against biotin with even more prominent and abundant labeling after treatment with the H2S precursor sodium hydrogen sulfide (NaHS) (Fig. 1A and fig. S1A). The three most prominent bands observed after NaHS treatment were glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (38 kD), β-tubulin (55 kD), and actin (43 kD), as identified by mass spectrometry, which has thus far identified 39 sulfhydrated proteins. Probing with antibodies to specific proteins, such as GAPDH, β-tubulin, and actin (Fig. 1C), revealed substantially more basal sulfhydration than was evident with the antibody directed against biotin (Fig. 1A), which may react less with particular proteins. DTT, which reduces disulfide bonds, reversed both the basal and the NaHS-induced sulfhydration, allowing MMTS to block the newly freed thiols (Fig. 1C). This implies that the modification is covalent and involves a sulfhydryl group. Reasons for the relative selectivity of MMTS for free, as opposed to modified, -SH groups are unclear. There was no signal without biotin-HPDP, indicating that the basal signal does not represent endogenous biotinylation of proteins (fig. S1B). The nature of the product of the reaction of biotin-HPDP with R-SSH is not established; it could be a disulfide or a trisulfide. In principle, biotin-HPDP could also react with polysulfides. Because sulfhydrated cysteines are highly reactive, they may tend to interact with dissolved oxygen present in the assay solutions. Consistent with this notion, we observed that using nitrogen gas to deoxygenate assay buffers enhances the basal sulfhydration signal in the liver (fig. S1C).
Fig. 1.
H2S covalently modifies proteins through S-sulfhydration of cysteine residues. (A) Liver lysates treated with 100 μM NaHS for 30 min at 37°C and subjected to the modified biotin switch assay with antibody against biotin (Anti-biotin Ab) to detect S-sulfhydration show numerous sulfhydrated proteins. (B) LC-MS/MS of a subset of the sulfhydrated proteins in (A) identifies 39 sulfhydrated proteins, including GAPDH, β-tubulin, and actin. (C) DTT treatment (1 mM) for 10 min reverses GAPDH, β-tubulin, and actin sulfhydration, detected with antibodies specific to each protein, implying a covalent sulfhydryl modification. (D) In HEK293 cells transfected with plasmids encoding CSE, exposure to 5 mM L-cysteine for 1 hour leads to GAPDH, β-tubulin, and actin sulfhydration, as assessed by the modified biotin switch assay with antibodies specific to each of the three proteins. Catalytically inactive CSE fails to sulfhydrate proteins.
CSE is a major source of H2S production in the liver (4–6). In human embryonic kidney (HEK) 293 cells, which lack endogenous CSE, transfection with plasmids expressing wild-type CSE, but not a non–PLP (pyridoxal 5′-phosphate)–binding catalytically inactive mutant CSE, elicited sulfhydration of GAPDH, β-tubulin, and actin (Fig. 1D and fig. S2). Liver displayed abundant CSE with negligible amounts of neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS), whereas nNOS and eNOS were relatively abundant in the brain (fig. S3B). Furthermore, we detected no alterations in GAPDH sulfhydration in the livers of nNOS−/−, eNOS−/−, and iNOS−/− mice compared to those of wild-type mice (fig. S3A), ruling out any involvement of S-nitrosylation in the modified biotin switch assay (that is, the assay performed without ascorbate) we used to detect sulfhydration. Thus, we conclude that a large number of proteins are basally sulfhydrated through H2S generated physiologically in the liver.
Sulfhydration physiologically modifies proteins in wild-type but not CSE−/− animals
H2S production by blood vessels is abolished in CSE−/− mice (6). We now show that CSE−/− liver fails to generate H2S, as measured with an H2S-selective electrode, with no significant changes in L-cysteine concentration compared to wild-type liver (Fig. 2A and fig. S4). CBS does not contribute noticeably to H2S generation in the liver, because the livers of CBS−/− mice displayed H2S production similar to those of wild-type animals (fig. S5). Endogenous sulfhydration of GAPDH, β-tubulin, and actin is also abolished in CSE−/− liver, showing that these proteins are physiologically sulfhydrated through CSE activity (Fig. 2B). Using densitometric analysis of samples sulfhydrated in the modified biotin switch assay and comparably sized aliquots of lysate that had not been subjected to the assay (“loads”) run on the same blot, we estimate that at least 10 to 25% of total liver GAPDH, β-tubulin, and actin are basally sulfhydrated. Of these, GAPDH shows the highest basal sulfhydration, with nearly 25% of GAPDH being sulfhydrated (Fig. 2C).
Fig. 2.
Sulfhydration is a physiologic posttranslational modification absent in CSE−/− animals. (A) Measurements of H2S production with an H2S-selective electrode indicate that CSE−/− liver cannot generate H2S. All results are presented as the mean ± SEM. ***P < 0.001. (B) Modified biotin switch assay in liver shows the presence of endogenous physiologic sulfhydration of GAPDH, β-tubulin, and actin in wild-type but not CSE−/− animals. (C) Densitometric analysis quantitating basal protein sulfhydration in liver. Data are the mean ± SEM for five to seven independent experiments with representative data shown in (B) and (D). (D) Modified biotin switch assay in liver lysates incubated with increasing exogenous doses of the CSE substrate L-cysteine for 30 min at 37°C shows a dose-dependent increase in sulfhydration in wild-type but not CSE−/− animals. The untreated lanes represent endogenous liver sulfhydration, showing a substantially larger signal for GAPDH than for β-tubulin or actin, consistent with the mean values depicted in (C). (E) Densitometric analysis of (D) quantitating changes in GAPDH sulfhydration with increasing exogenous doses of L-cysteine. Data are the mean ± SEM for five to seven independent experiments.
Addition of L-cysteine, the precursor of H2S, markedly increased sulfhydration of GAPDH, β-tubulin, and actin with half-maximal effects at about 0.6 to 1 mM (Fig. 2, D and E), comparable to physiologic concentrations of free L-cysteine in liver, which are about 0.45 mM (fig. S4). L-Cysteine failed to elicit sulfhydration of proteins in CSE−/− mice (Fig. 2D). Approximately 50% of GAPDH protein can be sulfhydrated in vitro within 30 min during cell-free catalysis by active CSE at these physiologic L-cysteine concentrations (fig. S6). Augmentation of protein sulfhydration by L-cysteine is also apparent in HEK293 cells expressing exogenous CSE. These cells contain ~46 μM basal L-cysteine, one-tenth of the physiologic levels found in the liver, and show little basal sulfhydration.
GAPDH is sulfhydrated at cysteine 150
We examined the functional effects of sulfhydration on GAPDH, the most basally sulfhydrated of the evaluated proteins. To further confirm protein S-sulfhydration, we carried out liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) on the full-length GAPDH protein immunoprecipitated from mouse liver lysate. These experiments showed that GAPDH is physiologically sulfhydrated uniquely at Cys150, the cysteine that is critical for its catalytic activity and which is physiologically S-nitrosylated (10) (Fig. 3A). This is consistent with LC-MS/MS performed on purified full-length wild-type human GAPDH protein treated with NaHS showing sulfhydration at Cys150 (Fig. 3B and fig. S7). The mass spectrometric signal for sulfhydrated GAPDH was abolished in DTT-treated samples (Fig. 3B).
Fig. 3.
Sulfhydration of GAPDH occurs at Cys150. (A) HPLC followed by LC-MS/MS on endogenous full-length GAPDH protein immunoprecipitated from mouse liver shows sulfhydration of Cys150 with an additional mass of ~32.058 daltons. m/z is the mass-to-charge ratio; the amino acid sequence surrounding Cys150 is shown at the bottom. (B) LC-MS/MS on purified full-length human GAPDH protein treated with 100 μM NaHS for 30 min at 37°C shows a mass shift consistent with Cys150 (152 in human) sulfhydration. Sulfhydration or sulfination could not be detected after treatment with 100 μM NaHS and 1 mM DTT or 500 μM H2O2, respectively. (C) Modified biotin switch assay in HEK293 cells treated with 100 μM NaHS for 30 min at 37°C. C150S mutant is not sulfhydrated. (D) Sulfhydration of GAPDH with radiolabeled [35S]cysteine and CSE at 37°C. GAPDH radiolabeling is reversed by the addition of 1 mM DTT or boiling of the CSE protein for 5 min. Data were quantified by Cherenkov scintillation counting. All results are the mean ± SEM. **P < 0.01. (E) Radiolabeling wild-type and C150S GAPDH with [35S]cysteine and CSE. C150S mutant is not labeled. All results are the mean ± SEM. ** P < 0.01.
Because cysteines can also be oxidized to cysteine-sulfinic acids (-SO2H), which have a mass ~0.067 dalton less than that of sulfhydrated cysteines, we performed high-resolution LC-MS/MS with mass accuracy of ~3 parts per million (ppm) to distinguish between these two modifications. We were unable to detect sulfination even after treatment with hydrogen peroxide (H2O2), a strong sulfinating agent (11) (Fig. 3B). We did observe the final end product of H2O2 oxidation, cysteine sulfonation (-SO3H), after treatment with H2O2 but not with NaHS (Fig. 3B). Under our experimental conditions, GAPDH Cys150 appears to preferentially undergo sulfonation by H2O2, with cysteine-sulfinic acid likely acting as a transient intermediate. This is consistent with previous studies wherein only sulfonated GAPDH could be detected after H2O2 treatment (10, 12).
We confirmed the importance of Cys150 by showing that sulfhydration of GAPDH in HEK293 cells treated with NaHS occurred with overexpressed wild-type GAPDH but not with a mutant form of GAPDH in which Cys150 was substituted with serine (GAPDH-C150S) (Fig. 3C). We established that L-cysteine mediates sulfhydration through H2S by incubating CSE with GAPDH in the presence of [35S]cysteine. This resulted in augmentation of 35S radiolabeling in wild-type GAPDH that was reversible with DTT and absent with catalytically inactive CSE (Fig. 3D). 35S radiolabeling was not apparent in the GAPDH-C150S mutant (Fig. 3E).
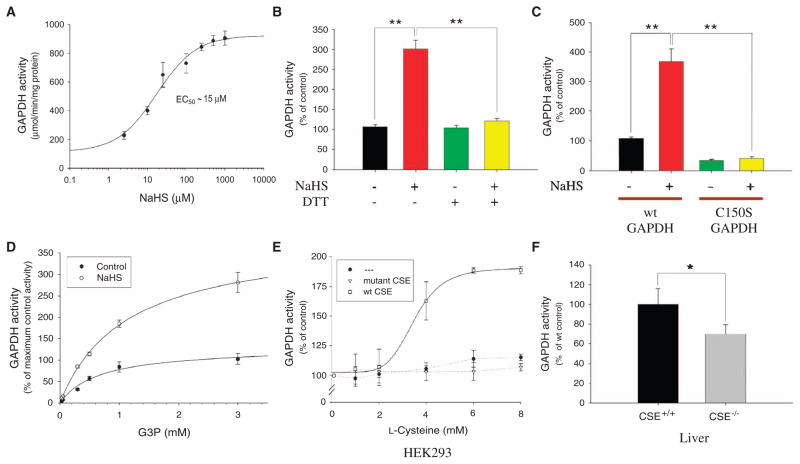
Sulfhydration physiologically increases GAPDH catalytic activity
S-Nitrosylation of GAPDH at Cys150 abolishes its catalytic activity (10). In contrast, incubation with NaHS markedly augmented GAPDH catalytic activity (Fig. 4A). Maximal enhancement of GAPDH activity at 0.1 to 1 mM NaHS was 700%, with half-maximal activation occurring at 15 μM NaHS. GAPDH activation by H2S was reversed by DTT, consistent with its mediation by sulfhydration (Fig. 4B). Although GAPDH activity is reduced in the C150S mutant, it is reliably detectable and is not increased by H2S, consistent with the H2S augmentation occurring through Cys150 (Fig. 4C). H2S increased the Vmax of GAPDH with no effect on Km (Fig. 4D). Physiologically generated H2S regulates GAPDH; in HEK293 cells, which lack endogenous CSE, L-cysteine failed to stimulate GAPDH activity (Fig. 4E). However, addition of L-cysteine to the culture media markedly increased GAPDH activity in cells expressing exogenous wild-type but not catalytically inactive CSE (Fig. 4E). To assess whether endogenous H2S generated by CSE physiologically regulates GAPDH, we monitored GAPDH activity in livers of CSE−/− mice (Fig. 4F). We observed a consistent 25 to 30% reduction in GAPDH activity in mutant mice compared to wild type, despite normal amounts of GAPDH protein (Fig. 2B).
Fig. 4.
Sulfhydration physiologically increases the catalytic activity of GAPDH. (A) GAPDH activity assayed in vitro at 37°C with increasing concentrations of NaHS. NaHS dose-dependently activates GAPDH. (B) DTT (1 mM) reverses in vitro GAPDH activation by 10 μM NaHS. All results are the mean ± SEM. **P < 0.01. (C) Wild-type versus C150S mutant GAPDH activity assayed in vitro with 15 μM NaHS. Wild-type (wt) but not C150S GAPDH is activated by NaHS. All results are the mean ± SEM. **P < 0.01. (D) GAPDH activity with increasing substrate G3P concentration with or without 10 μM NaHS. NaHS increases overall Vmax without affecting Km (~0.8 mM). (E) GAPDH activity in HEK293 cells transfected with nothing or plasmids encoding wild-type CSE or catalytically inactive CSE and incubated with increasing concentrations of L-cysteine in the media for 1 hour at 37°C. GAPDH is activated in a dose-dependent manner in the presence of wild-type CSE. (F) In vivo GAPDH activity from wild-type versus CSE−/− liver. Livers from CSE−/− mice show decreased GAPDH activity (n = 6 animals). All results are the mean ± SEM. *P < 0.05.
Sulfhydration enhances actin polymerization
Actin is sulfhydrated in vitro and in HEK293 cells with NaHS (Fig. 5, A and B), an effect reversed by treatment with DTT (Fig. 5A). Immunocytochemical analysis of the NaHS-treated cells reveals rearrangement of the actin cytoskeleton (Fig. 5C). These rearrangements appear to be present in all cells treated with the H2S donor. NaHS specifically enhances actin polymerization in vitro by more than 30%, an effect reversed by DTT (Fig. 5D), while having no substantial effect on actin depolymerization (Fig. 5E).
Fig. 5.
Sulfhydration enhances actin polymerization in vitro and in HEK293 cells. (A) Actin is sulfhydrated in vitro with 200 μM NaHS for 30 min at 37°C, an effect reversed by DTT (200 μM for 10 min). (B) NaHS (100 μM for 30 min at 37°C) sulfhydrates actin in HEK293 cells. (C) Immunocytochemical analysis of HEK293 cells treated with 100 μM NaHS for 1 hour at 37°C reveals rearrangement of the actin cytoskeleton (arrows show the extension of thin processes containing actin filaments in individual NaHS treated cells). Actin is stained in red. Scale bars, 20 μm. (D) NaHS (200 μM) enhances actin polymerization in vitro, an effect reversed by DTT (200 μM). (E) NaHS (200 μM) has no effect on actin depolymerization.
DISCUSSION
In summary, we have established that sulfhydration is a physiologic modification of many proteins that is mediated by H2S enzymatically generated from L-cysteine by CSE. Target proteins are sulfhydrated in the presence of CSE and cysteines. Presumably, CSE generates H2S to sulfhydrate the target rather than directly participating in the sulfhydration process, although it is possible that CSE binds to target proteins, analogous to the binding of NOS isoforms to S-nitrosylation targets (13). By combining densitometric analysis with the modified biotin switch assay, we estimate that 10 to 25% of endogenous GAPDH, β-tubulin, and actin are sulfhydrated in vivo. This contrasts with the much lower degree of protein S-nitrosylation thought to occur physiologically (9). Because of its greater abundance and chemical stability, sulfhydration can be readily demonstrated by mass spectrometry, which fails to identify the more labile S-nitrosylation. The mass spectrometric signal for sulfhydrated GAPDH in liver lysates was about 5% of native GAPDH. This value may represent an underestimate, because the more electronegative sulfhydrated species would be less attracted to the electronegative detector. In principle, H2S can disrupt disulfide bonds within proteins by acting as a reducing agent. However, our observations show that H2S primarily functions by sulfhydrating cysteines leading to the formation of persulfides (-SSH groups).
Sulfhydration of GAPDH physiologically augmented its catalytic activity, with substantial decreases in GAPDH activity evident in CSE−/− animals. Similarly, sulfhydration altered actin-dependent cytoskeletal rearrangements in HEK293 cells and directly enhanced actin polymerization in vitro, an effect reversed by DTT. This action is selective, because the rate of actin depolymerization was not affected by H2S donors. The EC50 (median effective concentration) of H2S for sulfhydrating protein targets is in the range of physiologic concentrations of H2S in most tissues (13).
Sulfhydration may mediate various reported physiologic actions of H2S. Physiologic relaxation of blood vessels through the endothelial-derived relaxing factor activity of H2S involves opening of adenosine 5′-triphosphate (ATP)–sensitive potassium channels (6, 14). ATP-sensitive potassium channels also mediate the cardioprotective actions of H2S, which are physiologically relevant because CSE inhibitors augment cardiac damage (15). The vasodilating actions of garlic reflect H2S acting on ATP-sensitive potassium channels (16). CSE inhibitors increase insulin secretion and H2S decreases its secretion through the same class of channels (3). We found that H2S sulfhydrates the Kir6.1 subunit of ATP-sensitive potassium channels exogenously expressed in HEK293 cells, an effect reversed by DTT (fig. S8).
H2S augments the life span of Caenorhabditis elegans through sirtuins (17), a process that may involve protein sulfhydration. Starvation (18) and glucocorticoid treatment (19) elicit a doubling of CSE activity, which in turn may lead to corresponding changes in sulfhydration of target proteins.
We have shown that a substantial number of proteins are physiologically sulfhydrated in the liver and that sulfhydration alters protein function. We propose sulfhydration as a physiologic signal that potentially influences a multitude of biological pathways. CSE is an abundant and ubiquitous protein, suggesting that H2S is physiologically generated in most organs of the body (4–6).
For a posttranslational modification to function in a signaling capacity, it must be tightly regulated. This is true for protein phosphorylation, which is controlled by both kinases and phosphatases, as well as for protein S-nitrosylation, which is regulated by both nitric oxide and the recently identified family of denitrosylating enzymes that includes the thioredoxins (20). Given the stable nature of protein S-sulfhydration and its chemical reversibility by DTT, similar desulfhydrating mechanisms could prevent the accumulation of cysteine persulfides. Another mode of regulation might involve competition between nitrosylation and sulfhydration for the same cysteine residues. Indeed, GAPDH, which can be nitrosylated (10) or sulfhydrated at Cys150 might represent one candidate for such competitive regulation.
MATERIALS AND METHODS
Animals
Biochemical experiments were performed on livers and brains removed from 1- to 2-month-old wild-type, CSE−/−, nNOS−/−, eNOS−/−, and iNOS−/− mice. Animals were maintained on a 12-hour light-dark cycle at a room temperature of 23°C, with free access to food and water. All animal use procedures were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Reagents
NaHS, L-cysteine, and glyceraldehyde-3-phosphate (G3P) as well as all other chemicals were purchased from Sigma. Antibody directed against GAPDH was purchased from Calbiochem, antibody to β-tubulin from Sigma, antibody to actin from Chemicon, antibodies to nNOS, eNOS, and iNOS from BD Biosciences, and antibody to Kir6.1 from Santa Cruz Biotechnology. Polyclonal antibody directed against CSE was prepared as follows: His-tagged CSE was produced in Escherichia coli, purified through Talon resin (Clontech), and used for antigen. Antisera were produced by a service of Research Genetics, and the sera were immunopurified. The Kir6.1 construct was obtained from Open Biosystems and the SUR2B construct was a gift from Y. Kurachi and H. Hibino (Osaka University, Japan). Both the Kir6.1 and SUR2B constructs were exogenously coexpressed in HEK293 cells to generate the ATP-sensitive potassium channel. Polyfect reagent from Qiagen was used to transfect cells with specific complementary DNA (cDNA) constructs.
Modified biotin switch (S-sulfhydration) assay
The assay was carried out as described previously (8), with modifications. Briefly, liver tissues or cells were homogenized in HEN buffer [250 mM Hepes-NaOH (pH 7.7), 1 mM EDTA, and 0.1 mM neocuproine] supplemented with 100 μM deferoxamine and centrifuged at 13,000g for 30 min at 4°C. Cell lysates (240 μg) or pure GAPDH protein (0.3 μg), treated with CSE, L-cysteine, or NaHS where indicated, were added to blocking buffer (HEN buffer adjusted to 2.5% SDS and 20 mM MMTS) at 50°C for 20 min with frequent vortexing. The MMTS was then removed by acetone and the proteins were precipitated at −20°C for 20 min. After acetone removal, the proteins were resuspended in HENS buffer (HEN buffer adjusted to 1% SDS). To the suspension was added 4 mM biotin-HPDP in dimethyl sulfoxide without ascorbic acid. After incubation for 3 hours at 25°C, biotinylated proteins were precipitated by streptavidin-agarose beads, which were then washed with HENS buffer. The biotinylated proteins were eluted by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and subjected to Western blot analysis. For quantitation of protein sulfhydration, samples were run on blots alongside total lysates (“loads”) and subjected to immunoblotting with antibodies specific to each protein. The sample to load ratio was then densitometrically analyzed with the software programs EagleSight 3.2 (Stratagene) and Odyssey 2.1 (Li-Cor).
Mass spectrometry analysis
Endogenous GAPDH protein was immunoprecipitated from 10 mg of mouse liver lysate with 4 μg of monoclonal antibody to GAPDH overnight at 4°C, after which 50 μl of protein G plus Agarose beads were added for another 2 hours. The beads were washed five times with buffer containing 20 mM tris-HCl (pH 7.4), 500 mM NaCl, and 0.1% Triton X-100. Some samples were run on an SDS-PAGE gel and Coomassie-stained to verify sample purity. The immunoprecipitated GAPDH protein bound to beads was directly subjected to tryptic digestion overnight at 37°C with 5 μg of trypsin in 20 mM ammonium bicarbonate. Samples were handled under nitrogen (N2) gas whenever possible and prevented from prolonged exposure to room air to avoid artifactual oxidation of cysteines, including the formation of disulfide bonds. The following morning, 1 μl of concentrated formic acid was added to quench the digestion; samples were then centrifuged at 13,000g for 10 min, and the supernatant was extracted and placed in 500-μl centrifuge tubes. For the pure human GAPDH protein, samples were treated with or without 100 μM NaHS or 500 μM H2O2 for 30 min at 37°C under nonreducing conditions before tryptic digestion. Samples were then evaporated in a speed vacuum and stored at −80°C. Before injection into the spectrometer, samples were dissolved in 8 μl of 2% acetonitrile with 0.1% trifluoroacetic acid. Samples were injected into the Thermo Finnegan LTQ Orbitrap XL mass spectrometer at 100,000 resolution at Johns Hopkins University School of Medicine and at Harvard Medical School mass spectrometry facilities. The instruments were calibrated with solutions of Ultramark, phosphoric acid, and bovine serum albumin (BSA) peptide standards. Mass accuracy around 3 ppm was obtained in both facilities, which was sufficient to distinguish cysteine sulfhydration (-SSH) from other oxidation reactions such as sulfination (-SO2H). Expected mass/charge ratio (m/z) represents a 2+ charged peptide.
[35S]Cysteine radioactivity assays
Purified CSE and GAPDH proteins were incubated with 1 mM L-cysteine and 40 μCi of [35S]cysteine (Perkin Elmer) without reducing agents in 10 mM potassium phosphate buffer (pH 7.4) and 15 μM PLP for 2 hours at 37°C. The reaction was then run on a nonreducing SDS-PAGE gel and Coomassie-stained, the protein load was assessed, and the radioactivity was quantitated by band excision and Cherenkov scintillation counting.
GAPDH activity assays
Purified GAPDH, HEK293 cell lysates, or liver lysates were incubated with nothing, NaHS, L-cysteine, NaCl, or H2O2 where indicated for 30 min at 37°C unless otherwise indicated in 300 μl of assay buffer [20 mM tris-HCl (pH 7.8), 100 mM NaCl, BSA (0.1 mg/ml), 1 mM NAD+ (nicotinamide adenine dinucleotide), 10 mM sodium pyrophosphate, 20 mM sodium arsenate, and 3 mM G3P], and the change in the rate of NAD+ to NADH (reduced form of NAD+) conversion was spectrophotometrically monitored in real time at 340 nm as described previously (10).
H2S measurements
H2S production was measured as described previously (6) with an H2S-selective electrode (Lazar Research Laboratories) on a Fisher Accumet Model 10 pH meter (Fisher Scientific) following the modified manufacturer’s directions. Standards were prepared from NaHS solution.
CSE activity assays
CSE activity was assessed by measuring H2S production as described previously (3). Briefly, the assay was carried out in a 250-μl reaction mixture containing 10 mM potassium phosphate buffer saline (pH 7.4), 10 mM L-cysteine, 15 μM PLP, and 10% (w/v) homogenate. High-performance liquid chromatography (HPLC) injection vials (2 ml) containing the reaction mixtures were flushed with N2 before being sealed. Reaction was initiated by transferring the vials from ice to a 37°C shaking water bath. After incubating at 37°C for 60 min, the reaction was terminated by injecting a mixture of 125 μl of 1% zinc acetate and 2.5 μl of 10 N NaOH trapping solution. The vials were horizontally shaken for another 60 min to ensure complete trapping of H2S released from the mixture. Then, 0.5 ml of water was added to each vial. Subsequently, 0.1 ml of 20 mM N,N-dimethyl-p-phenylenediamine sulfate in 7.2 M HCl was added, immediately followed by addition of 0.1 ml of 30 mM FeCl3·6H2O in 1.2 M HCl. Absorbance of the resulting solution at 670 nm was measured 20 min later with a spectrophotometer. H2S content was calculated against the calibration curve of standard H2S solutions prepared with NaHS. For in vitro experiments with purified CSE, enzyme activity was also assessed with the pyruvatelactate dehydrogenase assay system per manufacturer’s protocol (Sigma), which spectrophotometrically monitors the conversion of NADH to NAD+ at 340 nm.
L-Cysteine Measurements
L-Cysteine was measured by extracting liver or HEK293 cell samples in 2 N HCl, 1% tri-n-butyl phosphine (TBP) and then incubating at 95°C for 30 min. The samples were then cooled on ice for 1 hour and centrifuged at 13,500g for 30 min, after which the supernatants were collected and neutralized with equivalent amounts of NaOH, and the cysteine sulfhydryls were derivatized with the thiol-reactive compound 7-fluorobenz-2-oxa-1,3-diazole-4-sulfonamide (ABD-F) (Aldrich) in 0.1 M sodium borate buffer (pH 8.0) containing 1 mM EDTA and 1% TBP. The mixture was incubated at 50°C for 60 min, then cooled on ice and acidified to pH 2.0 with 10 to 12 N HCl. ABD-F becomes fluorescent only after reacting with thiols. Each sample was injected into a CROWNPAK CR(+) HPLC column (Chiral Technologies Inc.). Isocratic elution with perchloric acid (pH 1.5) was used for the separation. ABD-F–labeled L-cysteine was detected with a fluorescence detector (excitation λ = 374 nm and emission λ = 500 nm). Pure L-cysteine was used to calculate concentrations of the amino acid in actual samples.
Immunocytochemistry
HEK293 cells were incubated with 100 μM NaHS for 1 hour at 37°C after which cells were quickly rinsed with PBS and fixed with methanol for 20 min at −20°C. After blocking for 1 hour with 10% fetal bovine serum (FBS), the cells were incubated with antibody against actin overnight at 4°C. The cells were then washed with PBS, treated with secondary antibody for 1 hour, rinsed, and incubated with Vectashield mounting medium (Vector Labs) until confocal imaging, which was carried out in a Perkin-Elmer UltraView LCI confocal microscope.
Actin polymerization and depolymerization assays
To assay actin polymerization activity, we used an established pyrene-actin polymerization assay (21). Either 200 μM NaHS or 200 μM NaHS with 200 μM DTT was added to 10 μM pyrene-labeled muscle actin (Cytoskeleton Inc.). Using a spectrofluorometer (excitation λ = 360 nm and emission λ = 407 nm) (Perkin Elmer), we monitored fluorescence signals for 5 min at 30-s intervals and found no effects of NaHS on spontaneous actin polymerization. Next, we added 10-fold actin polymerization buffer [50 mM tris-HCl (pH 8.0), 500 mM KCl, 20 mM MgCl2, and 10 mM ATP] to each reaction and took fluorescence intensity readings every 30 s for 1 hour. For the actin depolymerization assay, we first prepared 20 μM F-actin stock. After diluting the F-actin stock to 4 μM with general actin buffer [5 mM tris-HCl (pH 8.0) and 0.2 mM CaCl2], we treated the stock with either NaHS or NaHS and DTT mixture as described above and immediately took fluorescence readings every 30 s for 1 hour. All experiments were performed at room temperature.
Protein purification
Mouse full-length CSE DNA was subcloned into pET-28c+ His vector (Novagen), which encodes a hexahistidine tag, and introduced into BL21 codon plus bacteria (Stratagene). Expression of CSE was induced by 0.5 mM isopropyl 1-thio-β-D-galactopyranoside (Sigma) for 12 hours at 30°C. Cells were collected by centrifugation and disrupted by sonication in medium containing 20 mM tris-HCl (pH 7.4), 15 μM PLP, 10 mM imidazole, and 400 mM NaCl. After addition of 1% Triton X-100, the suspension was cleared by centrifugation (40,000g for 15 min), and CSE was purified from the supernatant by binding to Talon resin (Clontech) according to the manufacturer’s instructions. Glutathione S-transferase (GST)–tagged GAPDH protein was prepared according to the manufacturer’s recommendations (Amersham Biosciences) and purified through glutathione-Sepharose (Amersham Biosciences) with the GST tag cleaved in the final purification step.
Cell culture
HEK293 cells were obtained from American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS, L-glutamine (300 μg/ml), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified atmosphere of 95% air and 5% CO2. Transfection of cells with Polyfect transfection reagent was carried out according to the manufacturer’s protocol.
Statistical Analysis
All data are expressed as the mean ± SEM of three independent experiments, each performed in triplicate unless otherwise indicated. Data were analyzed by unpaired Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary Material
Fig. S1. Modified biotin switch assay for protein S-sulfhydration.
Fig. S2. The non–PLP-binding CSE mutant is catalytically inactive.
Fig. S3. The modified biotin switch assay is specific for sulfhydration.
Fig. S4. L-Cysteine levels are comparable in wild-type and CSE−/− livers.
Fig. S5. Wild-type and CBS−/− livers show similar H2S production.
Fig. S6. GAPDH is substantially sulfhydrated during cell-free catalysis by CSE and L-cysteine.
Fig. S7. GAPDH sulfhydration at Cys150 with LC-MS/MS.
Fig. S8. H2S sulfhydrates ATP-sensitive potassium channels in HEK293 cells.
Acknowledgments
This study was supported by an NIH National Research Service Award (1 F30 MH074191-01A2) to A.K.M., a Medical Scientist Training Program Award (T32 GM007309) to M.M.G., an operating grant of The Canadian Institutes of Health Research to R.W., and a U. S. Public Health Service Grant (MH18501) and Research Scientist Award (DAOOO74) to S.H.S.
REFERENCES AND NOTES
- 1.Boehning D, Snyder SH. Novel neural modulators. Annu Rev Neurosci. 2003;26:105–131. doi: 10.1146/annurev.neuro.26.041002.131047. [DOI] [PubMed] [Google Scholar]
- 2.Blackstone E, Morrison M, Roth MB. H2S induces a suspended animation–like state in mice. Science. 2005;308:518. doi: 10.1126/science.1108581. [DOI] [PubMed] [Google Scholar]
- 3.Yang W, Yang G, Jia X, Wu L, Wang R. Activation of KATP channels by H2S in rat insulin-secreting cells and the underlying mechanisms. J Physiol. 2005;569:519–531. doi: 10.1113/jphysiol.2005.097642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabó C. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 2007;6:917–935. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Moore PK. Putative biological roles of hydrogen sulfide in health and disease: A breath of not so fresh air? Trends Pharmacol Sci. 2008;29:84–90. doi: 10.1016/j.tips.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, Wang R. H2S as a physiologic vasorelaxant: Hypertension in mice with deletion of cystathionine γ-lyase. Science. 2008;322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 8.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 9.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 10.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 11.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 12.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mustafa AK, Gadalla MM, Snyder SH. Signaling by gasotransmitters. Sci Signal. 2009;2:re2. doi: 10.1126/scisignal.268re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W, Zhang J, Lu Y, Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elrod JW, Calvert JW, Morrison J, Doeller JE, Kraus DW, Tao L, Jiao X, Scalia R, Kiss L, Szabó C, Kimura H, Chow CW, Lefer DJ. Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc Natl Acad Sci USA. 2007;104:15560–15565. doi: 10.1073/pnas.0705891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benavides GA, Squadrito GL, Mills RW, Patel HD, Isbell TS, Patel RP, Darley-Usmar VM, Doeller JE, Kraus DW. Hydrogen sulfide mediates the vasoactivity of garlic. Proc Natl Acad Sci USA. 2007;104:17977–17982. doi: 10.1073/pnas.0705710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller DL, Roth MB. Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:20618–20622. doi: 10.1073/pnas.0710191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson RC, Freedland RA. Factors affecting the rate of gluconeogenesis from L-cysteine in the perfused rat liver. J Nutr. 1976;106:1272–1278. doi: 10.1093/jn/106.9.1272. [DOI] [PubMed] [Google Scholar]
- 19.Heinonen K, Räihä NC. Induction of cystathionase in human foetal liver. Biochem J. 1974;144:607–609. doi: 10.1042/bj1440607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper JA, Pollard TD. Methods to measure actin polymerization. Methods Enzymol. 1982;85:182–210. doi: 10.1016/0076-6879(82)85021-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Modified biotin switch assay for protein S-sulfhydration.
Fig. S2. The non–PLP-binding CSE mutant is catalytically inactive.
Fig. S3. The modified biotin switch assay is specific for sulfhydration.
Fig. S4. L-Cysteine levels are comparable in wild-type and CSE−/− livers.
Fig. S5. Wild-type and CBS−/− livers show similar H2S production.
Fig. S6. GAPDH is substantially sulfhydrated during cell-free catalysis by CSE and L-cysteine.
Fig. S7. GAPDH sulfhydration at Cys150 with LC-MS/MS.
Fig. S8. H2S sulfhydrates ATP-sensitive potassium channels in HEK293 cells.