Abstract
It is well appreciated that several neurohormones and signaling cascades are activated that promote long-term deterioration of cardiac function and structure. Activation of the renin-angiotensin-aldosterone system (RAAS) and the adrenergic system is closely related to heart failure. Common gene variants that encode neurohormonal, adrenergic and intracellular proteins have been demonstrated to modulate the course and consequences of heart failure. However, the literature is replete with conflicting results and it remains uncertain as to whether particular gene variants predispose heart failure. Therefore, the main purpose of this review was to discuss the effects of single nucleotide polymorphisms (SNPs) that are located in genes encoding elements of the RAAS and the adrenergic system on the predisposition to and survival from heart failure. Most studies indicate that common SNPs encoding elements of the RAAS and the adrenergic system do not predispose individuals to heart failure. Conversely, it has been demonstrated that ARB1 Arg389Gly, GRK5 Gln41Leu, ACE I/D, CYP11B2 C-344T and AGTR1 A+1166C modulate pharmacological responses and have a considerable impact on cardiac-related survival. It should not be expected, however, that a single polymorphism determines survival, given that multiple gene products and environmental factors contribute to the pathogenesis of heart failure. Therefore, future studies should consider the interaction effects of multiple genes in populations that are as homogeneous as possible with respect to environmental characteristics.
Keywords: Cardiac failure, Polymorphisms, Susceptibility, Mortality
INTRODUCTION
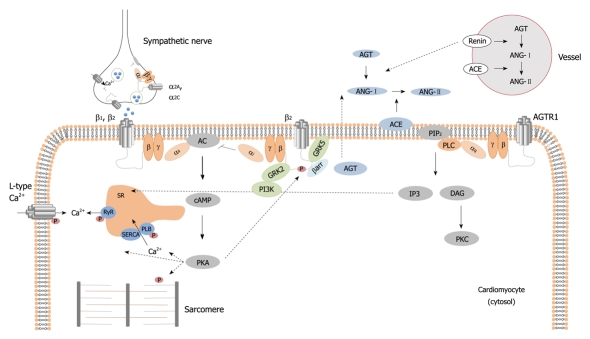
In recent years, the incidence of heart failure has continued to increase along with sustained elevated rates of morbidity and mortality[1]. It is now well appreciated that several neurohormonal and signaling cascades are activated that promote long-term deterioration of cardiac function and structure[2] (Figure 1). These changes, collectively referred to as cardiac remodeling, are modulated by genetic factors[3].
Figure 1.
Adrenergic and renin-angiotensin-aldosterone system signaling pathways. AC: Adenylyl cyclase; ACE: Angiotensin-converting enzyme; AGT: Angiotensinogen; ANG: Angiotensin; cAMP: Cyclic Adenosine Monophosphate; βarr: β-arrestin; DAG: Diaglycerol; GRK2: G-protein coupled receptor kinase 2; GRK5: G-protein coupled receptor kinase 5; IP3: Inositol trisphosphate; PI3K: Phosphoinositide 3-kinase; PIP2: Phosphatidylinositol 4,5-biphosphate; PKA: Protein kinase A; PKC: Protein kinase C; PLC: Phospholipase C; SR: Sarcoplasmic reticulum.
The renin-angiotensin-aldosterone system (RAAS) plays a pivotal role in the processes of heart failure. In response to sustained activation of the RAAS, the angiotensin-II receptors are deregulated in the human failing heart[4]. Several downstream intracellular signaling effectors are overexpressed and activated in tandem with cardiac hypertrophy[5]. The adrenergic system is also closely related to heart failure. Adrenergic receptors are deregulated in human heart failure in concert with impaired ventricular contraction and relaxation[6].
It seems that the molecular portrayal of heart failure is incomplete as molecular and genetic studies continue to recognize genes and signaling cascades that participate in the development and progression of heart failure[7]. In addition to the discovery of disease-causing (rare) mutations, common variants in genes that encode neurohormonal, adrenergic, intracellular and interstitial proteins have been demonstrated to modulate the course and consequences of heart failure. The literature, however, is replete with conflicting results and intense debate exists as to whether gene polymorphisms determine susceptibility to developing heart failure. Considering the prominent role of the RAAS and adrenergic receptors in the pathophysiology of heart failure, this review focuses on functional single nucleotide polymorphisms (SNPs) that appear to be related to the predisposition to and the survival from heart failure.
FUNCTIONAL PROPERTIES OF GENE POLYMORPHISMS
β-adrenergic receptors
The gene encoding the β1-adrenergic receptor is located on chromosome 10q24-26 and contains no introns. Several SNPs have been described in this gene, with two having been demonstrated to be functional and relevant for heart failure[8,9]. One of these polymorphisms is derived from a transition between the amino acids arginine and glycine in residue 389 (Arg389Gly)[10]. Given that this variant is located in the carboxyl terminal, it follows that variant alleles may have a distinct binding to G proteins and thus may have distinct signaling characteristics. The other variant is located at residue 49, wherein amino-acid serine is replaced by glycine (Ser49Gly)[11]. The functional properties of these SNPs have been demonstrated in transgenic animals as well as studies in vitro, ex vivo and in vivo. For example, numerous in vitro studies have demonstrated that cells expressing the human variant Arg389 have increased adenylyl cyclase activity in response to agonists compared to those expressing Gly389[10,12]. Upon agonist activation, the former variant appears more prone to desensitization and may predispose to heart failure under certain conditions; e.g. increased catecholamine stimulation[13]. These findings have been substantiated in numerous ex vivo and in vivo human and animal studies[10,14-16] in which mice expressing Arg389 have increased contractile responses to agonists and exhibited faster and greater desensitization upon activation[17]. Nonetheless, most human studies have shown identical hemodynamic responses to exercise in both variants[18,19]. While not in all studies, it has been demonstrated that Gly49 has similar characteristics to Arg389, showing increased contractile responses and desensitization upon stimulation with agonists[11,20]. Together, these data indicate that individuals who harbor Arg389 and Gly49 may have increased cardiac remodeling under adverse conditions.
The gene encoding the β2-adrenergic receptor is located on chromosome 5q31-32. Among numerous gene variants, two non-synonymous polymorphisms, namely Arg16Gly and Gln27Glu, have to some extent been related to cardiac functional changes[8,9]. Although consistent evidence has shown that these SNPs have no influence on agonist-mediated contractile responses, in vitro studies have reported that Gly16 and Gln27 are more prone to desensitization than Arg16 and Glu27[21]. Thus, these variants might be relevant to heart failure. However, some ex vivo and in vivo human studies showed that Gly16 is more resistant to agonist-mediated desensitization than Arg16[22]. The contrasting results are not easily explained, but may stem from the predominant desensitization of Gly16 from endogenous catecholamines[9,23]. In contrast, another SNP (Thr164Ile) has been shown to modulate cardiac contractile responses in vitro and in vivo. Consistent evidence indicates that Ile164 presents reduced basal and agonist-mediated intracellular effector activation, contractile response and heart rate as compared to Thr164[24-26]. However, most humans possess two copies of threonine, casting doubts as to whether this variant is relevant to heart failure.
α-adrenergic receptors
The α1-adrenergic receptors encompass three subtypes, the genes for which are located in different chromosomes. Nonetheless, only α1A and α1B receptors seem to be translated and are functional in the human heart[27]. Several gene variants have been reported, though many are uncommon or non-functional (for a review, see reference[28]). A common variant of α1A receptors, resulting from the substitution of arginine for cysteine, is located in the residue 347 (Arg347Cys) and has been expressed in vitro[29]. There were no differences with respect to antagonist and agonist binding affinities, intracellular calcium concentrations or receptor desensitization upon stimulation with noradrenaline. In contrast, another gene polymorphism located in the intracellular loop seems functional; the Gly247Arg variant enhances G-protein binding, inositol phosphate production and cellular growth[30]. Moreover, non-synonymous polymorphisms located in transmembrane domains have been reported to decrease ligand binding and receptor activation, including the Arg166Lys and Val311Ile[30]. To our knowledge, however, these three polymorphisms have never been studied in patients with heart failure.
The α2 adrenergic receptors also comprise three subtypes. The α2A and α2c are encoded by single genes and are both important in controlling noradrenaline release in pre-synaptic nerve terminals[31]. Among the α2A receptors, a particular gene polymorphism appears to be functional and has been described in the context of heart failure. This polymorphism is located in a cytoplasmic domain and occurs in amino acid 251, where an asparagine or lysine is present (Asn251Lys)[32]. Transfected cells with Lys251 have shown enhanced coupling to Gi-proteins, effective inhibition of adenylyl cyclase and activation of mitogen-activated protein kinases[32]. Thus, this variant could reduce noradrenaline release and confer protection under conditions of increased catecholamine stimulation, such as heart failure. In α2C receptors, a polymorphism has been demonstrated to be functional and related to heart failure. The Del322-325 derives from a 12 nucleic acid deletion and generates a receptor that lacks four amino acids (glycine, alanine, glycine, proline) in the third intracellular loop[33]. As a result, the generated receptors have reduced ligand-binding affinity, adrenaline-promoted coupling to Gi-proteins, inositol phosphate production and stimulation of mitogen-activated protein kinases[33].
G-protein receptor kinases
Besides second-messenger protein kinases, the particular class of G-protein coupled receptors kinases (GRK) modulates agonist-promoted desensitization and internalization of G-protein coupled receptors[34]. Among seven isoforms, GRK2 and GRK5 predominate in the myocardium and have been demonstrated to be important during heart failure[34]. While GRK2 showed no non-synonymous polymorphisms, four non-synonymous variants have been found in GRK5[35]. Of these, the substitution of leucine by glutamine in amino acid 41 (Gln41Leu) enhanced isoproterenol-promoted desensitization and decreased signaling of β1-adrenergic receptors. Consistent with these observations, the Leu41 allele has shown a protective effect against experimental cardiomyopathy induced by catecholamines. Nonetheless, this gene polymorphism is extremely uncommon in Caucasians[35].
RAAS POLYMORPHISMS AND HEART FAILURE
Activation of the RAAS is one of the earlier and critical steps in heart failure. Although important to maintaining circulatory homeostasis, unremitting activation imposes a significant load on the heart and activates an immense and intricate signaling pathway[36]. As a consequence, several elements of this system are deregulated in heart failure, including angiotensinogen, angiotensin-converting enzyme (ACE), angiotensin-II receptors and aldosterone[2,4].
Angiotensinogen
The gene encoding angiotensinogen is located on chromosome 1q42-q43 and contains various gene polymorphisms[37]. Two of these polymorphisms have been investigated particularly in heart failure[38]. A polymorphism on exon 2 causes the substitution of methionine to threonine in amino acid 235 (M235T). This polymorphism is in close linkage disequilibrium with another polymorphism in the promoter region, the -6 G/A variant, and is associated with plasma angiotensinogen concentrations[39]. Another polymorphism results from a threonine to methionine substitution in position 174. This polymorphism has been related to changes in cardiac function in patients with heart failure, although its functional implications remain unknown[38].
ACE
The ACE converts angiotensin-I into angiotensin-II, which activates angiotensin-II receptors for modulating various cardiovascular responses, including vasoconstriction and cardiac growth[36]. The gene that encodes ACE is located on chromosome 17q23. Despite the fact that more than 100 polymorphisms have been found in the ACE gene, a variant based on the presence (insertion) or absence (deletion) of 287 base pairs in intron 16 have been described and tested more than any other polymorphism[37]. An initial study demonstrated that the ACE I/D polymorphism accounts for more than 40% of the total variance in serum ACE, with subjects harboring D alleles having increased concentrations[40]. Nonetheless, its functional role in vivo is still under strong debate, as numerous studies have reported no association between ACE I/D genotypes and hypertension[41].
Aldosterone
Another important element is aldosterone, whose synthesis is stimulated by angiotensin-II[42]. Aldosterone is synthesized in the adrenal gland by aldosterone synthase, whose gene (CYP11B2) is located on chromosome 8q22[43]. A common polymorphism at position -344 within the promoter region (C-344T) has been described to be functional in vitro and determines concentrations of aldosterone[44,45]. In particular, the -344C allele has four times more binding affinity to the steroidogenic transcription factor 1 than the T allele and has been associated with increased production of aldosterone[45,46].
Angiotensin-II receptors
Two distinct subtypes of angiotensin-II receptors mediate the predominant actions of the renin-angiotensin system[47]. The gene encoding the type I receptor (AGT1R) is located on chromosomes 3q21-3q25. A particular gene polymorphism (A+1166C) in the 3’ untranslated region has been demonstrated to determine receptor expression and has been associated with hypertension. In particular, the presence of the +1166C allele seems to eliminate a particular microRNA (mir-155) binding site, preventing the receptors downregulation that occurs in the +1166A allele. The net result is increased receptor expression in +1166C[48]. Aligned with these observations, the +1166C allele has been associated with hypertension[49].
THE ROLE OF GENE POLYMORPHISMS ON THE PREDISPOSITION TO HEART FAILURE
To determine whether gene polymorphisms increase the chances of developing heart failure, several studies have investigated whether the proportion of functional alleles differs among affected and unaffected populations. Although contrasting results are abundant, most studies have found no differences in allele frequencies among patients and the general population (Tables 1 and 2).
Table 1.
Effect of adrenergic gene polymorphisms on the predisposition to patients with heart failure
| Gene/SNP | Ref. | Cases/controls | Ethnic group | Association | Risk allele frequency |
| β adrenergic receptor type 1 | |||||
| Arg389Gly | [50] | 201/141 | Mixed | No | 0.74 vs 0.76 |
| [51] | 256/230 | Italian | No | 0.69 vs 0.73 | |
| [52] | 189/378 | Italian | Yes | 0.74 vs 0.67 | |
| [53] | 91/119 | Japanese | No | 0.80 vs 0.81 | |
| [54] | 78/84 | African-American | No | 0.52 vs 0.56 | |
| [54] | 81/105 | Caucasians | No | 0.74 vs 0.76 | |
| [55] | 426/395 | French | No | 0.77 vs 0.75 | |
| [56] | 403/429 | South Africans | No | 0.70 vs 0.69 | |
| [57] | 260/230 | Italian | No | 0.69 vs 0.73 | |
| Ser49Gly | [50] | 201/141 | Mixed | No | 0.15 vs 0.15 |
| [58] | 184/77 | Swedish | No | 0.18 vs 0.13 | |
| [52] | 189/378 | Italian | Yes | 0.14 vs 0.08 | |
| [53] | 91/119 | Japanese | No | 0.16 vs 0.16 | |
| β adrenergic receptor type 2 | |||||
| Gly16Arg | [51] | 256/230 | Italian | No | 0.61 vs 0.60 |
| [52] | 189/378 | Italian | Yes | 0.67 vs 0.59 | |
| [59] | 259/212 | Mixed | No | 0.60 vs 0.63 | |
| [60] | 520/328 | Mixed | Yes | 0.62 vs 0.59 | |
| Gln27Glu | [51] | 256/230 | Italian | No | 0.32 vs 0.31 |
| [52] | 189/378 | Italian | No | 0.38 vs 0.33 | |
| [59] | 259/212 | Mixed | No | 0.44 vs 0.42 | |
| [60] | 520/328 | Mixed | No | 0.42 vs 0.40 | |
| [61] | 58/111 | Canadians | No | 0.41 vs 0.47 | |
| Thr164Ile | [52] | 189/378 | Italian | No | 0.02 vs 0.01 |
| [59] | 259/212 | Mixed | No | 0.02 vs 0.01 | |
| [60] | 520/328 | Mixed | No | 0.01 vs 0.01 | |
| 5’LC-Arg19Cys | [52] | 189/378 | Italian | No | 0.36 vs 0.31 |
| α adrenergic receptor type 2 | |||||
| Del 322-325 | [53] | 91/119 | Japanese | Yes | 0.04 vs 0.11 |
| [54] | 78/84 | African-Americans | Yes | 0.61 vs 0.41 | |
| [54] | 81/105 | Caucasians | No | 0.10 vs 0.04 | |
| [56] | 403/429 | South Africans | No | 1.00 vs 1.00 | |
| G-protein receptor kinase 5 | |||||
| Gln41Leu | [35] | 242/107 | African-Americans | No | 0.76 vs 0.77 |
| [35] | 568/406 | European-Americans | No | 0.98 vs 0.99 |
SNP: Single nucleotide polymorphism.
Table 2.
Effect of renin-angiotensin-aldosterone system gene polymorphisms on the predisposition to patients with heart failure
| Gene/SNP | Ref. | Cases/controls | Ethnic group | Association | Risk allele frequency |
| Angiotensin-converting enzyme | |||||
| I/D | [62] | 214/79 | Caucasian | Yes | 0.58 vs 0.56 |
| [63] | 193/77 | Swedish | No | 0.57 vs 0.56 | |
| [64] | 229/230 | Italian | No | 0.58 vs 0.60 | |
| [65] | 99/364 | Caucasian | No | 0.57 vs 0.54 | |
| [66] | 157/225 | South-Africans | No | 0.64 vs 0.69 | |
| [67] | 90/287 | Czech | No | 0.54 vs 0.57 | |
| [68] | 79/102 | Chinese Han | No | 0.43 vs 0.40 | |
| [69] | 104/183 | Chinese | No | 0.36 vs 0.37 | |
| [70] | 433/401 | French | No | 0.54 vs 0.57 | |
| [71] | 88/122 | Japanese | No | 0.39 vs 0.36 | |
| [61] | 58/111 | Canadians | No | 0.64 vs 0.62 | |
| Angiotensinogen | |||||
| M235T | [66] | 157/225 | South-Africans | No | 0.83 vs 0.87 |
| [72] | 158/200 | Czech | No | 0.51 vs 0.43 | |
| [72] | 40/63 (women) | Czech | Yes | 0.56 vs 0.39 | |
| [70] | 433/401 | French | No | 0.40 vs 0.43 | |
| [71] | 88/122 | Japanese | No | 0.80 vs 0.80 | |
| [61] | 58/111 | Canadians | Yes | 0.48 vs 0.31 | |
| T174M | [70] | 433/401 | French | No | 0.12 vs 0.14 |
| [71] | 88/122 | Japanese | No | 0.05 vs 0.10 | |
| [61] | 58/111 | Canadians | Yes | 0.18 vs 0.10 | |
| G(-6)A | [72] | 158/200 | Czech | No | 0.59 vs 0.58 |
| Angiotensin-II type 1 receptor | |||||
| A1166C | [70] | 433/401 | French | No | 0.28 vs 0.28 |
| [61] | 58/111 | Canadians | No | 0.31 vs 0.33 | |
| [73] | 193/77 | Swedish | No | 0.29 vs 0.30 | |
| A-153G | [70] | 433/401 | French | No | 0.16 vs 0.19 |
| Aldosterone | |||||
| T-344C | [66] | 157/225 | South-Africans | No | 0.21 vs 0.18 |
| [70] | 433/401 | French | No | 0.43 vs 0.46 | |
| [61] | 58/111 | Canadians | No | 0.39 vs 0.45 |
SNP: Single nucleotide polymorphism; T174M: Threonine to methionine substitution in position 174.
Adrenergic receptors and G-protein receptor kinases
Given their role in β1-adrenoceptors signaling and receptor desensitization, Arg389 and Gly49 alleles have been hypothesized to predispose heart failure. However, most studies to date have found no differences in allele frequencies between patients and healthy individuals[50-58]. Likewise, numerous investigations have demonstrated a similar genotype within β2 adrenergic receptors in heart failure patients and the general population[51,59]. On the other hand, two studies reported that Gly16 and Glu27 were more frequent in end-stage heart failure patients[52,60]. Given that these variants are more resistant to desensitization, patients could be more exposed to intracellular signaling and maladaptive cardiac hypertrophy. However, this association remains to be established. In addition, a foremost study advocated that genetic variation in α2C adrenoreceptors might predispose people to heart failure[54]. African Americans who are homozygous for the Del322-325 allele presented five times the odds for developing heart failure. The odds were augmented markedly among persons who were homozygous for both Del322-325 and Arg389. It makes reasonable sense that these variants predispose for heart failure by combining increased noradrenaline release with increased adrenergic signaling. However, more recent studies have not replicated these observations[53,56]. Furthermore, the GRK5 functional polymorphism also seems unrelated to heart failure predisposition. In a leading study, the proportion of Gln41 alleles was similar among African- and European-American patients and controls[35]. Together, these data show that most, if not all, polymorphisms play no role in the predisposition to heart failure. These observations have been substantiated in a recent large-scale genome-wide scan association study[74]. This study included more than 23 000 individuals, among whom almost 3000 developed heart failure during a 13-year follow-up, and assessed almost 2.5 million markers. Only two markers exceeded the genome-wide threshold for significance. One SNP was found in individuals with European ancestry and was located near the ubiquitin-specific protease gene 3. The other one was detected in individuals of African ancestry and was located close to the leucine-rich repeats and immunoglobulin-like domains 3 gene. Fourteen additional loci were identified, one of which is located in the GNA15 gene, which codes for a Gq-protein. The Gq-proteins are important mediators of α-adrenergic, endothelin and angiotensin-receptors signal transduction.
RAAS
The association between the ACE I/D polymorphism and heart failure was suggested in a leading study[62]. In that study, the proportion of ACE DD genotype was more than 50% higher in end-stage heart failure patients than in healthy controls. However, these observations have been called into question by numerous subsequent studies[63-69,75]. Likewise, the angiotensinogen M235 variant has not been associated with a predisposition to heart failure in most studies[61,66,70-72]. Similar results were reported for T174 and -6G/A variants[70-72]. One single study reported an association for these variants, although its small sample size precludes solid conclusions[61]. The aldosterone synthase polymorphism (-344C) is not more frequent in patients than controls[66,61,70], and neither are the AGT1R (+1116C) and AGT2R (G1675) polymorphisms[61,70,73]. Collectively, these data indicate that gene polymorphisms encoding elements of the RAAS also do not indicate predisposition for heart failure, at least in Caucasians.
THE ROLE OF GENE POLYMORPHISMS IN SURVIVAL FROM HEART FAILURE
Although most studies have shown that gene polymorphisms do not increase the risk for heart failure, functional gene polymorphisms might determine survival once heart failure develops. Stimulation of neurohormonal and interstitial proteins increases once the disease onsets. Hence, instead of predisposing to the disease, genetic variants might compromise heart function and survival by increasing function and expression of adverse proteins and/or by suppressing the favorable ones. However, even here the results are somewhat diverse and conflicting (Tables 3 and 4).
Table 3.
Effect of adrenergic gene polymorphisms on survival of patients with heart failure
| Ref. | Sample, design1 | Endpoints | Follow-up (mo) | Mortality rate (%) | SNP | Main findings |
| β adrenergic receptor type 1 | ||||||
| [15] | 10402 | AD, HZ | 48 | 19 | Arg389Gly | Increased survival in Arg389 homozygous treated with bucindolol |
| [58] | 1843 | AD, HT | 24-60 | 38 | Ser49Gly | Decreased survival in Ser49Ser patients |
| [76] | 6002 | AD, HZ | 7-17 | 26 | Arg389Gly | No association with endpoints |
| [50] | 2013 | AD, CD | 18-62 | 28 | Arg389Gly, Ser49Gly | Increased survival in Arg389 allele carriers on high dose beta-blockers |
| [77] | 4443 | CD, HT | 41 (median) | 25 | Arg389Gly, Ser49Gly | No association with endpoints |
| [78] | 2273 | AD, HT | 48 | 18 | Arg389Gly, Ser49Gly | No association with endpoints |
| [79] | 6373 | AD, HT | 35 (mean) | 23 | Arg389Gly, Ser49Gly | No association with endpoints |
| β adrenergic receptor type 2 | ||||||
| [59] | 2593 | AD, HT | 22 (mean) | Gly16Arg, Gln27Glu, Thr164Ile | Increased risk of death in Thr164Ile patients | |
| [77] | 4443 | CD, HT | 41 (median) | 25 | Gly16Arg, Thr164Ile | No association with endpoints |
| [78] | 2273 | AD, HT | 48 | 18 | Gly16Arg, Gln27Glu | Increased risk of death (haplotype) |
| [80] | 313 | AD, HT | 24 | 3 | Thr164Ile | Worsening HF in Thr164Ile patients |
| [81] | 4433 | AD | 36 (median) | Thr164Ile | Improved survival in Thr164Thr patients treated with beta-blockers | |
| α adrenergic receptor type 2 | ||||||
| [78] | 2273 | AD, HT | 48 | 18 | Del322-325 | No association with endpoints |
| [79] | 6373 | AD, HT | 35 (median) | 23 | Del322-325 | No association with endpoints |
| [82] | 3453 | AD, HT | 60 (mean) | 18 | Del322-325 | Reduced risk of death and end-points |
| G-protein kinase receptor 5 | ||||||
| [35] | 3752 | AD, HT | 30 (mean) | Gln41Leu | Increased survival in Leu41 African-American patients treated with beta-blockers. No impact on Caucasians | |
Study design;
Placebo controlled randomized trial;
Non-randomized, single group assignment. SNP: Single nucleotide polymorphism; AD: All cause mortality; CD: Cardiac mortality; HT: Heart transplantation; HZ: Hospitalizations.
Table 4.
Effect of renin-angiotensin-aldosterone system gene polymorphisms on survival of patients with heart failure
| Ref. | Sample, design1 | Endpoints | Follow-up (mo) | Mortality rate (%) | SNP | Main findings |
| Angiotensinogen | ||||||
| [75] | 822 | AD, HZ | 12 | 24 | M235T | No association with endpoints |
| [78] | 2273 | AD, HT | 48 | 18 | M235T | No association with endpoints |
| [83] | 4513 | AD | 48 | 49.7 | M235T, T174M | Increased mortality in 174M patients |
| Angiotensin-converting enzyme | ||||||
| [75] | 822 | AD, HZ | 12 | 24 | I/D | No association with endpoints |
| [73] | 1943 | AD, HT | 60 | 42 | I/D | Increased risk of death in DD patients |
| [78] | 2273 | AD, HT | 48 | 18 | I/D | No association with endpoints |
| [84] | 3283 | AD, HT | 3-38 | 23 | I/D | Decreased survival in D allele patients untreated with beta-blockers. No differences in treated patients |
| [85] | 4793 | AD, HT | 3-62 | 28.6 | I/D | Decreased survival in D allele patients untreated with β-blockers. No differences in treated patients and decreased impact with high dose ACE inhibitors |
| [86] | 3233 | AD, HZ | 10 (median) | 9.6 | I/D | Associated with severity of disease (NYHA class) |
| Angiotensin-II receptor type 1 | ||||||
| [73] | 1943 | AD, HT | 60 | 42 | A1166C | Not associated with end-points. Increased risk of mortality as haplotype (ACE DD) |
| [75] | 822 | AD, HZ | 12 | 24 | A1166C | No correlation with mortality rate |
| [78] | 2273 | AD, HT | 48 | 18 | A1166C | No association with endpoints |
| Aldosterone | ||||||
| [87] | 3542 | AD, HZ | 12 | 3.4 | -344 T/C | Decreased survival in C allele patients. Isosorbide dinitrate and hydralazine improved composite score in TT genotype but had no impact on C allele |
Study design;
Placebo controlled randomized trial;
Non-randomized, single group assignment. SNP: Single nucleotide polymorphism; AD: All cause mortality; CD: Cardiac mortality; HT: Heart transplantation; HZ: Hospitalizations; ACE: Angiotensin-converting enzyme.
Adrenergic receptors and G-protein receptor kinases
Several lines of evidence have led to the belief that genes encoding β1-receptors have no effect on clinical endpoints, including survival, hospitalization or heart transplantation[76-78]. Nonetheless, many clinical trials have not accounted for mortality for cardiac reasons and/or have not controlled for potential pharmacological confounding effects. For example, Liggett et al[15] demonstrated that patients who are homozygous for Arg389 and treated with bucindolol survive longer than patients treated with a placebo. In contrast, patients with Gly389 have not benefited from treatment with bucindolol. Biolo et al[50] also demonstrated that treatment dosage modulates survival in patients harboring Arg389. Once again, all patients carrying Gly389 survived irrespective of treatment conditions. These data indicate that medication attenuates the negative impact that variant Arg389 has on survival. In contrast, mounting evidence shows that Gly49 has no impact on survival or heart transplantation endpoints, irrespective of medication[77,79]. Moreover, most studies to date have reported that Gly16 and Glu27 are not associated with survival related to heart failure[59,77,79]. Nonetheless, it may be that, rather than single polymorphisms, the haplotype may determine the chances of survival. Indeed, one investigation reported that patients carrying two copies of Gly16 and Gln27 have an increased risk of adverse events[78].
A number of studies have reported that individuals carrying the Thr164Ile variant have a worse prognosis than those carrying the Thr164 homozygous variant[59,80]. Even though Thr164Thr patients have improved survival rates upon treatment with β-blockers, individuals who carry the Thr164Ile variant seem to present the opposite response[81].
Similarly, GRK5 gene variants are associated with different responses to medication and impact on survival. Liggett et al[35] showed that survival times were longer in Gln41 patients under treatment with β-blockers than medication naïve patients. Nonetheless, survival times were similar to Leu41 carriers who were not under treatment. These findings prompted the authors to conclude that the Leu41 variant provides similar effects as β-blockers (genetic β-blockade). A large prospective study supported these outcomes[88]. Among untreated African Americans, Leu41 carriers had longer survival times than those who were Gln41 homozygous. On the contrary, there were no differences in survival times among African Americans treated with β-blockers, indicating that medication attenuated the negative impact of the Gln41 variant. Even though GRK5 variants seem to be associated with pharmacological responses and survival rates in African-Americans, they are uncommon in Caucasians. However, since recent evidence points out that Caucasians who are homozygous for Gln41 and Gly389 have shown prolonged survival with treatment, as is the case with African Americans, future studies should consider gene-gene interaction effects.
Contrasting results have been reported for α2C adrenoceptor polymorphisms. The variant Del322-325 was surprisingly associated with a decreased event rate and reduced death rate in patients with dilated cardiomyopathy[82]. However, more than 90% of the patients were Caucasians, and similar to previous studies, had no individuals who were homozygous for the deletion variant. More recent studies disputed these observations, among which there was no association between this variant and survival related to heart failure[75,78,79]. It is not reasonable to expect that a single polymorphism will exert a marked influence on survival, given that multiple gene products and environmental factors contribute to the pathogenesis of the disease. Therefore, future studies should consider the combined effect of several genes involved in the progression of heart failure in populations that are as homogeneous as possible in regard to their environmental characteristics.
RAAS
Even though the RAAS genes are not associated with a predisposition for heart disease, numerous genes encoding elements of the RAAS have been associated with varied responses to pharmacological treatment and survival. One such example concerns the ACE I/D polymorphism. An initial investigation reported that survival was lower in patients harboring the DD genotype[63] and numerous subsequent studies have supported these observations. McNamara et al[84,85] demonstrated that survival was lower in untreated DD patients, albeit similar among those who received β-blockers and ACE inhibitors. Hence, in tandem with other polymorphisms, the adverse effects induced by the ACE DD genotype seem to be attenuated by standard heart failure medication. However, other studies have not replicated these findings[75,78,86]. The same group showed that the aldosterone -344C polymorphism has a scaled impact on survival, being considerably poorer in CC genotype patients[87]. Unlike other polymorphisms, TT patients, rather than CC, were the ones who benefited the most with isosorbide dinitrate and hydralazine. Therefore, individuals with the CC genotype might be at particular risk of death once they develop heart failure. Once more, allele frequencies were different between African-Americans and Caucasians.
The angiotensin-II receptor polymorphisms also are expected to have a negative impact upon survival following heart failure, because their sustained activation has serious cellular and cardiovascular consequences. Nonetheless, the results are contradictory. Evidence exists that 1166C is associated with a more severe disease condition and increased risk of death when combined with ACE DD[73]. However, in a recent study, survival and heart transplantation endpoints were not associated with either 1166C or 1166A[78]. In addition, studies are unanimous in demonstrating that the angiotensinogen M235T polymorphism does not influence survival from heart failure[75,78,83]. On the other hand, the M174 allele was associated with increased mortality[83]. Overall, angiotensinogen polymorphisms seem to have a slight effect on survival, however, thus far, studies have included somewhat small samples and are of insufficient number for drawing definitive conclusions. In addition, some of these studies have not taken into consideration the potential attenuating effects that medication has on certain gene variants.
CONCLUSION
Although gene products related to the RAAS and the adrenergic system are strongly implicated in the pathogenesis of heart failure, functional genetic variations that enhance or suppress their function and/or expression do not seem to predispose the development of heart failure. These observations are not entirely unexpected as the production of these end products markedly increases once heart failure onsets. In contrast, when the disease develops, genetic variants that adversely modify the function/expression of proteins are expected to lead to a worse outcome and possibly to poorer prognosis. Indeed, numerous studies have demonstrated that some SNPs not only modify responses to medication but also have implications for survival related to heart failure. This has been the case for ARB1 Arg389Gly, GRK5 Gln41Leu, ACE I/D, CYP11B2 C-344T and AGTR1 A+1166C.
However, conflicting results abound in the literature, wherein positive associations reported in initial studies often were not supported by subsequent investigations. Several reasons may explain these discrepant results, including limitations in design, technical procedures (mistyping) and analysis (unconditional). Numerous studies have included small sample sizes, which may compromise the power to detect common small effects in genetic association studies of multifactorial traits, such as heart failure[38]. In addition, numerous studies have not matched or grouped sample cohorts according to the severity of heart failure. With respect to survival, many studies were retrospective and have not controlled for potential confounding pharmacological effects. As outlined previously, standard heart failure medications (e.g. β-blockers and ACE inhibitors) appear to offset the adverse impact that some gene variants exert on survival. Furthermore, several studies involved various SNPs and multiple testing may generate false-positive results[8]. On the other hand, the impact of one SNP on survival is expected to be small and can be counteracted by the presence of other SNPs, at least in theory. Given that multiple gene products and environmental factors contribute to heart failure, future studies should consider studying haplotypes and the interacting effects of multiple genes implicated in the pathogenesis of heart failure. Future studies should be carried out in populations that are as homogeneous as possible regarding etiology, gender, race and environmental characteristics.
Footnotes
Peer reviewer: Folkert Wouter Asselbergs, MD, PhD, Department of Cardiology, University Medical Center Groningen, Jan Lutmastraat 29A, 9718 LB Groningen, The Netherlands
S- Editor Cheng JX L- Editor Lutze M E- Editor Zheng XM
References
- 1.Bleumink GS, Knetsch AM, Sturkenboom MC, Straus SM, Hofman A, Deckers JW, Witteman JC, Stricker BH. Quantifying the heart failure epidemic: prevalence, incidence rate, lifetime risk and prognosis of heart failure The Rotterdam Study. Eur Heart J. 2004;25:1614–1619. doi: 10.1016/j.ehj.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 2.Mann DL, Bristow MR. Mechanisms and models in heart failure: the biomechanical model and beyond. Circulation. 2005;111:2837–2849. doi: 10.1161/CIRCULATIONAHA.104.500546. [DOI] [PubMed] [Google Scholar]
- 3.Cohn JN, Ferrari R, Sharpe N. Cardiac remodeling--concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. Behalf of an International Forum on Cardiac Remodeling. J Am Coll Cardiol. 2000;35:569–582. doi: 10.1016/s0735-1097(99)00630-0. [DOI] [PubMed] [Google Scholar]
- 4.Haywood GA, Gullestad L, Katsuya T, Hutchinson HG, Pratt RE, Horiuchi M, Fowler MB. AT1 and AT2 angiotensin receptor gene expression in human heart failure. Circulation. 1997;95:1201–1206. doi: 10.1161/01.cir.95.5.1201. [DOI] [PubMed] [Google Scholar]
- 5.Haq S, Choukroun G, Lim H, Tymitz KM, del Monte F, Gwathmey J, Grazette L, Michael A, Hajjar R, Force T, et al. Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation. 2001;103:670–677. doi: 10.1161/01.cir.103.5.670. [DOI] [PubMed] [Google Scholar]
- 6.Bristow MR, Ginsburg R, Minobe W, Cubicciotti RS, Sageman WS, Lurie K, Billingham ME, Harrison DC, Stinson EB. Decreased catecholamine sensitivity and beta-adrenergic-receptor density in failing human hearts. N Engl J Med. 1982;307:205–211. doi: 10.1056/NEJM198207223070401. [DOI] [PubMed] [Google Scholar]
- 7.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat Rev Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 8.Muthumala A, Drenos F, Elliott PM, Humphries SE. Role of beta adrenergic receptor polymorphisms in heart failure: systematic review and meta-analysis. Eur J Heart Fail. 2008;10:3–13. doi: 10.1016/j.ejheart.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Brodde OE. Beta-1 and beta-2 adrenoceptor polymorphisms: functional importance, impact on cardiovascular diseases and drug responses. Pharmacol Ther. 2008;117:1–29. doi: 10.1016/j.pharmthera.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem. 1999;274:12670–12674. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- 11.Rathz DA, Brown KM, Kramer LA, Liggett SB. Amino acid 49 polymorphisms of the human beta1-adrenergic receptor affect agonist-promoted trafficking. J Cardiovasc Pharmacol. 2002;39:155–160. doi: 10.1097/00005344-200202000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Joseph SS, Lynham JA, Grace AA, Colledge WH, Kaumann AJ. Markedly reduced effects of (-)-isoprenaline but not of (-)-CGP12177 and unchanged affinity of beta-blockers at Gly389-beta1-adrenoceptors compared to Arg389-beta1-adrenoceptors. Br J Pharmacol. 2004;142:51–56. doi: 10.1038/sj.bjp.0705753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rathz DA, Gregory KN, Fang Y, Brown KM, Liggett SB. Hierarchy of polymorphic variation and desensitization permutations relative to beta 1- and beta 2-adrenergic receptor signaling. J Biol Chem. 2003;278:10784–10789. doi: 10.1074/jbc.M206054200. [DOI] [PubMed] [Google Scholar]
- 14.Sandilands AJ, O'Shaughnessy KM, Brown MJ. Greater inotropic and cyclic AMP responses evoked by noradrenaline through Arg389 beta 1-adrenoceptors versus Gly389 beta 1-adrenoceptors in isolated human atrial myocardium. Br J Pharmacol. 2003;138:386–392. doi: 10.1038/sj.bjp.0705030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruck H, Leineweber K, Temme T, Weber M, Heusch G, Philipp T, Brodde OE. The Arg389Gly beta1-adrenoceptor polymorphism and catecholamine effects on plasma-renin activity. J Am Coll Cardiol. 2005;46:2111–2115. doi: 10.1016/j.jacc.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 17.Mialet Perez J, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, Dorn GW, Liggett SB. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med. 2003;9:1300–1305. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 18.Büscher R, Belger H, Eilmes KJ, Tellkamp R, Radke J, Dhein S, Hoyer PF, Michel MC, Insel PA, Brodde OE. In-vivo studies do not support a major functional role for the Gly389Arg beta 1-adrenoceptor polymorphism in humans. Pharmacogenetics. 2001;11:199–205. doi: 10.1097/00008571-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Xie HG, Dishy V, Sofowora G, Kim RB, Landau R, Smiley RM, Zhou HH, Wood AJ, Harris P, Stein CM. Arg389Gly beta 1-adrenoceptor polymorphism varies in frequency among different ethnic groups but does not alter response in vivo. Pharmacogenetics. 2001;11:191–197. doi: 10.1097/00008571-200104000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Levin MC, Marullo S, Muntaner O, Andersson B, Magnusson Y. The myocardium-protective Gly-49 variant of the beta 1-adrenergic receptor exhibits constitutive activity and increased desensitization and down-regulation. J Biol Chem. 2002;277:30429–30435. doi: 10.1074/jbc.M200681200. [DOI] [PubMed] [Google Scholar]
- 21.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 22.Bruck H, Leineweber K, Park J, Weber M, Heusch G, Philipp T, Brodde OE. Human beta2-adrenergic receptor gene haplotypes and venodilation in vivo. Clin Pharmacol Ther. 2005;78:232–238. doi: 10.1016/j.clpt.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Liggett SB. The pharmacogenetics of beta2-adrenergic receptors: relevance to asthma. J Allergy Clin Immunol. 2000;105:S487–S492. doi: 10.1016/s0091-6749(00)90048-4. [DOI] [PubMed] [Google Scholar]
- 24.Brodde OE, Büscher R, Tellkamp R, Radke J, Dhein S, Insel PA. Blunted cardiac responses to receptor activation in subjects with Thr164Ile beta(2)-adrenoceptors. Circulation. 2001;103:1048–1050. doi: 10.1161/01.cir.103.8.1048. [DOI] [PubMed] [Google Scholar]
- 25.Green SA, Cole G, Jacinto M, Innis M, Liggett SB. A polymorphism of the human beta 2-adrenergic receptor within the fourth transmembrane domain alters ligand binding and functional properties of the receptor. J Biol Chem. 1993;268:23116–23121. [PubMed] [Google Scholar]
- 26.Turki J, Lorenz JN, Green SA, Donnelly ET, Jacinto M, Liggett SB. Myocardial signaling defects and impaired cardiac function of a human beta 2-adrenergic receptor polymorphism expressed in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10483–10488. doi: 10.1073/pnas.93.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen BC, Swigart PM, De Marco T, Hoopes C, Simpson PC. {alpha}1-Adrenergic receptor subtypes in nonfailing and failing human myocardium. Circ Heart Fail. 2009;2:654–663. doi: 10.1161/CIRCHEARTFAILURE.108.846212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaak S, Mialet-Perez J, Flordellis C, Paris H. Genetic variation of human adrenergic receptors: from molecular and functional properties to clinical and pharmacogenetic implications. Curr Top Med Chem. 2007;7:217–231. doi: 10.2174/156802607779318163. [DOI] [PubMed] [Google Scholar]
- 29.Shibata K, Hirasawa A, Moriyama N, Kawabe K, Ogawa S, Tsujimoto G. Alpha 1a-adrenoceptor polymorphism: pharmacological characterization and association with benign prostatic hypertrophy. Br J Pharmacol. 1996;118:1403–1408. doi: 10.1111/j.1476-5381.1996.tb15552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei B, Morris DP, Smith MP, Svetkey LP, Newman MF, Rotter JI, Buchanan TA, Beckstrom-Sternberg SM, Green ED, Schwinn DA. Novel human alpha1a-adrenoceptor single nucleotide polymorphisms alter receptor pharmacology and biological function. Naunyn Schmiedebergs Arch Pharmacol. 2005;371:229–239. doi: 10.1007/s00210-005-1019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hein L, Altman JD, Kobilka BK. Two functionally distinct alpha2-adrenergic receptors regulate sympathetic neurotransmission. Nature. 1999;402:181–184. doi: 10.1038/46040. [DOI] [PubMed] [Google Scholar]
- 32.Small KM, Forbes SL, Brown KM, Liggett SB. An asn to lys polymorphism in the third intracellular loop of the human alpha 2A-adrenergic receptor imparts enhanced agonist-promoted Gi coupling. J Biol Chem. 2000;275:38518–38523. doi: 10.1074/jbc.M004550200. [DOI] [PubMed] [Google Scholar]
- 33.Small KM, Forbes SL, Rahman FF, Bridges KM, Liggett SB. A four amino acid deletion polymorphism in the third intracellular loop of the human alpha 2C-adrenergic receptor confers impaired coupling to multiple effectors. J Biol Chem. 2000;275:23059–23064. doi: 10.1074/jbc.M000796200. [DOI] [PubMed] [Google Scholar]
- 34.Dorn GW 2nd. GRK mythology: G-protein receptor kinases in cardiovascular disease. J Mol Med. 2009;87:455–463. doi: 10.1007/s00109-009-0450-7. [DOI] [PubMed] [Google Scholar]
- 35.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinh DT, Frauman AG, Johnston CI, Fabiani ME. Angiotensin receptors: distribution, signalling and function. Clin Sci (Lond) 2001;100:481–492. [PubMed] [Google Scholar]
- 37.Jeunemaitre X. Genetics of the human renin angiotensin system. J Mol Med. 2008;86:637–641. doi: 10.1007/s00109-008-0344-0. [DOI] [PubMed] [Google Scholar]
- 38.Bleumink GS, Schut AF, Sturkenboom MC, Deckers JW, van Duijn CM, Stricker BH. Genetic polymorphisms and heart failure. Genet Med. 2004;6:465–474. doi: 10.1097/01.gim.0000144061.70494.95. [DOI] [PubMed] [Google Scholar]
- 39.Jeunemaitre X, Gimenez-Roqueplo AP, Célérier J, Corvol P. Angiotensinogen variants and human hypertension. Curr Hypertens Rep. 1999;1:31–41. doi: 10.1007/s11906-999-0071-0. [DOI] [PubMed] [Google Scholar]
- 40.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudnicki M, Mayer G. Significance of genetic polymorphisms of the renin-angiotensin-aldosterone system in cardiovascular and renal disease. Pharmacogenomics. 2009;10:463–476. doi: 10.2217/14622416.10.3.463. [DOI] [PubMed] [Google Scholar]
- 42.Laragh JH, Angers M, Kelly WG, Lieberman S. Hypotensive agents and pressor substances. The effect of epinephrine, norepinephrine, angiotensin II, and others on the secretory rate of aldosterone in man. JAMA. 1960;174:234–240. doi: 10.1001/jama.1960.03030030014003. [DOI] [PubMed] [Google Scholar]
- 43.Curnow KM, Tusie-Luna MT, Pascoe L, Natarajan R, Gu JL, Nadler JL, White PC. The product of the CYP11B2 gene is required for aldosterone biosynthesis in the human adrenal cortex. Mol Endocrinol. 1991;5:1513–1522. doi: 10.1210/mend-5-10-1513. [DOI] [PubMed] [Google Scholar]
- 44.White PC, Hautanen A, Kupari M. Aldosterone synthase (CYP11B2) polymorphisms and cardiovascular function. J Steroid Biochem Mol Biol. 1999;69:409–412. doi: 10.1016/s0960-0760(99)00071-0. [DOI] [PubMed] [Google Scholar]
- 45.White PC, Slutsker L. Haplotype analysis of CYP11B2. Endocr Res. 1995;21:437–442. doi: 10.3109/07435809509030459. [DOI] [PubMed] [Google Scholar]
- 46.Brand E, Chatelain N, Mulatero P, Féry I, Curnow K, Jeunemaitre X, Corvol P, Pascoe L, Soubrier F. Structural analysis and evaluation of the aldosterone synthase gene in hypertension. Hypertension. 1998;32:198–204. doi: 10.1161/01.hyp.32.2.198. [DOI] [PubMed] [Google Scholar]
- 47.de Gasparo M, Catt KJ, Inagami T, Wright JW, Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol Rev. 2000;52:415–472. [PubMed] [Google Scholar]
- 48.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3' untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonnardeaux A, Davies E, Jeunemaitre X, Féry I, Charru A, Clauser E, Tiret L, Cambien F, Corvol P, Soubrier F. Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension. 1994;24:63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- 50.Biolo A, Clausell N, Santos KG, Salvaro R, Ashton-Prolla P, Borges A, Rohde LE. Impact of beta1-adrenergic receptor polymorphisms on susceptibility to heart failure, arrhythmogenesis, prognosis, and response to beta-blocker therapy. Am J Cardiol. 2008;102:726–732. doi: 10.1016/j.amjcard.2008.04.070. [DOI] [PubMed] [Google Scholar]
- 51.Covolo L, Gelatti U, Metra M, Nodari S, Piccichè A, Pezzali N, Zani C, Alberti A, Donato F, Nardi G, et al. Role of beta1- and beta2-adrenoceptor polymorphisms in heart failure: a case-control study. Eur Heart J. 2004;25:1534–1541. doi: 10.1016/j.ehj.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 52.Forleo C, Sorrentino S, Guida P, Romito R, De Tommasi E, Iacoviello M, Pitzalis M. Beta1- and beta2-adrenergic receptor polymorphisms affect susceptibility to idiopathic dilated cardiomyopathy. J Cardiovasc Med (Hagerstown) 2007;8:589–595. doi: 10.2459/01.JCM.0000281710.51304.03. [DOI] [PubMed] [Google Scholar]
- 53.Nonen S, Okamoto H, Akino M, Matsui Y, Fujio Y, Yoshiyama M, Takemoto Y, Yoshikawa J, Azuma J, Kitabatake A. No positive association between adrenergic receptor variants of alpha2cDel322-325, beta1Ser49, beta1Arg389 and the risk for heart failure in the Japanese population. Br J Clin Pharmacol. 2005;60:414–417. doi: 10.1111/j.1365-2125.2005.02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- 55.Tesson F, Charron P, Peuchmaurd M, Nicaud V, Cambien F, Tiret L, Poirier O, Desnos M, Jullières Y, Amouyel P, et al. Characterization of a unique genetic variant in the beta1-adrenoceptor gene and evaluation of its role in idiopathic dilated cardiomyopathy. CARDIGENE Group. J Mol Cell Cardiol. 1999;31:1025–1032. doi: 10.1006/jmcc.1999.0947. [DOI] [PubMed] [Google Scholar]
- 56.Woodiwiss AJ, Badenhorst D, Sliwa K, Brooksbank R, Essop R, Sareli P, Norton GR. Beta1- and alpha2c-adrenoreceptor variants as predictors of clinical aspects of dilated cardiomyopathy in people of African ancestry. Cardiovasc J Afr. 2008;19:188–193. [PMC free article] [PubMed] [Google Scholar]
- 57.Metra M, Zani C, Covolo L, Nodari S, Pezzali N, Gelatti U, Donato F, Nardi G, Dei Cas L. Role of beta1- and alpha2c-adrenergic receptor polymorphisms and their combination in heart failure: a case-control study. Eur J Heart Fail. 2006;8:131–135. doi: 10.1016/j.ejheart.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 58.Börjesson M, Magnusson Y, Hjalmarson A, Andersson B. A novel polymorphism in the gene coding for the beta(1)-adrenergic receptor associated with survival in patients with heart failure. Eur Heart J. 2000;21:1853–1858. doi: 10.1053/euhj.1999.1994. [DOI] [PubMed] [Google Scholar]
- 59.Liggett SB, Wagoner LE, Craft LL, Hornung RW, Hoit BD, McIntosh TC, Walsh RA. The Ile164 beta2-adrenergic receptor polymorphism adversely affects the outcome of congestive heart failure. J Clin Invest. 1998;102:1534–1539. doi: 10.1172/JCI4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leineweber K, Tenderich G, Wolf C, Wagner S, Zittermann A, Elter-Schulz M, Moog R, Müller N, Jakob HG, Körfer R, et al. Is there a role of the Thr164Ile-beta(2)-adrenoceptor polymorphism for the outcome of chronic heart failure? Basic Res Cardiol. 2006;101:479–484. doi: 10.1007/s00395-006-0601-8. [DOI] [PubMed] [Google Scholar]
- 61.Zakrzewski-Jakubiak M, de Denus S, Dubé MP, Bélanger F, White M, Turgeon J. Ten renin-angiotensin system-related gene polymorphisms in maximally treated Canadian Caucasian patients with heart failure. Br J Clin Pharmacol. 2008;65:742–751. doi: 10.1111/j.1365-2125.2007.03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raynolds MV, Bristow MR, Bush EW, Abraham WT, Lowes BD, Zisman LS, Taft CS, Perryman MB. Angiotensin-converting enzyme DD genotype in patients with ischaemic or idiopathic dilated cardiomyopathy. Lancet. 1993;342:1073–1075. doi: 10.1016/0140-6736(93)92061-w. [DOI] [PubMed] [Google Scholar]
- 63.Andersson B, Sylvén C. The DD genotype of the angiotensin-converting enzyme gene is associated with increased mortality in idiopathic heart failure. J Am Coll Cardiol. 1996;28:162–167. doi: 10.1016/0735-1097(96)00098-8. [DOI] [PubMed] [Google Scholar]
- 64.Covolo L, Gelatti U, Metra M, Donato F, Nodari S, Pezzali N, Dei Cas L, Nardi G. Angiotensin-converting-enzyme gene polymorphism and heart failure: a case-control study. Biomarkers. 2003;8:429–436. doi: 10.1080/13547500310001599052. [DOI] [PubMed] [Google Scholar]
- 65.Montgomery HE, Keeling PJ, Goldman JH, Humphries SE, Talmud PJ, McKenna WJ. Lack of association between the insertion/deletion polymorphism of the angiotensin-converting enzyme gene and idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 1995;25:1627–1631. doi: 10.1016/0735-1097(95)00109-h. [DOI] [PubMed] [Google Scholar]
- 66.Tiago AD, Badenhorst D, Skudicky D, Woodiwiss AJ, Candy GP, Brooksbank R, Sliwa K, Sareli P, Norton GR. An aldosterone synthase gene variant is associated with improvement in left ventricular ejection fraction in dilated cardiomyopathy. Cardiovasc Res. 2002;54:584–589. doi: 10.1016/s0008-6363(02)00281-x. [DOI] [PubMed] [Google Scholar]
- 67.Vancura V, Hubácek J, Málek I, Gebauerová M, Pitha J, Dorazilová Z, Langová M, Zelízko M, Poledne R. Does angiotensin-converting enzyme polymorphism influence the clinical manifestation and progression of heart failure in patients with dilated cardiomyopathy? Am J Cardiol. 1999;83:461–462, A10. doi: 10.1016/s0002-9149(98)00889-3. [DOI] [PubMed] [Google Scholar]
- 68.Huang W, Xie C, Zhou H, Yang T, Sun M. Association of the angiotensin-converting enzyme gene polymorphism with chronic heart failure in Chinese Han patients. Eur J Heart Fail. 2004;6:23–27. doi: 10.1016/j.ejheart.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Sanderson JE, Young RP, Yu CM, Chan S, Critchley JA, Woo KS. Lack of association between insertion/deletion polymorphism of the angiotensin-converting enzyme gene and end-stage heart failure due to ischemic or idiopathic dilate cardiomyopathy in the Chinese. Am J Cardiol. 1996;77:1008–1010. doi: 10.1016/s0002-9149(97)89160-6. [DOI] [PubMed] [Google Scholar]
- 70.Tiret L, Mallet C, Poirier O, Nicaud V, Millaire A, Bouhour JB, Roizès G, Desnos M, Dorent R, Schwartz K, et al. Lack of association between polymorphisms of eight candidate genes and idiopathic dilated cardiomyopathy: the CARDIGENE study. J Am Coll Cardiol. 2000;35:29–35. doi: 10.1016/s0735-1097(99)00522-7. [DOI] [PubMed] [Google Scholar]
- 71.Yamada Y, Ichihara S, Fujimura T, Yokota M. Lack of association of polymorphisms of the angiotensin converting enzyme and angiotensinogen genes with nonfamilial hypertrophic or dilated cardiomyopathy. Am J Hypertens. 1997;10:921–928. doi: 10.1016/s0895-7061(97)00112-x. [DOI] [PubMed] [Google Scholar]
- 72.Goldbergova M, Spinarova L, Spinar J, Toman J, Vasku A, Vacha J. Association of two angiotensinogen gene polymorphisms, M235T and G(-6)A, with chronic heart failure. Int J Cardiol. 2003;89:267–272. doi: 10.1016/s0167-5273(02)00506-5. [DOI] [PubMed] [Google Scholar]
- 73.Andersson B, Blange I, Sylvén C. Angiotensin-II type 1 receptor gene polymorphism and long-term survival in patients with idiopathic congestive heart failure. Eur J Heart Fail. 1999;1:363–369. doi: 10.1016/s1388-9842(99)00045-8. [DOI] [PubMed] [Google Scholar]
- 74.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, Cupples LA, Dehghan A, Lumley T, Rosamond WD, et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–266. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanderson JE, Yu CM, Young RP, Shum IO, Wei S, Arumanayagam M, Woo KS. Influence of gene polymorphisms of the renin-angiotensin system on clinical outcome in heart failure among the Chinese. Am Heart J. 1999;137:653–657. doi: 10.1016/s0002-8703(99)70218-8. [DOI] [PubMed] [Google Scholar]
- 76.White HL, de Boer RA, Maqbool A, Greenwood D, van Veldhuisen DJ, Cuthbert R, Ball SG, Hall AS, Balmforth AJ. An evaluation of the beta-1 adrenergic receptor Arg389Gly polymorphism in individuals with heart failure: a MERIT-HF sub-study. Eur J Heart Fail. 2003;5:463–468. doi: 10.1016/s1388-9842(03)00044-8. [DOI] [PubMed] [Google Scholar]
- 77.de Groote P, Lamblin N, Helbecque N, Mouquet F, Mc Fadden E, Hermant X, Amouyel P, Dallongeville J, Bauters C. The impact of beta-adrenoreceptor gene polymorphisms on survival in patients with congestive heart failure. Eur J Heart Fail. 2005;7:966–973. doi: 10.1016/j.ejheart.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 78.Shin J, Lobmeyer MT, Gong Y, Zineh I, Langaee TY, Yarandi H, Schofield RS, Aranda JM Jr, Hill JA, Pauly DF, et al. Relation of beta(2)-adrenoceptor haplotype to risk of death and heart transplantation in patients with heart failure. Am J Cardiol. 2007;99:250–255. doi: 10.1016/j.amjcard.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 79.Sehnert AJ, Daniels SE, Elashoff M, Wingrove JA, Burrow CR, Horne B, Muhlestein JB, Donahue M, Liggett SB, Anderson JL, et al. Lack of association between adrenergic receptor genotypes and survival in heart failure patients treated with carvedilol or metoprolol. J Am Coll Cardiol. 2008;52:644–651. doi: 10.1016/j.jacc.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 80.Barbato E, Penicka M, Delrue L, Van Durme F, De Bruyne B, Goethals M, Wijns W, Vanderheyden M, Bartunek J. Thr164Ile polymorphism of beta2-adrenergic receptor negatively modulates cardiac contractility: implications for prognosis in patients with idiopathic dilated cardiomyopathy. Heart. 2007;93:856–861. doi: 10.1136/hrt.2006.091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Littlejohn MD, Palmer BR, Richards AM, Frampton CM, Pilbrow AP, Troughton RW, Cameron VA, Kennedy MA. Ile164 variant of beta2-adrenoceptor does not influence outcome in heart failure but may interact with beta blocker treatment. Eur J Heart Fail. 2008;10:55–59. doi: 10.1016/j.ejheart.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 82.Regitz-Zagrosek V, Hocher B, Bettmann M, Brede M, Hadamek K, Gerstner C, Lehmkuhl HB, Hetzer R, Hein L. Alpha2C-adrenoceptor polymorphism is associated with improved event-free survival in patients with dilated cardiomyopathy. Eur Heart J. 2006;27:454–459. doi: 10.1093/eurheartj/ehi659. [DOI] [PubMed] [Google Scholar]
- 83.Pilbrow AP, Palmer BR, Frampton CM, Yandle TG, Troughton RW, Campbell E, Skelton L, Lainchbury JG, Richards AM, Cameron VA. Angiotensinogen M235T and T174M gene polymorphisms in combination doubles the risk of mortality in heart failure. Hypertension. 2007;49:322–327. doi: 10.1161/01.HYP.0000253061.30170.68. [DOI] [PubMed] [Google Scholar]
- 84.McNamara DM, Holubkov R, Janosko K, Palmer A, Wang JJ, MacGowan GA, Murali S, Rosenblum WD, London B, Feldman AM. Pharmacogenetic interactions between beta-blocker therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. Circulation. 2001;103:1644–1648. doi: 10.1161/01.cir.103.12.1644. [DOI] [PubMed] [Google Scholar]
- 85.McNamara DM, Holubkov R, Postava L, Janosko K, MacGowan GA, Mathier M, Murali S, Feldman AM, London B. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Am Coll Cardiol. 2004;44:2019–2026. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 86.Fatini C, Sticchi E, Marcucci R, Said AA, Del Pace S, Verdiani V, Nozzoli C, Gensini GF, Abbate R. ACE insertion/deletion, but not -240A>T polymorphism, modulates the severity in heart failure. J Investig Med. 2008;56:1004–1010. doi: 10.2310/JIM.0b013e31818e8028. [DOI] [PubMed] [Google Scholar]
- 87.McNamara DM, Tam SW, Sabolinski ML, Tobelmann P, Janosko K, Taylor AL, Cohn JN, Feldman AM, Worcel M. Aldosterone synthase promoter polymorphism predicts outcome in African Americans with heart failure: results from the A-HeFT Trial. J Am Coll Cardiol. 2006;48:1277–1282. doi: 10.1016/j.jacc.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 88.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, Dorn GW 2nd. Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]