Abstract
Reactive oxygen species (ROS) mediate apoptosis in many different cell types. We have previously shown that the antioxidant Mn(III) tetrakis(5,10,15,20-benzoic acid)porphyrin (MnTBAP) decreased intracellular ROS and prevented the apoptosis of activated T cells in vitro. To determine the mechanism(s) by which MnTBAP afforded such protection, we used Affymetrix (Santa Clara, CA) gene arrays to compare gene expression in T cells activated with staphylococcal enterotoxin B in vivo then cultured with or without MnTBAP. This analysis showed that the antioxidant increased the expression of Bcl-2, an antiapoptotic molecule whose levels are normally decreased by T cell activation. Culture with MnTBAP revealed a tight inverse correlation between the levels of Bcl-2 and ROS within T cells. In vivo, production of ROS in activated T cells occurred before Bcl-2 down-regulation. Furthermore, MnTBAP's ability to prevent death required the expression of Bcl-2 in most T cells. Finally, neither ROS production nor the effects on Bcl-2 expression required Bim, the Bcl-2 antagonist that mediates the death of activated T cells in vivo. Taken together, our results suggest that ROS sensitize T cells to apoptosis by decreasing expression of Bcl-2.
Reactive oxygen species (ROS) are known to regulate cell death in a variety of cell types. Differential effects of ROS on cell death are observed depending on the level of ROS within the cell (1). High levels of ROS can lead to lipid peroxidation, damage to cellular membranes, inactivation of caspase enzymes, and necrotic cell death (1). In contrast, low levels of ROS have been shown to activate protein kinases and phosphatases, mobilize Ca2+ stores, activate or inactivate transcription factors, and lead to apoptotic cell death (1). Thus, high levels of ROS probably kill cells by means of direct damage, whereas low levels of ROS probably mediate apoptosis indirectly through their effects on protein kinases and/or phosphatases, transcription factors, and the subsequent effects of these molecules on gene expression. However, little has been done to assess the extent to which ROS affect gene expression in cells undergoing apoptosis.
It has been known for some time that Bcl-2 overexpression can protect cells from apoptosis mediated by ROS (2). However, the mechanism by which Bcl-2 prevents ROS-induced apoptosis is unknown. Bcl-2 itself does not possess antioxidant activity; rather, it may act indirectly to increase the levels and/or activities of endogenous antioxidants (e.g., glutathione or superoxide dismutase) within cells (3–5). Thus, overexpression of Bcl-2 may allow cells to cope better with the effects of ROS, possibly by allowing increases in endogenous antioxidant enzymes.
An alternative but not mutually exclusive hypothesis is that ROS act to down-regulate endogenous Bcl-2 levels within cells. Because levels of Bcl-2 within cells are critical to antiapoptotic activity, decreasing Bcl-2 could be a mechanism to sensitize cells to apoptosis. By detoxifying ROS, antioxidants may therefore reverse the ROS-induced decline in Bcl-2 and prevent apoptosis. Several studies suggest a reciprocal relationship between ROS and Bcl-2 levels within cells (6–8). Thus, in divergent cell types, decreases in ROS correlate with increases in Bcl-2 levels and vice versa.
Recently we showed that, in vivo, activated T cells decrease their levels of Bcl-2 and become susceptible to the proapoptotic effects of Bim (9). We also showed that culture of these activated T cells with a synthetic catalytic antioxidant, Mn(III) tetrakis(5,10,15,20-benzoic acid)porphyrin (MnTBAP), lowered superoxide levels and prevented apoptosis (10). Here, we show that culture of T cells with MnTBAP reverses the decline in Bcl-2. The corollary of this finding is that ROS are responsible for Bcl-2 down-regulation within activated T cells. Taken together, these results suggest a “two-signal” model for activated T cell apoptosis. The first signal is driven by ROS down-regulation of Bcl-2, which is necessary, but not sufficient, to complete apoptosis. The second signal requires the expression of Bim.
Methods
T Cell Purification for Microarray Analysis. T cells were activated in C57BL/10 (n = 16) mice by i.v. injection of 150 μg of staphylococcal enterotoxin B (SEB; Toxin Technology, Sarasota, FL). Mice were killed 48 h later, and lymph nodes cells were cultured with or without 150 μM MnTBAP for 7 h at 37°C and then stained with fluorescently labeled antibodies to CD4, CD8, Vβ8, and I-Ab (Pharmingen). CD4+ CD8+ Vβ8+ I-Ab- cells were sorted by using a high-speed MoFlo flow cytometer (Cytomation, Fort Collins, CO). RNA extraction, cDNA synthesis, and gene microarray analysis were performed as described (11, 12)
Mice. C57BL/10 and C57BL/6J (BL/6) mice were purchased from The Jackson Laboratory. Bim-/- mice were a kind gift from Philippe Bouillet and Andreas Strasser (The Walter and Eliza Hall Institute of Medical Research, Melbourne) and were mated as heterozygotes; genotypes of the offspring were screened by PCR as described (9). Heterozygous Bcl-2+/- mice were purchased from The Jackson Laboratory (Bcl-2tm1sjk) and mated with Bim+/- mice. Genotypes of the offspring were determined by PCR. Vβ8.2+ T cell antigen receptor (TCR) transgenic (VβDO) mice were described (13).
Real-Time PCR. T cells were activated in VβDO mice by i.v. injection of 150 μg of SEB. Mice were killed 48 h later, and lymph node cells were cultured with or without 150 μM MnTBAP and then stained with antibodies to B220 and I-Ab and sorted for B220- I-Ab- cells by using a MoFlo cell sorter. RNA extraction, cDNA synthesis, and real-time RT-PCR were performed as described (9, 12). Primers and probes used for each gene were designed by using PRIMER EXPRESS software (Applied Biosystems) and synthesized by the Molecular Resource Center (Denver, CO).
Flow Cytometry. For intracellular stains, cells were stained with fluorescent antibodies against CD4, CD8, and Vβ8.x (Pharmingen), then washed, permeabilized with 0.03% saponin (Sigma), stained with anti-Bcl-2 or anti-Bcl-xL antibodies (Pharmingen), washed, and stained with secondary antibodies. Data were collected on a FACSCalibur flow cytometer and analyzed with CELLQUEST software (Becton Dickinson). T cell death was assessed by staining with anti-Vβ8.x-FITC and propidium iodide (0.5 μg/ml) as described (10). Percent inhibition of cell death was calculated as follows: [(percent dead without MnTBAP - percent dead with MnTBAP)/percent dead without MnTBAP] × 100. Superoxide levels were determined by using dihydroethidium as described (10).
Production and Use of Retroviruses. pMSCV-IRES-GFP (MiG, a plasmid derived from the murine stem cell virus containing an internal ribosome entry sequence followed by a GFP cassette) was a kind gift from William Sha (University of California, Berkeley), and pCLEco (a plasmid encoding gag-pol-env cDNAs) was a kind gift from Inder Verma (The Salk Institute, La Jolla, CA) (14). pMSCV-IRES-Thy1.1 (MiT) was derived from MiG as described (12). Mouse cDNAs encoding catalase, Mn superoxide dismutase (MnSOD), and Bcl-2 were cloned by means of PCR amplification from activated T cell cDNA, and inserts were confirmed by restriction digestion and DNA sequencing. MnSOD Q143A was generated from MiT MnSOD by site-directed mutagenesis. Plasmid DNA was transformed and amplified in DH5α Escherichia coli and purified by using the QIAfilter Mega kit (Qiagen, Valencia, CA).
Retroviruses were produced by cotransfection of 293 human embryonic kidney cells with pCLEco and the MiT plasmid of interest by using calcium phosphate as described (9, 12). After transduction, cells were stained with various fluorescently labeled antibodies, all of which were supplied by Pharmingen, and analyzed by flow cytometry. Live and dead cells were distinguished by their forward/side scatter properties as described (12, 15).
Results
Properties of Cells Used for Gene Array Analysis. Vβ8+ T cells were activated by injection of SEB into C57BL/10 mice, isolated 2 days later, cultured for 7 h, and purified from MnTBAP-treated or control-treated cultures by using high-speed cell sorting. After sorting, both treated and control T cells were roughly 98% Vβ8+ (Table 1). For both groups, 0.18% of the sorted cells were I-Ab+ and the remaining non-Vβ8+ cells were mostly resting T cells expressing a Vβ other than Vβ8. To determine whether MnTBAP had the desired effect on the T cells in this experiment, samples from the two groups of T cells were cultured for an additional 5 h (total of 12 h) and assessed for cell death. Indeed, culture with MnTBAP significantly decreased activated T cell death (Table 1; P < 0.00001, two-sample t test; MINITAB for Windows).
Table 1. Properties of sorted T cells used for Affymetrix GeneChip analysis.
| Activated T cells cultured | Vβ8, % | I-Ab, % | Vβ8+ cells that died,* % | Transcript levels for GAPDH |
|---|---|---|---|---|
| Control | 97.75 ± 0.47 | 0.18 ± 0.11 | 59.35 ± 3.82 | 16,285 ± 2,551 |
| MnTBAP | 98.49 ± 0.34 | 0.18 ± 0.14 | 23.57 ± 2.24 | 16,540 ± 2,574 |
Sixteen C57BL/10 mice were injected i.v. with 150 μg of SEB and killed 48 h later. Lymph node cells were then cultured with either 150 μM MnTBPAP or diluent control for 7 h. A fraction of cells were cultured for 5 h more and analyzed for cell death. Results show the mean percentage of cells ± SD. Cells from lymph nodes were sorted for live Vβ8+ CD4+ CD8+ I-Ab- cells by cell sorting. After sorting, cells were analyzed for Vβ8+ or I-Ab+, and results show the mean percentage of positive cells ± SD. The remaining sorted cells were used for RNA isolation and gene array analysis; the mean hybridization intensity for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase ± SD is shown.
Results are of Vβ8+ cells that died after culture
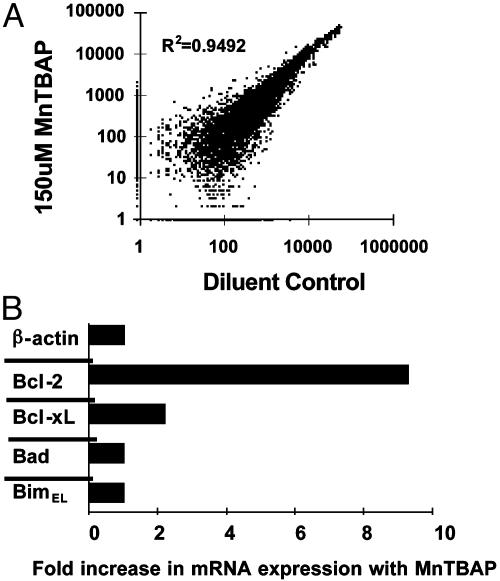
RNA was prepared from the sorted cells and analyzed by using Affymetrix Murine 11K gene arrays as described (11, 12). The hybridization intensity values for the housekeeping gene glyceraldehyde-phosphate dehydrogenase were not significantly different between MnTBAP-treated and -untreated cells (Table 1). Also, expression of most of the genes/ESTs on the GeneChip was not affected by MnTBAP treatment (Fig. 1A). Thus, according to the controls tested, overall RNA levels in the two cell populations were similar.
Fig. 1.
Comparison of gene expression between activated T cells treated with and without MnTBAP. (A)Vβ8+ T cells were activated in mice by injection of 150 μg of SEB, removed after 48 h, and cultured with or without MnTBAP for 7 h; RNA derived from sorted cells was then hybridized with Affymetrix 11K gene microarrays. The plot compares raw hybridization intensity values for all genes. R2 denotes the correlation coefficient between the two samples. (B) MnTBAP increases the levels of RNA for Bcl-2 and Bcl-xL but not for Bad or BimEL in activated T cells. Lymph node T cells from SEB-injected VβDO mice were cultured for 8 h with or without 150 μM MnTBAP and then negatively sorted, after which RNA was extracted. Real-time RT-PCR was performed in triplicate for the indicated genes. β-Actin was used to normalize the total amounts of cDNA in the two samples. Results were then normalized to the values in the cDNA from control-activated T cells cultured without MnTBAP and expressed as the fold increase in mRNA expression with MnTBAP. Similar results were obtained in two independent experiments.
Control of Gene Expression in Activated T Cells by ROS. Hybridization values below 100 previously gave us unreliable data (16). So here we included only those genes with hybridization intensity values above 100 (i.e., we excluded genes whose expression increased with MnTBAP that were below 100 in the MnTBAP data set and excluded genes whose expression decreased with MnTBAP below 100 in the SEB data set). These cutoffs gave a conservative estimate of genes whose expression was altered by ROS and revealed that expression of 407 genes/ESTs was decreased, whereas expression of 182 genes/ESTs was increased after MnTBAP treatment (Table 2, which is published as supporting information on the PNAS web site).
We next eliminated the ESTs from consideration, thus reducing the number of genes to 80 with increased expression and 188 with decreased expression after culture with MnTBAP. These genes were then ordered into subsets based on function (Tables 3 and 4, which are published as supporting information on the PNAS web site). Expression of two genes whose products are involved in the production of ROS was decreased by MnTBAP, ubiquinol–cytochrome c reductase core protein 1, and P450 oxidoreductase (17–19). Three other genes whose expression was decreased by MnTBAP were those for MnSOD, heat shock factor 1, and 8-oxo-dGTPase, which are induced by ROS and protect cells against ROS damage (20–23). Moreover, the genes for heat shock factor 1 and 8-oxo-dGTPase are known to be induced during T cell activation, an event that increases ROS (11). Because MnTBAP reduces the intracellular levels of ROS, these results suggest that decreasing ROS levels leads to feedback inhibition of enzymes involved in the production of ROS and of enzymes that function to protect cells from this damage.
The simplest explanations for the antiapoptotic effects of MnTBAP on activated T cells is that MnTBAP caused down-regulation of a proapoptotic factor and/or caused up-regulation of an antiapoptotic factor. We found evidence supporting both of these possibilities. MnTBAP decreased the expression of two proapoptotic genes, namely PKCδ and death-associated kinase 3 (Table 4) (24, 25). PKCδ is of particular interest because it has been implicated in ROS-mediated apoptosis (26). However, PKC inhibitors failed to prevent activated T cell apoptosis in vitro (data not shown). Expression of several proapoptotic genes, including Fas, Fas ligand, tumor necrosis factor α, Bax, and Bad, was not affected by MnTBAP (data not shown).
On the other hand, MnTBAP increased the expression of a major antiapoptotic gene, Bcl-2 (Table 3). MnTBAP also decreased the expression of three antiapoptotic genes, including the glucocorticoid-induced leucine zipper, inhibitor of apoptosis protein 3, and interleukin-6 (Table 4) (27–29). Thus, MnTBAP decreased expression of certain proapoptotic genes and both increased and decreased the expression of certain antiapoptotic genes.
MnTBAP Increases Levels of Bcl-2 mRNA and Protein. We confirmed the Bcl-2 gene array results by using real-time RT-PCR. MnTBAP did not affect the transcript levels for β-actin, BimEL, and Bad (Fig. 1B). However, relative to nontreated cells, MnTBAP increased the expression of Bcl-2 roughly 9-fold and that of Bcl-xL roughly 2-fold (Fig. 1B). Additionally, culture of T cells with actinomycin D blocked the MnTBAP-induced increase in Bcl-2, whereas cycloheximide did not (data not shown). Thus, the MnTBAP-induced increase in Bcl-2 was transcriptionally regulated and was similar whether measured by gene microarray or by real-time RT-PCR.
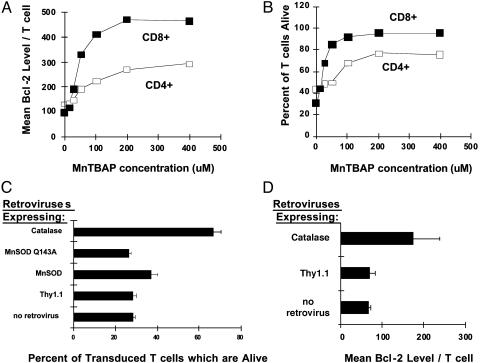
We used flow cytometry to determine whether the MnTBAP also increased expression of Bcl-2 protein. MnTBAP induced a dose-dependent increase in Bcl-2 levels in activated CD4+ and CD8+ T cells (Fig. 2A). MnTBAP did not significantly alter the levels of Bcl-xL protein (data not shown). The increase in Bcl-2 levels correlated directly with an increase in the survival of both activated CD4+ and CD8+ T cells (Fig. 2B). Thus, MnTBAP increased the levels of Bcl-2 mRNA and protein within T cells.
Fig. 2.
MnTBAP and antioxidant enzymes prevent activated T cell death and increase Bcl-2 protein levels. Vβ8+ T cells were activated in C57BL/6 mice (n = 3) by means of injection with SEB. Forty-eight hours later, mice were killed, and single-cell suspensions from lymph nodes of individual mice were cultured with the indicated doses of MnTBAP for 18 h at 37°C. After culture, cells were stained and analyzed. (A) Bcl-2 levels in CD4+ Vβ8+ (□) or CD8+ Vβ8+ (▪) T cells were measured as described in Methods. Results show the mean fluorescence intensity of the anti-Bcl-2 signal ± SEM. Error bars are hidden by the symbols. (B) The percentage of dead CD8- Vβ8+ (mainly CD4+) (□) or CD8+ Vβ8+ (▪) T cells was measured by staining cells with propidium iodide and fluorescent antibodies against Vβ8 and CD8. Results show the mean percent of cells dead ± SEM. Error bars are hidden by the symbols. (C) In a separate experiment, T cells were activated in vivo by injecting VβDO mice with 100 μg of SEB and isolated from lymph nodes 24 h later. Activated T cells were transduced with retroviruses encoding cDNAs for MnSOD, the inactive mutant MnSOD Q143A, or catalase. Twenty-four hours after transduction, T cells were stained with anti-Thy1.1-phycoerythrin (a marker of retrovirally transduced cells) and anti-CD8-Cy-Chrome (Pharmingen) and gated by light scatter for live versus dead cells (12, 15). Results show the mean percent of CD8+ Thy1.1+ cells alive ± SD. Cell-death prevention with MnSOD was significant over both MnSOD Q143A (P < 0.034, Student's two-sample t test) and MiT vector alone (P < 0.025), whereas cell-death prevention with catalase was significant over MiT vector alone (P < 0.002). Similar results were obtained with CD4+ T cells. This experiment is representative of five independent experiments with similar results. (D) T cells were activated in vivo by injecting VβDO mice with 100 μg of SEB and were isolated from lymph nodes 24 h later. Either control Thy1.1-expressing retrovirus or retrovirus encoding catalase cDNA were used to transduce T cells. Bcl-2 levels were measured in retrovirally transduced cells by flow cytometry 24 h later. Results show the mean fluorescence intensity for the anti-Bcl-2 signal within CD8+ T cells ± SD. This experiment is representative of two independent experiments with similar results.
Overexpression of Catalase Increases Bcl-2 and Prevents Cell Death. To determine whether the antioxidant effects of MnTBAP were responsible for T cell survival and Bcl-2 up-regulation, we transduced activated T cells with retroviruses expressing cDNAs for MnSOD and catalase, two proteins whose activities are mimicked by MnTBAP (30). MnSOD detoxifies superoxide, and catalase detoxifies hydrogen peroxide. As expected, both MnSOD and catalase, but not vector alone or an inactive mutant of MnSOD (MnSOD Q143A) (31), prevented the death of a significant number of activated CD8+ T cells (Fig. 2C). Retroviral delivery of catalase was substantially better at rescuing CD8+ T cells from death than was MnSOD. Similar results were observed for activated CD4+ T cells (data not shown). Furthermore, retroviral overexpression of catalase led to an increase in the levels of Bcl-2 within T cells (Fig. 2D). Taken together, these data suggest that ROS are a mediator of Bcl-2 suppression in activated T cells.
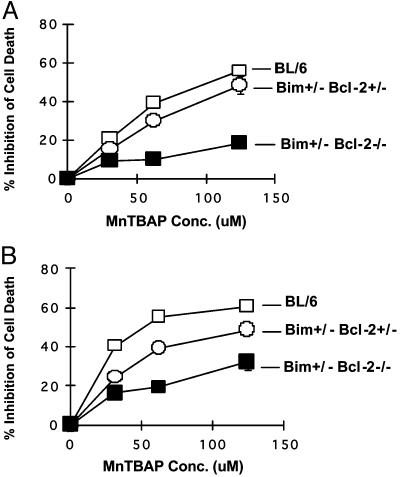
Bcl-2 Up-Regulation by MnTBAP Is Required for T Cell Survival. We next determined the requirement of Bcl-2 in MnTBAP-mediated survival. Unfortunately, Bcl-2-/- mice do not survive beyond 2 weeks of age, and SEB is lethal to them (data not shown). However, we took advantage of Bim+/- Bcl-2-/- mice, which are viable (32). We cultured SEB-activated T cells from BL/6, Bim+/- Bcl-2+/-,or Bim+/- Bcl-2-/- with various doses of MnTBAP and measured cell death. MnTBAP significantly inhibited the death of both CD4+ and CD8+ T cells from both BL/6 and Bim+/- Bcl-2+/- mice (Fig. 3). In contrast, a large percentage of both CD4+ and CD8+ T cells from Bim+/- Bcl-2-/- required Bcl-2 for MnTBAP-mediated survival (Fig. 3). Similar results were obtained by using a synthetic Bcl-2 antagonist, 2-methoxyantimycin A3 (data not shown) (33). Thus, the effects of MnTBAP on the survival of most SEB-activated T cells requires Bcl-2.
Fig. 3.
The rescuing activity of MnTBAP requires Bcl-2. Groups (n = 3) of BL/6, Bim+/- Bcl-2+/-,or Bim+/- Bcl-2-/- mice were injected i.v. with 100 μg of SEB. After 48 h, lymph node T cells from individual mice were cultured with the indicated doses of MnTBAP. After 18 h in vitro, cell death was measured by flow cytometry after staining cells with propidium iodide and antibodies to Vβ8 and CD8 or CD4. Results show the percent inhibition of cell death in either CD4+ Vβ8+ (A) or CD8+ Vβ8+ (B) T cells from either BL/6(□) Bim+/- Bcl-2+/- (○) or Bim+/- Bcl-2-/- (▪) mice ± SD. Error bars are hidden by the symbols.
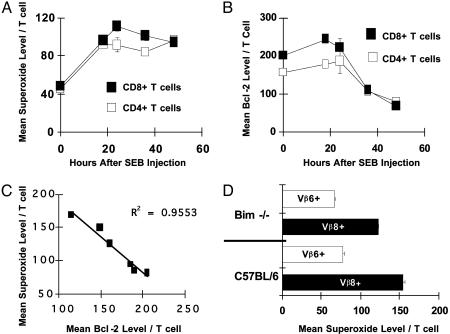
ROS Production in Vivo Precedes Bcl-2 Down-Regulation and Is Not Mediated by Bim. Increased levels of ROS in activated T cells could be caused by decreased levels of Bcl-2, or ROS could themselves lead to decreased Bcl-2 expression (34). The MnTBAP data described here favor the latter idea, but to support this notion we measured whether increases in ROS occurred before or after decreases in Bcl-2. Before stimulation in vivo with SEB, T cells contained low levels of superoxide (Fig. 4A) (10). Eighteen hours after SEB injection, superoxide levels within activated T cells were elevated >2-fold within both CD4+ and CD8+ T cells (Fig. 4A) and persisted within activated T cells as long as 48 h after SEB injection. In contrast, Bcl-2 levels in activated T cells were slightly increased at 18 h and began to decrease between 24 and 36 h after SEB administration (Fig. 4 A and B). Thus, increases in ROS precede Bcl-2 down-regulation and, therefore, cannot be explained by decreased levels of Bcl-2. Further, we correlated the levels of Bcl-2 and ROS within activated T cells. As ROS levels decreased by means of the actions of MnTBAP, levels of Bcl-2 increased in a tight linear inverse manner (Fig. 4C).
Fig. 4.
ROS production precedes Bcl-2 down-regulation and is not driven by Bim. (A) Groups of C57BL/6 mice (n = 3 per time point) were injected i.v. with 150 μg of SEB and killed at various time points after injection. A noninjected C57BL/6 mouse was used as a control for time point 0. Superoxide levels within Vβ8+ CD4+ (□) or Vβ8+ CD8+ (▪) T cells were measured by flow cytometry as described (10). Results show the mean superoxide level per T cell ± SEM. Error bars are hidden by the symbols. (B) Bcl-2 levels were measured, and results show the mean fluorescence intensity of the anti-Bcl-2 signal in CD4+ Vβ8+ (□) or CD8+ Vβ8+ (▪) T cells ± SEM. Error bars are hidden by the symbols. (C) A C57BL/6 mouse was injected i.v. with SEB and killed 48 h later. Lymph nodes cells were cultured in six replicate wells with various doses of MnTBAP to manipulate the level of superoxide. Eighteen hours after culture was initiated, triplicate sets of cells were stained for superoxide content or Bcl-2 (9, 10). Data from various MnTBAP doses were used to plot mean superoxide level per T cell against mean Bcl-2 level per T cell, and linear regression analysis was performed. Results are representative of two independent experiments. (D) Groups of either Bim-/- or C57BL/6 mice (n = 3 per group) were injected i.v. with 150 μg of SEB; 48 h later, superoxide levels within either Vβ8+ (activated) or Vβ6+ (nonactivated) T cells were measured by flow cytometry by staining cells with 5 μM dihydroethidium and anti-Vβ8-FITC or anti-Vβ6-FITC. Results show the mean superoxide level per Vβ8+ or Vβ6+ T cell ± SD.
Because Bim is required for SEB-driven T cell death (9) and because it acts on mitochondria, we postulated that it might cause ROS production. However, activated (Vβ8+) T cells from Bim-/- and C57BL/6 mice had similarly increased levels of superoxide relative to nonstimulated Vβ6+ T cells in the same culture (Fig. 4D). Furthermore, Bcl-2 levels within Bim-/- T cells were decreased after activation with SEB (9). Thus, Bim does not cause ROS production or Bcl-2 down-regulation within activated T cells after SEB activation in vivo. Taken together, these results strongly suggest that ROS causes Bcl-2 down-regulation within activated T cells.
Discussion
When T cells respond to antigen, they are activated; T cells then proliferate and differentiate, and then most of the responding cells die by apoptosis. Our recent reports have identified crucial players involved in activated T cell apoptosis, in vitro and in vivo, namely ROS and Bim (9, 10, 35). Our findings here not only illustrate the spectrum of genes controlled by ROS in activated T cells, but also point to a signaling pathway used by activated T cells to bring about their demise, their internal induction of apoptosis by means of the down-regulation of Bcl-2. This suppression of Bcl-2 expression increases the overall Bim/Bcl-2 ratio within the cell, which is critical for T cell survival in vivo (9). Thus, signals controlling Bcl-2 levels within cells are critical to our understanding of how activated T cells die in vivo.
It has been known for some time that antioxidants can prevent activated T cell death (36, 37). Recently, two reports have shown that antioxidants can prevent Fas-driven activation-induced cell death (38, 39). In both of these reports, the effect of the antioxidant was to inhibit Fas ligand up-regulation after TCR stimulation. Here, we report that antioxidants can also prevent a form of activated T cell death that is independent of Fas and tumor necrosis factor receptor signaling (9, 10). In this case, the antioxidant-driven survival signal works by up-regulating Bcl-2. Bcl-2 and Fas control separate pathways to death in activated T cells (40). Thus, antioxidants may operate by controlling both Fas-dependent and Fas-independent activated T cell autonomous death (35).
The mechanism whereby antioxidants increase Bcl-2 transcription is currently unclear. We presume that MnTBAP functions by preventing the inactivation of a transcription factor or a signaling molecule by ROS. It is possible that some nonantioxidant property of MnTBAP controls the expression of Bcl-2 (41); however, our results with catalase and MnSOD overexpression argue against that possibility. Another possible explanation is that MnTBAP induces cytokines that might be responsible for Bcl-2 up-regulation and prevention of death (29, 42–44). We think this possibility is unlikely for several reasons: (i) the aforementioned results with catalase; (ii) we failed to observe any increased mRNA levels for any of the cytokines (e.g., common γ-chain cytokines) known to up-regulate Bcl-2 and prevent activated T cell death (Table 2); and (iii) we cultured MnTBAP-treated T cells with antibodies against IL-2, IL-4, IL-7, and IL-2Rβ (to inhibit IL-15), and treatment with these antibodies failed to block the MnTBAP-induced survival of activated T cells (data not shown). Thus, it is likely that the transcriptional up-regulation of Bcl-2 by MnTBAP is caused directly by MnTBAP's ability to lower ROS.
Candidate signaling pathways for this effect of MnTBAP include those involving cAMP response element-binding protein (CREB), NF-κB, and phosphatidylinositol 3-kinase (PI-3K). All of these pathways are known to up-regulate Bcl-2 and are modulated by redox signaling (45–49). CREB is an interesting candidate, because signaling through CREB is known to up-regulate Bcl-2 in B cells and T cells (50, 51) and ROS inactivation of CREB can decrease Bcl-2 expression in neurons (8). Alternatively, NF-κB may be involved in the control of Bcl-2 expression in activated T cells. NF-κB is known to repress Bcl-2 expression and is also activated by ROS (52). However, we have failed to observe a role for NF-κB in suppression of Bcl-2 expression by using NF-κB inhibitors (data not shown). Finally, PI-3K/Akt signaling is a known pathway involved in Bcl-2 up-regulation in T cells (45, 46). Endogenous antagonists of PI-3K can be activated by ROS, and inactivation of ROS leads to increased PI-3K signaling and prevention of apoptosis (53). Furthermore, targeted deletion of a major PI-3K antagonist, PTEN, results in protection of T cells from SEB-induced deletion in vivo; however, Bcl-2 levels in the activated T cells of these mice were not investigated (54). We have found that pretreatment of T cells with a PI-3K inhibitor prevents MnTBAP-induced Bcl-2 up-regulation (data not shown). Future work is necessary to investigate this problem in more detail.
In light of these results, we propose that the reason for Bcl-2 suppression is to greatly decrease the half-life of effector T cells and thus rid animals of cells that might otherwise be dangerous once infection has been cleared. Indeed, Bcl-2-deficient T cells have exceedingly shortened half-lives in vivo (55). This decrease in half-life sets the default pathway to death unless the T cell receives survival stimuli required for memory cell formation. This hypothesis is supported by recent data from Kaech et al. (56), who showed that few memory cells exist at the peak of the response and that memory cells accumulate during the cell deletion. Thus, it is probably more beneficial to the organism to rid itself of effector T cells than to acquire memory T cells.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI-17134, AI-18785, AI-22295, and AI-52225 (to P.M.) and by start-up funds and a Trustee Research Grant from Cincinnati Children's Hospital Research Foundation (to D.A.H).
Abbreviations: ROS, reactive oxygen species; MnTBAP, Mn(III) tetrakis(5,10,15,20-benzoic acid)porphyrin; SEB, staphylococcal enterotoxin B; MiT, plasmid murine stem cell virus internal ribosomal entry site Thy1.1; TCR, T cell antigen receptor; VβDO, Vβ8.2+ TCR transgenic; MnSOD, Mn superoxide dismutase; PI-3K, phosphatidylinositol 3-kinase.
References
- 1.Kannan, K. & Jain, S. (2000) Pathophysiology 7, 153-163. [DOI] [PubMed] [Google Scholar]
- 2.Hockenbery, D. M., Oltvai, Z. N., Yin, X. M., Milliman, C. L. & Korsmeyer, S. J. (1993) Cell 75, 241-251. [DOI] [PubMed] [Google Scholar]
- 3.Voehringer, D. W. & Meyn, R. E. (2000) Antioxid. Redox Signal. 2, 537-550. [DOI] [PubMed] [Google Scholar]
- 4.Lee, M., Hyun, D. H., Marshall, K. A., Ellerby, L. M., Bredesen, D. E., Jenner, P. & Halliwell, B. (2001) Free Radical Biol. Med. 31, 1550-1559. [DOI] [PubMed] [Google Scholar]
- 5.Ellerby, L. M., Ellerby, H. M., Park, S. M., Holleran, A. L., Murphy, A. N., Fiskum, G., Kane, D. J., Testa, M. P., Kayalar, C. & Bredesen, D. E. (1996) J. Neurochem. 67, 1259-1267. [DOI] [PubMed] [Google Scholar]
- 6.Maulik, N., Engelman, R. M., Rousou, J. A., Flack, J. E., III, Deaton, D. & Das, D. K. (1999) Circulation 100, II369-II375. [DOI] [PubMed] [Google Scholar]
- 7.Chang, W. K., Yang, K. D., Chuang, H., Jan, J. T. & Shaio, M. F. (2002) Clin. Immunol. 104, 151-160. [DOI] [PubMed] [Google Scholar]
- 8.Pugazhenthi, S., Nesterova, A., Jambal, P., Audesirk, G., Kern, M., Cabell, L., Eves, E., Rosner, M. R., Boxer, L. M. & Reusch, J. E. B. (2003) J. Neurochem. 84, 982-996. [DOI] [PubMed] [Google Scholar]
- 9.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Bouillet, P., Strasser, A., Kappler, J. & Marrack, P. (2002) Immunity 16, 759-767. [DOI] [PubMed] [Google Scholar]
- 10.Hildeman, D. A., Mitchell, T., Teague, T. K., Henson, P., Day, B. J., Kappler, J. & Marrack, P. C. (1999) Immunity 10, 735-744. [DOI] [PubMed] [Google Scholar]
- 11.Teague, T. K., Hildeman, D., Kedl, R. M., Mitchell, T., Rees, W., Schaefer, B. C., Bender, J., Kappler, J. & Marrack, P. (1999) Proc. Natl. Acad. Sci. USA 96, 12691-12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell, T. C., Hildeman, D., Kedl, R. M., Teague, T. K., Schaefer, B. C., White, J., Zhu, Y., Kappler, J. & Marrack, P. (2001) Nat. Immunol. 2, 397-402. [DOI] [PubMed] [Google Scholar]
- 13.Fenton, R. G., Marrack, P., Kappler, J. W., Kanagawa, O. & Seidman, J. G. (1988) Science 241, 1089-1092. [DOI] [PubMed] [Google Scholar]
- 14.Naviaux, R. K., Costanzi, E., Haas, M. & Verma, I. M. (1996) J. Virol. 70, 5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrack, P., Kappler, J. & Mitchell, T. (1999) J. Exp. Med. 189, 521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrack, P., Mitchell, T., Hildeman, D., Kedl, R., Teague, T. K., Bender, J., Rees, W., Schaefer, B. C. & Kappler, J. (2000) Curr. Opin. Immunol. 12, 206-209. [DOI] [PubMed] [Google Scholar]
- 17.Turrens, J. F., Alexandre, A. & Lehninger, A. L. (1985) Arch. Biochem. Biophys. 237, 408-414. [DOI] [PubMed] [Google Scholar]
- 18.Sprong, C., Janssen, Y. M. & Borm, P. J. (1991) Med. Hypotheses 34, 296-299. [DOI] [PubMed] [Google Scholar]
- 19.Kuthan, H. & Ullrich, V. (1982) Eur. J. Biochem. 126, 583-588. [DOI] [PubMed] [Google Scholar]
- 20.Kops, G. J., Dansen, T. B., Polderman, P. E., Saarloos, I., Wirtz, K. W., Coffer, P. J., Huang, T. T., Bos, J. L., Medema, R. H. & Burgering, B. M. (2002) Nature 419, 316-321. [DOI] [PubMed] [Google Scholar]
- 21.Lee, B. S., Chen, J., Angelidis, C., Jurivich, D. A. & Morimoto, R. I. (1995) Proc. Natl. Acad. Sci. USA 92, 7207-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohrdanz, E., Schmuck, G., Ohler, S. & Kahl, R. (2001) Brain Res. 900, 128-136. [DOI] [PubMed] [Google Scholar]
- 23.Tsutsui, H., Ide, T., Shiomi, T., Kang, D., Hayashidani, S., Suematsu, N., Wen, J., Utsumi, H., Hamasaki, N. & Takeshita, A. (2001) Circulation 104, 2883-2885. [DOI] [PubMed] [Google Scholar]
- 24.Scheel-Toellner, D., Pilling, D., Akbar, A. N., Hardie, D., Lombardi, G., Salmon, M. & Lord, J. M. (1999) Eur. J. Immunol. 29, 2603-2612. [DOI] [PubMed] [Google Scholar]
- 25.Kissil, J. L., Deiss, L. P., Bayewitch, M., Raveh, T., Khaspekov, G. & Kimchi, A. (1995) J. Biol. Chem. 270, 27932-27936. [DOI] [PubMed] [Google Scholar]
- 26.Majumder, P. K., Mishra, N. C., Sun, X., Bharti, A., Kharbanda, S., Saxena, S. & Kufe, D. (2001) Cell Growth Differ. 12, 465-470. [PubMed] [Google Scholar]
- 27.D'Adamio, F., Zollo, O., Moraca, R., Ayroldi, E., Bruscoli, S., Bartoli, A., Cannarile, L., Migliorati, G. & Riccardi, C. (1997) Immunity 7, 803-812. [DOI] [PubMed] [Google Scholar]
- 28.Farahani, R., Fong, W. G., Korneluk, R. G. & MacKenzie, A. E. (1997) Genomics 42, 514-518. [DOI] [PubMed] [Google Scholar]
- 29.Teague, T. K., Marrack, P., Kappler, J. W. & Vella, A. T. (1997) J. Immunol. 158, 5791-5796. [PubMed] [Google Scholar]
- 30.Patel, M. & Day, B. J. (1999) Trends Pharmacol. Sci. 20, 359-364. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh, Y., Guan, Y., Tu, C., Bratt, P. J., Angerhofer, A., Lepock, J. R., Hickey, M. J., Tainer, J. A., Nick, H. S. & Silverman, D. N. (1998) Biochemistry 37, 4731-4739. [DOI] [PubMed] [Google Scholar]
- 32.Bouillet, P., Cory, S., Zhang, L. C., Strasser, A. & Adams, J. M. (2001) Dev. Cell 1, 645-653. [DOI] [PubMed] [Google Scholar]
- 33.Tzung, S., Kim, K., Basanez, G., Giedt, C., Simon, J., Zimmerberg, J., Zhang, K. & Hockenbery, D. M. (2001) Nat. Cell Biol. 3, 183-191. [DOI] [PubMed] [Google Scholar]
- 34.Hochman, A., Sternin, H., Gorodin, S., Korsmeyer, S., Ziv, I., Melamed, E. & Offen, D. (1998) J. Neurochem. 71, 741-748. [DOI] [PubMed] [Google Scholar]
- 35.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Kappler, J. & Marrack, P. (2002) Curr. Opin. Immunol. 14, 354-359. [DOI] [PubMed] [Google Scholar]
- 36.Sandstrom, P. A. & Buttke, T. M. (1993) Proc. Natl. Acad. Sci. USA 90, 4708-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandstrom, P. A., Mannie, M. D. & Buttke, T. M. (1994) J. Leukocyte Biol. 55, 221-226. [DOI] [PubMed] [Google Scholar]
- 38.Bauer, M. K., Vogt, M., Los, M., Siegel, J., Wesselborg, S. & Schulze-Osthoff, K. (1998) J. Biol. Chem. 273, 8048-8055. [DOI] [PubMed] [Google Scholar]
- 39.Devadas, S., Zaritskaya, L., Rhee, S. G., Oberley, L. & Williams, M. S. (2002) J. Exp. Med. 195, 59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strasser, A., Harris, A. W., Huang, D. C., Krammer, P. H. & Cory, S. (1995) EMBO J. 14, 6136-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konorev, E. A., Kotamraju, S., Zhao, H., Kalivendi, S., Joseph, J. & Kalyanaraman, B. (2002) Free Radical Biol. Med. 33, 988-997. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell, T., Kappler, J. & Marrack, P. (1999) J. Immunol. 162, 4527-4535. [PubMed] [Google Scholar]
- 43.Vella, A. T., Dow, S., Potter, T. A., Kappler, J. & Marrack, P. (1998) Proc. Natl. Acad. Sci. USA 95, 3810-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vella, A., Teague, T. K., Ihle, J., Kappler, J. & Marrack, P. (1997) J. Exp. Med. 186, 325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahmed, N. N., Grimes, H. L., Bellacosa, A., Chan, T. O. & Tsichlis, P. N. (1997) Proc. Natl. Acad. Sci. USA 94, 3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly, E., Won, A., Refaeli, Y. & Van Parijs, L. (2002) J. Immunol. 168, 597-603. [DOI] [PubMed] [Google Scholar]
- 47.Pugazhenthi, S., Nesterova, A., Sable, C., Heidenreich, K. A., Boxer, L. M., Heasley, L. E. & Reusch, J. E. (2000) J. Biol. Chem. 275, 10761-10766. [DOI] [PubMed] [Google Scholar]
- 48.Sohur, U. S., Dixit, M. N., Chen, C. L., Byrom, M. W. & Kerr, L. A. (1999) Gene Expression 8, 219-229. [PMC free article] [PubMed] [Google Scholar]
- 49.Watson, P. A., Nesterova, A., Burant, C. F., Klemm, D. J. & Reusch, J. E. (2001) J. Biol. Chem. 276, 46142-46150. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, B. E., Mochon, E. & Boxer, L. M. (1996) Mol. Cell. Biol. 16, 5546-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, F., Rincon, M., Flavell, R. A. & Aune, T. M. (2000) J. Immunol. 165, 1762-1770. [DOI] [PubMed] [Google Scholar]
- 52.Matsushita, H., Morishita, R., Nata, T., Aoki, M., Nakagami, H., Taniyama, Y., Yamamoto, K., Higaki, J., Yasufumi, K. & Ogihara, T. (2000) Circ. Res. 86, 974-981. [DOI] [PubMed] [Google Scholar]
- 53.Gardai, S., Whitlock, B. B., Helgason, C., Ambruso, D., Fadok, V., Bratton, D. & Henson, P. M. (2002) J. Biol. Chem. 277, 5236-5246. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki, A., Yamaguchi, M. T., Ohteki, T., Sasaki, T., Kaisho, T., Kimura, Y., Yoshida, R., Wakeham, A., Higuchi, T., Fukumoto, M., Tsubata, T., et al. (2001) Immunity 14, 523-534. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama, K., Nakayama, K., Negishi, I., Kuida, K., Shinkai, Y., Louie, M. C., Fields, L. E., Lucas, P. J., Stewart, V., Alt, F. W. & Loh, D. Y. (1993) Science 261, 1584-1588. [DOI] [PubMed] [Google Scholar]
- 56.Kaech, S., Hemby, S., Kersh, E. & Ahmed, R. (2002) Cell 111, 837-851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.