Abstract
Oocytes contain a maternal store of the histone variant MacroH2A, which is eliminated from zygotes shortly after fertilization. Preimplantation embryos then execute three cell divisions without MacroH2A before the onset of embryonic MacroH2A expression at the 16-cell stage. During subsequent development, MacroH2A is expressed in most cells, where it is assembled into facultative heterochromatin. Because differentiated cells contain heterochromatin rich in MacroH2A, we investigated the fate of MacroH2A during somatic cell nuclear transfer (SCNT). The results show that MacroH2A is rapidly eliminated from the chromosomes of transplanted somatic cell nuclei by a process in which MacroH2A is first stripped from chromosomes, and then degraded. Furthermore, MacroH2A is eliminated from transplanted nuclei by a mechanism requiring intact microtubules and nuclear envelope break down. Preimplantation SCNT embryos express endogenous MacroH2A once they reach the morula stage, similar to the timing observed in embryos produced by natural fertilization. We also show that the ability to reprogram somatic cell heterochromatin by SCNT is tied to the developmental stage of recipient cell cytoplasm because enucleated zygotes fail to support depletion of MacroH2A from transplanted somatic nuclei. Together, the results indicate that nuclear reprogramming by SCNT utilizes the same chromatin remodeling mechanisms that act upon the genome immediately after fertilization.
Introduction
Recent successes in mammalian cloning using differentiated somatic nuclei indicate that the ooplasm of metaphase II oocytes contains activities that can erase most of the epigenetic memory of cellular differentiation. After nuclear transfer, extensive chromatin remodeling events result in increased accessibility to DNA (Byrne et al., 2003; Hansis et al., 2004; Kikyo et al., 2000; Simonsson and Gurdon, 2004), and are the underlying basis for the epigenetic regulation of gene expression that governs reprogramming during somatic cell nuclear transfer (SCNT). In nuclear transfer experiments, exchange of chromatin proteins has been demonstrated following injection of Xenopus oocytes with human or Xenopus somatic nuclei, which lose 80–90% of preradiolabeled nuclear proteins and incorporate oocyte proteins (Gurdon et al., 1979). In mammalian SCNT, rapid nuclear protein exchange also occurs; for example, histone H1 of somatic donor nuclei is replaced by the oocyte-specific H1 linker histone (H1FOO) within minutes of nuclear transfer (Gao et al., 2004). Hence, successful cloning of mammals seems to rely on factors present in the oocyte to execute exchange of chromatin proteins. In general, mammalian clones do not develop when somatic nuclei are transferred into enucleated blastomeres from preimplantation embryos (McGrath and Solter, 1984; Robl et al., 1987; Wakayama et al., 2000), and it is generally believed that the cytoplasm of these recipient cells lacks the components necessary to carry out epigenetic reprogramming. However, a recent report shows that the reprogramming activities are transiently available in mitotic zygotes, which lack intact nuclear envelopes (Egli et al., 2007). An understanding of the subcellular location and developmental timing for which reprogramming activities are available is emerging, but detailed molecular mechanisms that function in oocyte-mediated reprogramming are not currently available.
MacroH2A is a unique histone variant, consisting of an N-terminal region that closely resembles conventional histone H2A, and a nonhistone domain at its carboxyl end that constitutes almost two-thirds of the molecular protein mass (Pehrson and Fried, 1992). The MacroH2A NHD has been shown to impede SWI/SNF nucleosome remodeling (Angelov et al., 2003), and recent evidence has suggested its involvement in the recruitment of histone deacetylases (Chakravarthy et al., 2005). MacroH2A colocalizes with centromeric heterochromatin (Costanzi et al., 2000), is present in the developing XY body in early pachytene spermatocytes (Hoyer-Fender et al., 2000), and has been shown to coalesce into the macrochromatin body (MCB), which defines the inactive X chromosome of female mammals (Costanzi and Pehrson, 1998).
Maternal store histone variants such as H1oo (also known as H1FOO) likely regulate gene expression during during oocyte maturation and the initial stages of preimplanation development (Tanaka et al., 2001). Maternal-store H1FOO is rapidly replaced with the somatic linker histone H1 after fertilization or nuclear transfer (Gao et al., 2004). Another recent study shows that a high percentage of nuclei in one-cell SCNT mouse embryos have improperly remodeled heterochromatin, which is alleviated in part by treatment with Trichostatin A (Maalouf et al., 2009), a finding that also indicates that chromatin reorganization is critical for normal preimplantation development and successfully SCNT. A growing body of evidence indicates that the mechanisms that mediate nuclear reprogramming during SCNT are epigenetic in nature (Armstrong et al., 2006), and that constitutive heterochromatin is actively managed during mammalian preimplantation development (Probst and Almouzni, 2008). Our group has shown that oocytic MacroH2A levels drop to undetectable levels soon after metaphase II (MII)-arrested oocytes are fertilized, and do not reappear until the morula stage (Chang et al., 2005). Until now, the behavior of MacroH2A during SCNT has not been studied. Here, we demonstrate that ooplasm contains a potent activity that efficiently strips MacroH2A from chromatin when somatic nuclei are transplanted into enucleated MII oocytes. Furthermore, our results define the developmental timing and subcellular location of the responsible chromatin remodeling activity. Last, we provide mechanistic insights into the process whereby MacroH2A is removed from the chromatin of transplanted nuclei.
Materials and Methods
Chemicals
Unless otherwise indicated, all chemicals were purchased from Sigma Chemical Co. (St. Louis, MO, USA).
Animals and recovery of oocytes and embryos
B2D6F1 mice (Charles River Laboratories, Wilmington, MA, USA) were used to obtain MII stage oocytes, cumulus cells, and zygotes. All animals were handled under the guidelines of the University of Connecticut Institutional Animal Care and Use Committee (IACUC). Superovulation was induced with 5 IU of equine chorionic gonadotropin (eCG), followed by 5 IU of human chorionic gonadotropin (hCG) 48 h later. Oocytes were flushed from oviducts 14 h after hCG treatment and were freed of cumulus cells by brief exposure to 100 IU/mL of hyaluronidase at 37°C with gentle pipetting. Oocytes were incubated in Chatot, Ziomek, Bavister (CZB) medium (Chatot et al., 1989) before nuclear transfer.
To collect zygotes and early embryos, females were paired with CD-1 males after superovulation, and were inspected the following morning for copulation plugs. Zygotes and embryos at the four- and eight-cell stage were flushed from oviducts 22 and 68 h after mating or hCG treatments, respectively, and were briefly incubated in KSOM + AA medium (Specialty Media, Phillipsburg, NJ, USA) before use.
Enucleation of MII oocytes and zygotes
Mature MII oocytes and zygotes were enucleated with a glass pipette (10–12-μm inner diameter for oocytes and 18 μm for zygotes) in HEPES-buffered CZB medium supplemented with 5.5 mM glucose and cytochalasin B (2.5 μg/mL for oocytes and 5.0 μg/mL for zygotes). After enucleation, the cytoplasts of MII oocytes and zygotes were washed thoroughly in CZB medium to remove any remaining cytochalasin B.
Cumulus cell nucleus injection
Cumulus cells obtained in the course of ooctye collection (above) were resuspended with 3% (w/v) polyvinyl pyrolidone (PVP; Mr 360 × 103) dissolved in HEPES-buffered CZB. To obtain nuclei, cumulus cells were drawn in and out of the injection pipette (inner diameter, 6 μm), which was loaded with mercury, and attached to a piezo-driving unit (PMAS-CT150, Prime Tech, Japan), until the cell membrane was broken and separated from the nucleus. Nuclei were injected into the enucleated oocytes and zygotes by piezo-driven microinjection. Injected oocytes were cultured in CZB medium for 1–3 h and subjected to oocyte activation.
Embryonic stem cell nucleus injection
Embryonic stem (ES) cells were cultured in DMEM supplemented with 15% fetal bovine srum (FBS) in the presence of leukemia inhibitory factor (LIF). ES cells were trypsinized and resuspended in 3% (w/v) polyvinyl pyrolidone (PVP; Mr 360 × 103) dissolved in HEPES-buffered CZB. To obtain ES cell nuclei, an ES cell was drawn in and out of the injection pipette (inner diameter, 4–5 μm), which was loaded with mercury and attached to a piezo-driving unit (PMAS-CT150, Prime Tech), until the cell membrane was broken and separated from the nucleus. Nuclei were injected into the enucleated oocyte by piezo-driven microinjection. Injected oocytes were cultured in CZB medium for 1–3 h and subjected to oocyte activation.
Four- and eight-cell stage embryonic nucleus injection
To isolate blastomeres from four- and eight-cell embryos, the zona pellucida was first dissolved with acidic phosphate-buffered saline (PBS) (pH 2.5), and individual blastomeres were dissociated by gentle pipetting of the zona-free embryos in Ca2+- and Mg2+-free PBS. Blastomere nuclei were obtained by drawing the blastomeres in and out of the injection pipette (inner diameter, 11 μm) until the cell membrane was broken and separated from the nucleus. Nuclei were then injected into the enucleated oocytes and zygotes using piezo-driven microinjection. Injected oocytes were cultured in CZB medium for 1 or 3 h.
Oocyte activation and embryo culture
Micromanipulated oocytes were subjected to oocyte activation by exposure to activation medium (Ca2+-free CZB medium, 10 mM SrCl2, and 5 μg/mL cytochalasin B) for 6 h and then incubated in KSOM + AA medium (Specialty Media, Phillipsburg, NJ, USA).
Antibodies
The rabbit antimouse MacroH2A antibody has been described previously (Ma et al., 2005). Mouse monoclonal anti-γ-tubulin (Sigma, cat. no. T-6557, diluted 1:50) and mouse monoclonal anti-α-tubulin (Sigma, cat. no. T-5168, diluted 1:200) were used in combination with anti-MacroH2A (1:1000). Anti-MacroH2A was detected using FITC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Inc., West Grove, PA, 1:200). Mouse α-tubulin or γ-tubulin antibodies were detected with donkey antimouse TRITC-conjugated IgG (Jackson ImmunoResearch Inc., 1:200).
Immunofluorescence and cell imaging
Preparation of oocytes and embryos for immunofluorescence and subsequent analysis with confocal microscopy has been described previously (Chang et al., 2005). Individual confocal sections were electronically merged so that all nuclei were visible in a single image.
Results
Somatic MacroH2A rapidly dissociates from chromosomes after nuclear transfer
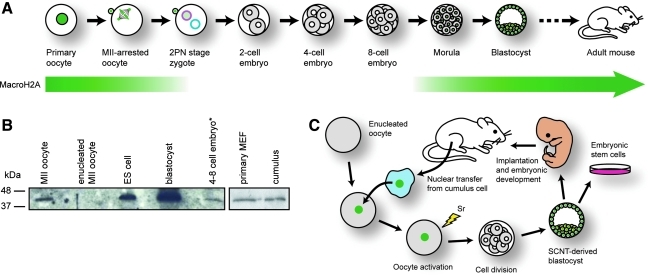
We previously demonstrated that oocytes contain a maternal store of MacroH2A, which is reduced to undetectable levels soon after fertilization and is absent from the two-, four-, and eight-cell stage embryo before expression of MacroH2A of embryonic origin, which commences at the morula stage (Chang et al., 2005); (summarized in Fig. 1A). We assayed the expression of macroH2A by Western analysis of extracts from pooled MII oocytes, enucleated MII oocytes, ES cells, and cleavage stage embryos (Fig. 1B; left panel). We also found that cumulus cells express MacroH2A at levels comparable to other somatic cells, in this case, primary mouse embryonic fibroblasts (Fig. 1B; right panel). Because embryos produced by natural fertilization rapidly eliminate MacroH2A soon after fertilization, we were interested in determining the fate of MacroH2A residing in the heterochromatin of differentiated somatic cells, when such nuclei are subjected to somatic cell nuclear transfer (SCNT; Fig. 1C).
FIG. 1.
MacroH2A distribution during early mammalian development. (A) MacroH2A (green) is present in the germinal vesicle of primary oocytes and is associated with condensed chromosomes and the first polar body in metaphase II (MII)-arrested oocytes. Upon fertilization, some maternal store MacroH2A is extruded with the second polar body and the remainder is removed prior to formation of the maternal pronucleus (pink). MacroH2A is absent from the paternal pronucleus (blue). Cell division proceeds in the absence of MacroH2A until the morula stage, where MacroH2A expression begins and is sustained throughout the remainder of development. See (Chang et al. 2005) for details. (B) A MacroH2A-specific antibody was used in Western analysis to examine MacroH2A levels in whole-cell lysates from MII oocytes (n = 100), enucleated oocytes (n = 100), embryonic stem (ES) cells (n = 2000), blastocysts (n = 100), and four-cell stage embryos (n = 100) (left panel), as well as mouse embryonic fibroblasts (MEF) (n = 2000) and cumulus cells (n = 2000) (right panel). The asterisk denotes that the faint signal in the last lane may result from residual MacroH2A contained in polar bodies. (C) Summary of experimental design and rationale to investigate the fate of somatic MacroH2A (green nuclei) during somatic cell nuclear transfer (SCNT). A MacroH2A-positive nucleus from a somatic cell—in this case, a cumulus cell—is injected into an enucleated oocyte. Following oocyte activation, successive cell divisions produce a cloned blastocyst, which can be used to obtain embryonic stem cells or can be implanted into a female mouse to produce a reproductive clone. This study investigates the fate of somatic cell MacroH2A dynamics in embryo culture to the blastocyst stage.
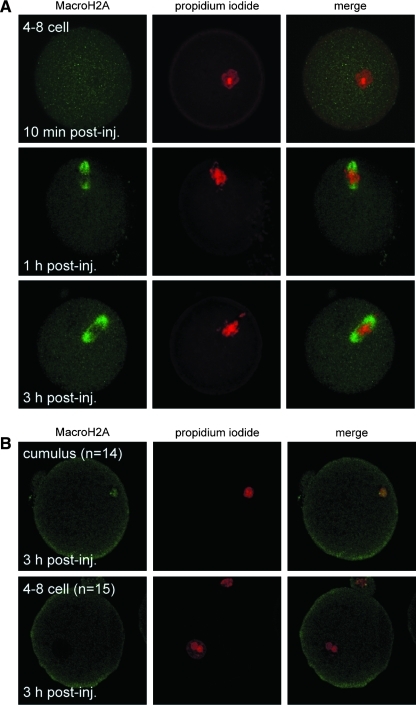
To investigate the fate of MacroH2A during nuclear transfer, cumulus cell nuclei were injected into enucleated and intact MII-stage oocytes. Injected oocytes were fixed at 10 min, 1 h, and 3 h following cumulus cell nuclear transfer. Of 25 injected oocytes examined, all had readily detectable MacroH2A in the nuclei. Ten minutes after injection, the MacroH2A signal (green) was still present in the transferred cumulus nuclei (Fig. 2, top row). After 1 h, the cumulus cell chromosomes (red) condensed, and MacroH2A became noncoincident with chromosomes and was localized to two foci resembling mitotic spindle poles (Fig. 2, middle row). Three hours following injection, chromosomes had separated into an anaphase configuration, and the majority of intracellular MacroH2A was spatially distinct from condensed chromosomes and localized to two pole-like foci (Fig. 2, bottom row). To determine whether the chromatin dynamics of MacroH2A was restricted to highly differentiated nuclei, we also injected enucleated oocytes with nuclei from ES cells and found that 1 h after nuclear transfer most of the MacroH2A was located in the regions resembling spindle poles (data not shown). These results show that during the SCNT process MacroH2A dissociates from chromosomes of somatic cell origin soon after they condense.
FIG. 2.
Intracellular distribution of MacroH2A following nuclear transfer. Enucleated metaphase II oocytes (n = 25) were injected with cumulus cell nuclei and observed by indirect immunofluorescence at 10 min (top row), 1 h (middle row), and 3 h (bottom row) postinjection. Injected oocytes were stained with a MacroH2A-specific antibody (left column) and propidium iodide (PI) to visualize DNA (middle column). Green (MacroH2A) and red (PI) signals were merged to show colocalization (right column). Representative images are shown.
MacroH2A is depleted from chromosomes and localizes to spindle poles during SCNT in a process dependent upon intact microtubules
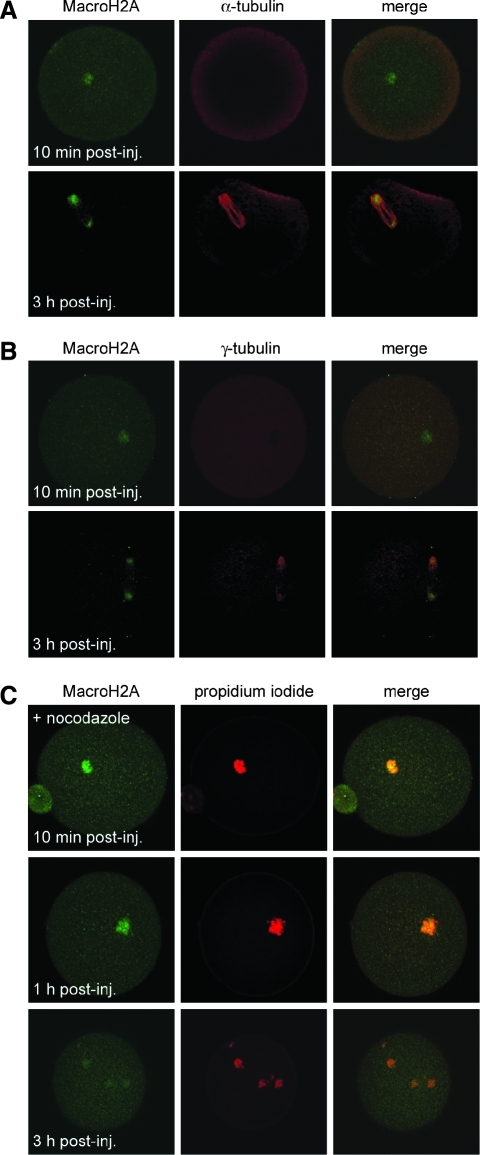
Our results demonstrate that MacroH2A dissociates from chromosomes of transferred somatic nuclei and becomes localized to two foci that, based on their positions relative to condensed chromosomes, resemble meiosis II spindle poles. To investigate if these structures were indeed spindle poles, we performed coimmunofluorescence on oocytes at 10 min and 3 h after nuclear transfer using antibodies specific for MacroH2A and either α-tubulin (to stain microtubules) or γ-tubulin (to stain microtubule organizing centers (MTOCs) of spindle poles). After 10 min, the majority of MacroH2A was confined to the interior of transplanted somatic cell nuclei (Fig. 3A and B, top rows). After 3 h, MacroH2A was dissociated from chromosomes and resided at the poles of the first SCNT spindle made apparent by staining for α-tubulin (Fig. 3A). The redistribution of somatic MacroH2A to spindle poles was further confirmed by the finding that MacroH2A becomes coincident with γ-tubulin, a marker of MTOCs, by 3 h post-SCNT (Fig. 3B). We also used a second fixation method to assess the integrity of the spindle apparatus in preimplantation embryos, which yielded similar results (Suppl. Fig. 1).
FIG. 3.
MacroH2A associates with microtubules and aggregates at centrosomes in SCNT oocytes. To investigate intracellular MacroH2A distribution with respect to the spindle apparatus, cumulus cell nucleus-injected oocytes were subjected to coimmunofluorescence using an antibody against MacroH2A and antibodies specific to α-tubulin (A) or γ-tubulin (B) at 10 min (top row) and 3 h (bottom row) postinjection. Green (MacroH2A) and red (α- or γ-tubulin) signals were merged to show colocalization (right column). (C) Treatment of SCNT oocytes with nocodazole prevents the redistribution of MacroH2A (green) from chromosomes (PI) to microtubules.
To investigate the functional involvement of microtubules for MacroH2A dynamics during SCNT, we treated SCNT embryos with nocodazole to prevent microtubule assembly. After 10 min, chromosomes were condensed and confined to the somatic cell nuclei (Fig. 3C, top row). After 1 h, condensed chromosomes remained in a tight cluster (Fig. 3C, middle row). After 3 h, nuclear envelope breakdown was complete, but chromosomes remained condensed and were dispersed randomly throughout the ooplasm (Fig. 3C, bottom row). At all three time points, MacroH2A remained tightly associated with chromosomes, indicating that intact microtubules are required for the dissociation of MacroH2A from chromosomes of somatic cell origin.
Chromatin dynamics of MacroH2A in SCNT embryos after oocyte activation
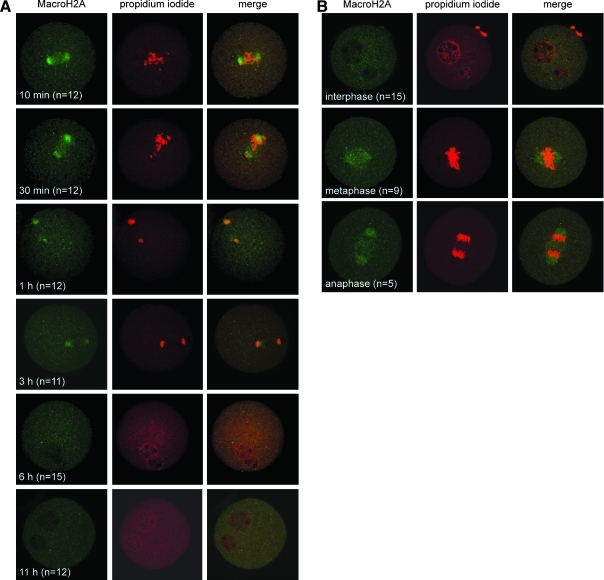
We next wished to investigate the behavior of MacroH2A following activation of the SCNT embryos. For this, cumulus cell nucleus-injected oocytes were cultured for 1–3 h in vitro and subjected to activation by strontium (to initiate the first cell cycle) and cytochalasin B (to prevent extrusion of the second polar body) (see Materials and Methods for details). Activated oocytes were then fixed for immunofluorescence at 10 min, 0.5, 1, 3, 6, and 11 h after treatment. At 10 (Fig. 4A, top row) and 30 min (Fig. 4A, second row) postactivation, the condensed cumulus cell chromosomes were randomly scattered along the newly assembled spindle, but the majority of MacroH2A signal was already located at spindle poles. By 1 h (Fig. 4A, third row) and 3 h (Fig. 4A, fourth row) postactivation, we observed an anaphase/telophase configuration of chromosomes and a progressive fading of the MacroH2A signal, which could be due to either diffusion of MacroH2A into the cytoplasm or active elimination of MacroH2A. After 6 h, pronuclei formed and chromosomes underwent progressive decondensation, while MacroH2A foci remained absent (Fig. 4A, fifth and sixth row).
FIG. 4.
MacroH2A distribution in activated SCNT zygotes. (A) Cumulus cell nucleus-injected oocytes were activated with 10 mM SrCl2 and 5 μg/mL Cytochalasin B and observed by indirect immunofluorescence at 10 min, 30 min, 1 h, 3 h, 6 h, and 11 h postactivation. Activated oocytes were stained with a MacroH2A-specific antibody (left column) and propidium iodide (PI) to visualize DNA (middle column). Green (MacroH2A) and red (PI) signals were merged to show colocalization (right column). The number of cells (n) observed at each interval is indicated. (B) Cumulus cell nucleus-injected oocytes were visualized 16 h after activation to examine MacroH2A distribution during the first mitotic division. Indirect immunofluorescence was used to detect MacroH2A during interphase (top row), metaphase (middle row), and anaphase (bottom row). Oocytes were stained with a MacroH2A-specific antibody (left column) and propidium iodide (PI) to visualize DNA (middle column). Green (MacroH2A) and red (PI) signals were merged to show colocalization (right column). The number of cells (n) observed at each interval is indicated.
As pronuclear envelopes formed, we noted that the interior of the enlarged pronuclei lacked MacroH2A, whereas a diffuse particulate haze of signal, possibly representing a residual pool of cytosolic MacroH2A, became detectable prior to fusion of the pronuclei (Fig. 4B, top row). During the first embryonic mitosis, we again detected MacroH2A signal at spindle poles, albeit at low levels, when the replicated chromosomes aligned on the metaphase plate (Fig. 4B, middle row), a pattern that persisted to the first anaphase (Fig. 4B, bottom row).
Dynamics of MacroH2A in cleavage-stage SCNT embryos
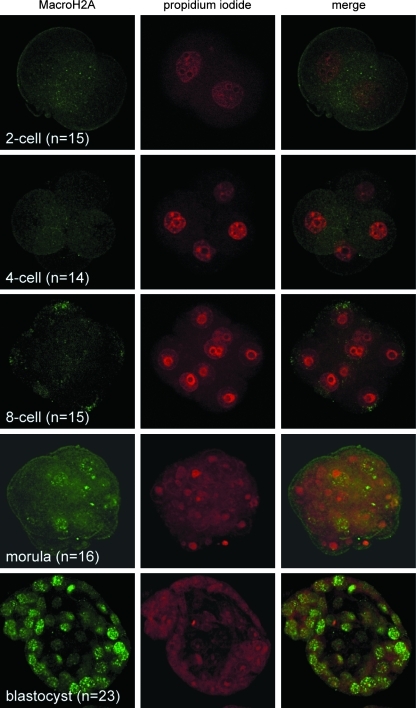
In mouse embryos produced by fertilization in vivo, MacroH2A is undetectable in two-cell to eight-cell embryos, but expression is initiated at the morula stage and persists throughout the remainder of development (Chang et al., 2005). To address whether MacroH2A exhibits similar dynamics in SCNT embryos, we fixed embryos derived from cumulus cell nuclear transfer at the two-, four-, and eight-cell stage, as well as at the morula and blastocyst stages. Similar to naturally produced embryos, we failed to detect MacroH2A in the nuclei of two- (Fig. 5, top row), four- (Fig. 5, second row), and eight-cell (Fig. 5, third row) stage embryos. All SCNT embryos at the morula (Fig. 5, fourth row), and blastocyst (Fig. 5, bottom row) stages contained nuclear MacroH2A, concurrent with the stage at which MacroH2A is expressed in developing embryos produced by natural fertilization (Chang et al., 2005). These observations show that the pattern of MacroH2A distribution in cloned embryos assumes that of natural embryos, suggesting that MacroH2A expression is properly repressed in SCNT embryos as early as the two-cell stage.
FIG. 5.
Distribution of MacroH2A during preimplantation development of cloned mouse embryos. Embryos formed following cumulus cell nuclear transfer were observed by indirect immunofluorescence at the two-cell (top row), four-cell (second row), eight-cell (third row), morula (fourth row), and blastocyst (fifth row) stages. Cloned embryos were stained with a MacroH2A-specific antibody (left column) and propidium iodide (PI) to visualize DNA (middle column). Green (MacroH2A) and red (PI) signals were merged to show colocalization (right column). The number of embryos (n) observed at each interval is indicated.
Reprogramming activities that act on MacroH2A during SCNT are developmentally restricted
Maternal stores of MacroH2A are rapidly depleted shortly after fertilization (Chang et al., 2005) and immediately after nuclear transfer, as demonstrated in this study. Furthermore, MacroH2A repression is maintained in two-, four-, and eight-cell SCNT embryos, prior to the onset of embryonic MacroH2A expression in SCNT morulae, a timing that is identical to that observed in natural embryos. We also typically observed a low-level particulate haze of MacroH2A signal after its elimination from chromosomes, which is excluded from pronuclei. However, this signal appeared to transiently reassociate with the first mitotic spindle upon nuclear envelope breakdown during the first embryonic mitosis (Fig. 4). To further explore the apparent affinity of the ooplasmic signal for the mitotic spindle apparatus, we injected nuclei from blastomeres at the four-cell stage (which are devoid of MacroH2A) into enucleated oocytes. One hour after injection, we observed an enrichment of MacroH2A in the region of the spindle and condensed chromosomes (Fig. 6A, middle row), a pattern that was maintained at least until 3 h postinjection (Fig. 6A, bottom row).
FIG. 6.
The ability to remodel MacroH2A heterochromatin is dependent on the developmental stage of the embryo. (A) MacroH2A associates with the spindle after injection of a nucleus from a four-cell stage embryo. Injected oocytes were stained 10 min (top row), 1 h (middle row), and 3 h (bottom row) postinjection using a MacroH2A-specific antibody (left column) and propidium iodide (PI) to visualize DNA (middle column). Green (MacroH2A) and red (PI) signals were merged to show colocalization (right column). (B) The enucleated zygote (i.e., both pronuclei have been removed) is incapable of eliminating MacroH2A following nuclear transfer. Enucleated zygotes were injected with nuclei from a cumulus cell (top row) or four-cell stage embryo (bottom row) and were analyzed 3 h postinjection by indirect immunofluorescence with a MacroH2A-specific antibody (left column) and propidium iodide (PI) to visualize DNA (middle column). Green (MacroH2A) and red (PI) signals were merged to show colocalization (right column). The number of zygotes observed (n) at each interval is indicated.
We next examined whether zygotes retain the capacity to remodel MacroH2A. For this, we injected cumulus cell nuclei into zygotes, from which the genetic material was removed. Three hours after injection, cumulus cell nuclei were still intact and retained their complement of MacroH2A (Fig. 6B, top row). We also injected enucleated zygotes with nuclei from blastomeres at the four-cell stage, which are devoid of MacroH2A. In this experiment, we again observed a failure of the enucleated zygotes to support breakdown of the nuclear envelopes of the transplanted nuclei (Fig. 6B, bottom row). Together, the results suggest that the activities responsible for MacroH2A remodeling are restricted to oocytes, and that nuclear envelope breakdown is a requirement for MacroH2A elimination from somatic chromatin.
Discussion
SCNT provides an excellent system for the investigation of epigenetic reprogramming mechanisms because it is rapid, highly efficient, and amenable to investigation using the tools of cell biology. During SCNT, the epigenetic status of highly differentiated cells is reversed to a totipotent state in a process that is complete within hours. The targeted formation of heterochromatin is essential for mammalian development, because inappropriate gene expression must be repressed as cellular lineages are established. The histone variant MacroH2A is assembled into a wide range of heterochromatin, including the sex chromatin of the inactive X chromosome (Costanzi and Pehrson, 1998; Turner et al., 2001), as development unfolds. In addition, MarcroH2A generally represses transcription initiation (Angelov et al., 2003; Doyen et al., 2006; Perche et al., 2000) and renders chromatin resistant to remodeling in somatic cells (Angelov et al., 2003). We reasoned that the investigation of MacroH2A during SCNT might yield insights into mechanisms that lead to the erasure of heterochromatin present within differentiated cells after their nuclei are transplanted into enucleated oocytes. This approach was made even more attractive by our previous finding that preimplantation embryos produced by natural fertilization rapidly eliminate a maternal store of MacroH2A upon fertilization and then develop to the morula stage in the absence of MacroH2A (Chang et al., 2005). The results presented in this study show that oocytes contain a chromatin-remodeling activity that rapidly strips MacroH2A from the chromosomes of somatic nuclei. Furthermore, we find that somatic heterochromatin (as marked by MacroH2A content) is erased within 6 h of nuclear transfer, with kinetics that are comparable to those which dictate the elimination of maternal-store macroH2A during normal preimplantation development (Chang et al., 2005).
We have shown that, following SCNT, MacroH2A is efficiently stripped from somatic chromosomes prior to localization to microtubules and aggregation at centrosomes. Importantly, we demonstrate that microtubule integrity is necessary for relocalization of MacroH2A to the centrosome, suggesting that MacroH2A is actively transported along spindle fibers. The domain structure of MacroH2A provides useful insights into its reorganization from chromosomes to the spindle. The nonhistone domain of MacroH2A contains a macro domain, which strongly binds ADP-ribose (Karras et al., 2005). ADP-ribose is assembled into branched polymers (poly ADP-ribose, or PAR) by poly-ADP-ribose polymerases (PARPs) (D'Amours et al., 1999). Interestingly, the spindle features concentrations of PAR and several PARPs localize to the spindle. Furthermore, hydrolysis of PAR leads to spindle disassembly (Chang et al., 2004). Because the spindle is rich in PAR, it may be that MacroH2A, once free from chromosomes, aggregates at centrosomes via interactions between the macrodomain and spindle-associated PAR. Determining the mechanism by which MacroH2A (and possibly other components of heterochromatin) are stripped from chromosomes and loaded onto microtubules will be of critical importance to understanding this process and its role in developmental reprogramming.
Our results are supported by the findings of others, which demonstrate MacroH2A relocalization to the centrosome in both ES cells and somatic cells (Chadwick and Willard, 2002; Mermoud et al., 2001; Rasmussen et al., 2000). Indeed, the centrosome may function as a storage compartment for MacroH2A until it is required for processes such as cell differentiation and X chromosome inactivation (Mermoud et al., 2001; Rasmussen et al., 2000), whereupon it is recruited to chromosomes to help silence X-linked and autosomal genes. However, centrosomal aggregation of MacroH2A is also significant since the centrosome is a major site of cellular protease activity (Corboy et al., 2005; Gordon, 2002), which has been implicated as a site of MacroH2A proteolysis in somatic cells (Chadwick and Willard, 2002). Treatment of cells with lactacystin, a potent inhibitor of the 20S proteasome, interfered with MacroH2A depletion (Chadwick and Willard, 2002), suggesting that MacroH2A turnover is mediated primarily, if not exclusively, by the ubiquitin–proteasome degradation pathway. This is particularly relevant for preimplantation development, given that MacroH2A levels are virtually nonexistent during the two-, four-, and eight-cell stages of naturally fertilized and SCNT-derived embryos (Chang et al., 2005); this study). Further studies will be required to determine whether proteasomal activity is responsible for elimination of MacroH2A during this period, and also to elucidate the signaling events which trigger this process.
The data show that activities exist within MII ooplasm that rapidly and thoroughly deplete MacroH2A from somatic cell chromatin soon after nuclear transfer. However, it seems clear that this activity is closely tied to the developmental stage of the recipient cell. When we transferred cumulus cell nuclei or blastomere nuclei from four-cell preimplantation embryos into MII oocytes, nuclear envelop breakdown (NEBD) readily occurred. However, transferred nuclei failed to undergo NEBD after injection into enucleated zygotes, and these failed to reorganize somatic MacroH2A content. Although cleavage-stage blastomeres lack macroH2A, we were surprised to observe spindle-associated MacroH2A signal soon after NEBD, which increased over 3 h. One possibility is that the MacroH2A gene may undergo a burst of transcription after transfer. Alternatively, MII ooplasm may contain maternal MacroH2A transcripts or low levels of dispersed particulate MacroH2A that is recruited to the spindle apparatus once it is assembled. A second, more likely possibility parallels the situation we observed during the first mitosis of SCNT embryos (Fig. 4B), where a residual pool of diffuse MacroH2A is recruited to the first mitotic spindle once the spindle is assembled and accessible following NEBD.
Our results show that somatic cell heterochromatin, as marked by MacroH2A, is rapidly disassembled by MII ooplasm as soon as it is accessible following nuclear transplantation. In addition, we note that by the time pronuclei are formed in SCNT embryos, the configuration of MacroH2A is identical to that observed in embryos produced by normal fertilization. Based on the timing and rapidity of MacroH2A reprogramming during SCNT, it seems likely that an activity that normally remodels maternal chromatin is co-opted for use during the SCNT process. Furthermore, because oocytes contain a maternal store of MacroH2A, which is rapidly depleted shortly after fertilization, it seems likely that this same mechanism is used to strip MacroH2A from chromosomes of transplanted somatic nuclei. We also find it interesting that developing oocytes and somatic cells both seem to contain significant amounts of heterochromatin, and that both are extensively remodeled in the fertilized oocyte. These similarities suggest that the mature oocyte is indeed a highly differentiated cell, but one that is uniquely poised to execute dramatic chromatin remodeling events, including the erasure of heterochromatin following fertilization.
Supplementary Material
Footnotes
The first two authors contributed equally to this work.
Acknowledgments
This article was supported by NIH Grant RO1AG023687 (T.R.) and USDA grant ARS 58-1265-7-028 (X.T.). We thank Therese Doherty for proofreading.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Angelov D. Molla A. Perche P.Y., et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- Armstrong L. Lako M. Dean W., et al. Epigenetic modification is central to genome reprogramming in somatic cell nuclear transfer. Stem Cells. 2006;24:805–814. doi: 10.1634/stemcells.2005-0350. [DOI] [PubMed] [Google Scholar]
- Byrne J.A. Simonsson S. Western P.S., et al. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- Chadwick B.P. Willard H.F. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S. Gundimella S.K. Caron C., et al. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C. Ma Y. Jacobs S., et al. A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev. Biol. 2005;278:367–380. doi: 10.1016/j.ydbio.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Chang P. Jacobson M.K. Mitchison T.J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Chatot C.L. Ziomek C.A. Bavister B.D., et al. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- Corboy M.J. Thomas P.J. Wigley W.C. Aggresome formation. Methods Mol. Biol. 2005;301:305–327. doi: 10.1385/1-59259-895-1:305. [DOI] [PubMed] [Google Scholar]
- Costanzi C. Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Costanzi C. Stein P. Worrad D.M., et al. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- D'Amours D. Desnoyers S. D'Silva I., et al. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Doyen C.M. An W. Angelov D. Bondarenko V., et al. Mechanism of polymerase II transcription repression by the histone varient macroH2A. Mol. Cell. Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D. Rosains J. Birkhoff G., et al. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- Gao S. Chung Y.G. Parseghian M.H., et al. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev. Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Gordon C. The intracellular localization of the proteasome. Curr. Top. Microbiol. Immunol. 2002;268:175–184. doi: 10.1007/978-3-642-59414-4_7. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B. Laskey R.A. De Robertis E.M., et al. Reprogramming of transplanted nuclei in amphibia. Int. Rev. Cytol. Suppl. 1979:161–178. doi: 10.1016/s0074-7696(08)60902-x. [DOI] [PubMed] [Google Scholar]
- Hansis C. Barreto G. Maltry N., et al. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr. Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Herman B. Langevin M.A. Albertini D.F. The effects of taxol on the organization of the cytoskeleton in cultured ovarian granulosa cells. Eur. J. Cell Biol. 1983;31:34–45. [PubMed] [Google Scholar]
- Hoyer-Fender S. Costanzi C. Pehrson J.R. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 2000;258:254–260. doi: 10.1006/excr.2000.4951. [DOI] [PubMed] [Google Scholar]
- Karras G.I. Kustatscher G. Buhecha H.R., et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1120. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikyo N. Wade P.A. Guschin D., et al. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science. 2000;289:2360–2362. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- Ma Y. Jacobs S.B. Jackson-Grusby L., et al. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J. Cell Sci. 2005;118:1607–1616. doi: 10.1242/jcs.02291. [DOI] [PubMed] [Google Scholar]
- Maalouf W.E. Liu Z. Brochard V. Renard J.P. Debey P. Beaujean N. Zink D. Trichostatin A treatment of cloned mouse embryos improves constitutive heterochromatin remodeling as well as developmental potential to term. BMC Dev. Biol. 2009;9:11. doi: 10.1186/1471-213X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. Solter D. Inability of mouse blastomere nuclei transferred to enucleated zygotes to support development in vitro. Science. 1984;226:1317–1319. doi: 10.1126/science.6542249. [DOI] [PubMed] [Google Scholar]
- Mermoud J.E. Tassin A.M. Pehrson J.R., et al. Centrosomal association of histone macroH2A1.2 in embryonic stem cells and somatic cells. Exp. Cell Res. 2001;268:245–251. doi: 10.1006/excr.2001.5277. [DOI] [PubMed] [Google Scholar]
- Pehrson J.R. Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Perche P.Y. Vourc'h C. Konecny L., et al. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 2000;10:1531–1534. doi: 10.1016/s0960-9822(00)00832-0. [DOI] [PubMed] [Google Scholar]
- Probst A.V. Almouzni G. Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation. 2008;76:15–23. doi: 10.1111/j.1432-0436.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen T.P. Mastrangelo M.A. Eden A., et al. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol. 2000;150:1189–1198. doi: 10.1083/jcb.150.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robl J.M. Prather R. Barnes F., et al. Nuclear transplantation in bovine embryos. J. Anim. Sci. 1987;64:642–647. doi: 10.2527/jas1987.642642x. [DOI] [PubMed] [Google Scholar]
- Simonsson S. Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- Tanaka M. Hennebold J.D. Macfarlane J., et al. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- Turner J.M. Burgoyne P.S. Singh P.B. M31 and macroH2A1.2 colocalize at the pseudoautosomal region during mouse melosis. J. Cell. Sci. 2001;114:3367–3375. doi: 10.1242/jcs.114.18.3367. [DOI] [PubMed] [Google Scholar]
- Wakayama T. Tateno H. Mombaerts P., et al. Nuclear transfer into mouse zygotes. Nat. Genet. 2000;24:108–109. doi: 10.1038/72749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.