Abstract
Multiple sclerosis (MS) is a chronic neurological disease of unknown etiology, but a genetic basis for the disease is undisputed. We have reported that CD24 is required for the pathogenicity of autoreactive T cells in experimental autoimmune encephalomyelitis, the mouse model of MS. Here we investigate the contribution of CD24 to MS by studying single-nucleotide polymorphism in the ORF among 242 MS patients and 207 population controls. This single-nucleotide polymorphism results in replacement of alanine (CD24a) with valine (CD24v) in the mature protein. We found that the CD24v/v renders a >2-fold increase in the relative risk of MS in the general population (P = 0.023). Among familial MS, the CD24v allele is preferentially transmitted into affected individuals (P = 0.017). Furthermore, 50% of CD24v/v patients with expanded disability status scale 6.0 reached the milestone in 5 years, whereas the CD24a/v (P = 0.00037) and CD24a/a (P = 0.0016) patients did so in 16 and 13 years, respectively. Moreover, our data suggest that the CD24v/v patients expressed higher levels of CD24 on peripheral blood T cells than did the CD24a/a patients. Transfection with CD24a and CD24v cDNA demonstrated that the CD24v allele can be expressed at higher efficiency than the CD24a alleles. Thus, CD24 polymorphism is a genetic modifier for susceptibility and progression of MS in the central Ohio cohort that we studied, perhaps by affecting the efficiency of CD24 expression on the cell surface.
Keywords: single-nucleotide polymorphism, disease susceptibility, autoimmunity, costimulatory molecules, T lymphocytes
Multiple sclerosis (MS) is a chronic disorder in the CNS that affects ≈0.1% of Caucasians of northern European origin (1). The incidence of MS is increased among family members of affected individuals. The concordance rate of the identical twins can be as high as 30% (1–3). The HLA loci is perhaps the most important genetic element for MS susceptibility, because the HLA-DR2 allele has been identified as the most important susceptibility gene among Caucasians (4–10). Several additional loci have also been proposed (8–12).
One of the whole-genome scans suggested a linkage disequilibrium in distal 6q (8) whose identity has not been revealed. An interesting candidate in the region is CD24 (13), which we showed to be essential for the induction of experimental autoimmune encephalomyelitis (EAE) in mice (13). CD24 is a glycosylphosphatidylinositol (GPI)-anchored cell surface protein with expression in a variety of cell types that can participate in the pathogenesis of MS, including activated T cells (14, 15), B cells (16), macrophages (17), dendritic cells (18), and local antigen-presenting cells in the CNS, such as vascular endothelial cells, astrocytes, and microglia (our unpublished observation). It is well established that in the mouse CD24 mediates a CD28-independent costimulatory pathway that promotes activation of CD4 and CD8 T cells (16–21). In addition, CD24 has been shown to modulate the very late antigen 4–fibronectin/vascular cell adhesion molecule 1 interaction (22), which is required for the migration of T cells to the CNS and therefore the development of EAE in the mouse (23). We have recently demonstrated that CD24 is required for the development of EAE in the mouse (13). Interestingly, CD24 controls a checkpoint of EAE pathogenesis after the autoreactive T cells are produced (13).
Because MS patients have a high frequency of autoreactive T cells, molecules that control events after T cell activation, such as CD24, will have a unique advantage as therapeutic target over those that are involved in early events. It is therefore of great importance to determine whether CD24 is relevant for the development of MS. Interestingly, the human CD24 gene has a single-nucleotide polymorphism (SNP), resulting in a nonconservative replacement of an amino acid (from alanine in CD24a to valine in CD24v) immediately preceding the putative cleavage site for the GPI anchor (ω-1 position) (24). The existence of such a SNP provided an opportunity to address the relevance of the CD24 gene in human MS susceptibility. Here we show that the CD24v/v genotype is associated with increased risk and more rapid progression of MS. Because the CD24v is more efficiently expressed than CD24a, the CD24 SNP may influence MS pathogenesis by affecting the expression of CD24. To our knowledge, this is the first SNP to have a significant impact on MS susceptibility.
Materials and Methods
Human Subjects. All sample collection and experimentation were approved by the Institutional Review Board, and informed consent from all participants was obtained before sample collection. Patients with definite MS, as diagnosed by K.R. at the Ohio State University Multiple Sclerosis Center according to the McDonald criteria (25), were offered the opportunity to participate. Consenting family members with or without MS provided blood samples as well. When family members were in other sites, samples were obtained by a local physician or nurse and transported or mailed to our center. Ascertainment of presence or absence of MS amongst the relatives was by history only, and relatives who provided blood samples were not subject to neurological evaluation or MRI at our center. Of the 498 samples that yielded valid genotyping information, 242 were from MS patients and 256 were from the non-MS relatives. Only multiplex families were used for association analysis.
The clinical diagnosis of MS type and the expanded disability status scale (EDSS) (26) were determined by two of the authors (K.R. and N.R.). The time of first onset and the time when the patients were first prescribed a walking aid (EDSS 6.0) was determined retrospectively by analysis of case record.
Leftover blood samples from the American Red Cross (Columbus, OH) were used as a population control. A total of 207 samples were selected on basis of availability only over a 1-year period. It is therefore expected that the genetic distribution resembles that of the central Ohio population from which most of the patients and their family members were recruited.
PCR Amplification and Restriction Fragment Length Polymorphism Analysis of the CD24 Gene. The reported SNP for CD24 is a replacement of T at nucleotide 226 by C (T→C) in the coding region of exon 2 (GenBank accession no. NM_013230), which results in a substitution of Ala at amino acid 57 by Val near the GPI anchorage site of the mature protein. The genomic DNA was isolated from ≈5 × 106 human peripheral blood leukocytes (PBL) by using the QIAamp DNA Blood Minikit (Qiagen, Valencia, CA). DNA fragments bearing this SNP site were amplified by PCR by using a forward primer (TTG TTG CCA CTT GGC ATT TTT GAG GC) and a reverse primer (GGA TTG GGT TTA GAA GAT GGG GAA A). The PCR conditions were as follows: 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, for 35 cycles. The predicted CD24 PCR fragment is 453 bp long. The T→C change yielded a BstXI restriction enzyme site at nucleotide 215, which allowed us to differentiate these two different CD24 alleles by restriction fragment length polymorphism analysis. Briefly, an aliquot of CD24 PCR products was digested with BstXI for 16 h at 50°C. The digested products were then separated in a 2.5% agarose gel. The predicted digestion pattern is as follows: PCR products of C226 allele are cut into two small fragments (317 and 136 bp), whereas those of the T226 are completely resistant. A combination of the two types of the products at close to 50% levels indicates the heterozygosity of the subject.
Molecular Cloning and Expression of CD24a and CD24v cDNA. The CD24 cDNA was amplified from PBL or CD24v/v and CD24a/a individuals by RT-PCR. The primers used were GGCCAAGCTTATGGGCAGAGCAATGGTG (forward, CD24F.H3) and ATCCCTCGAGTTAAGAGTAGAGATGCAG (reverse, CD24R.XhoI). The PCR products (256 bp) were digested with HindIII/XhoI and then cloned into the pCDNA3 expression vector at the HindIII/XhoI site, thus generating plasmid pCDNA3-CD24A and pCDNA3-CD24V. The sequence of CD24 cDNA inserts was confirmed by DNA sequencing. To test the expression efficiency of the two CD24 alleles, we transfected varying concentrations of the plasmids into the Chinese hamster ovary (CHO) cells using FuGENE 6 as described (27). Three days after transfection, the cell surface expression of the CD24 was determined by flow cytometry with saturating amounts of anti-CD24 antibodies.
Flow Cytometry. Expression of human and mouse CD24 was determined by flow cytometry by using fluorochrome-labeled anti-human CD24 (Pharmingen). PBL were isolated from fresh blood samples and stained with saturating amounts of anti-CD24 antibodies in conjunction with anti-CD3 antibodies to mark the T cells among the PBL.
Statistical Analysis
Case-Control Population Study. MS patients and normal controls were examined for significant differences in their genotype distributions in the CD24 SNP at the population level. Most of the cases and the control subjects were from central Ohio, reflecting, at least to some extent, a similarity in the disease and control populations. Pearson's χ2 test (28) was used to perform the homogeneity test between the two distributions of the genotypes. In addition, we performed further tests to compare the frequencies of CD24v/v genotype between the cases and controls, again using the χ2 tests, but with Yates' correction. Because the number of individuals falling into each of the three genotypes in both the cases and controls is fairly large, the χ2 tests should yield valid estimates of the P values.
Association Test for Transmission Disequilibrium of the v Allele. Because results from population studies can be affected by population admixture and stratification, we also carried out transmission disequilibrium test (TDT) using family data. Families with at least two MS patients (multiplex families) were ascertained for our genetic analysis to determine whether, in families that exhibit evidence of familial aggregation, the v allele in the CD24 SNP is transmitted preferentially to MS patients.
Two types of informative nuclear families were extracted from the multiplex families and included in our analysis. The type I families (trios) are those in which there is one MS patient and both parental genotypes are available, with at least one being heterozygous. The type II families (sibships) are those in which both affected and unaffected siblings are available with at least two different genotypes in the sibship. For a family that can be either type I or type II, it is classified to be a type I family following the recommendation of Spielman and Ewens (29).
A combined TDT (for type I families) and sibship TDT (STDT) (for type II families) test, as suggested by Spielman and Ewens (29), but with a Monte Carlo procedure for estimating the P value, was used. Specifically, let XTDT denote the total number of v alleles transmitted to the MS patients from heterozygous parents in the type I families. Let XSTDT denote the total number of v alleles among the affected siblings in the type II families. Then Xobs = XTDT + XSTDT is the observed test statistic for all informative families combined. Although one could estimate the P value using normal asymptotic as suggested in Spielman and Ewens (29), we opted for the Monte Carlo procedure described here to avoid the need to rely on an asymptotic distribution with a moderate sample size.
To estimate the P value of the test, 1,000,000 replicated data sets, under the null hypothesis that the CD24 SNP is unlinked to an MS locus, were generated as follows. For each type I family, we randomly selected one of the two alleles in each parent to make up the new genotype of the patient, whereas the parental genotypes are unchanged. For each type II family, we followed the scheme of Spielman and Ewens (29) by simply permuting the affection status of the individuals in the sibship. For each simulated replicate, a test statistic X was computed. The P value was taken to be the proportion of the X values that were equal to or greater than the observed statistic, Xobs, in the actual data. This Monte Carlo estimate of the P value should be very close to the true P value given the large number of replicates performed.
Comparison of Survival Curves. Patients with MS severity reaching EDSS 6.0 or higher were classified into three groups according to their CD24 genotypes. To assess whether MS progression is different among patients with different genotypes, we first estimated the survival curve, using the Kaplan–Meier method, for each of the three groups, two of which had right censored data. Then the estimated Kaplan–Meier survival curves were compared by using the log-rank test (30). Here, survival was taken to mean that a patient had not reached EDSS 6.0 yet, and the time span was measured by the number of years lapsed since the first symptom.
Results
CD24v/v Genotype and Increased MS Risk in a Population Study. We obtained 207 unused blood samples from the American Red Cross in Columbus and 242 samples of MS patients for the distribution of CD24 genotypes. The demography of the normal control population was not collected among the American Red Cross samples but was assumed to reflect the general demography of the central Ohio population. Moreover, the distribution of the CD24 genotype among our control population was similar to what was reported in a small population analysis in Europe (24). Among the 242 MS samples, 233 were Caucasian, 7 were African-American, 1 was Hispanic, and 1 was Asian. The race distribution of the samples reflected both the demography of the central Ohio population and the higher incidence of MS among the Caucasian, but not selective, recruitment.
As shown in Fig. 1a, the CD24 genotype can be distinguished by digesting the PCR products of CD24 with BstXI. The CD24a/a products were completely resistant to the digestion, whereas the CD24v/v products cleaved into two fragments of 317 and 136 bp. Partial digestion of 50% or less indicated the CD24a/v genotype. We therefore used this method to genotype the DNA isolated from leukocytes of normal population control and MS patients. The distributions of the genotypes among normal (CD24a/a, 109; CD24a/v, 85; CD24v/v, 13) and MS (CD24a/a, 113; CD24a/v, 97; CD24v/v, 32) were compared by the χ2 test. It was revealed that the distribution of CD24 genotypes among the MS patients appeared to differ significantly from that of the normal controls (P = 0.048). The difference was significant among the CD24v/v genotype (6.3% in control vs. 13.2% in MS, P = 0.023). The increased risk among the CD24v/v individuals of ≈2-fold suggests that the CD24 gene may be a modifier for MS susceptibility. Although some of the patients were related, they were treated as independent samples in the tests.
Fig. 1.
Distribution of CD24 genotypes among MS patients and the normal population control. (a) The reported SNP of the CD24 gene and its resulted amino acid replacement. Note that the alanine (A) to valine (V) change occurs immediately preceding the site (ω) for the GPI cleavage. (b) Example of genotyping by PCR followed by restriction enzyme digestion. The samples are from normal donors. The genotypes of the individuals are marked in the lanes. (c) Distribution of CD24 genotypes among normal population control (open bars) and MS patients (filled bars). The data are based on analysis of 207 normal control and 242 MS patients. The distributions of the genotypes are as follows: normal (CD24a/a, 109; CD24a/v, 85; CD24v/v, 13) and MS (CD24a/a, 113; CD24a/v, 97; CD24v/v, 32). The P values are shown.
Association of the CD24v Allele with MS in a Family Study. Eleven trios (type I families) and 18 sibships (type II families) from the multiplex families were extracted (see Fig. 2 for an example of each of these two types of families). Three of the type I families and one of the type II families are from the same extended pedigree. However, the three type I families are only distantly related, so they can be treated as independent for our purpose and are included in our TDT analysis (yielding a total of 28 informative nuclear families). Among the 11 trios, there were 15 heterozygous parents with the genotype CD24a/v, of which 13 transmitted the v allele to their affected children. The contribution to the overall test statistic was thus XTDT = 13, much larger than the expected value of 7.5. Among the 17 sibships, the total number of v alleles among the affected siblings was XTDT = 20, still larger than the expected value of 18.57, although the discrepancy between the observed and the expected was not as striking as in the trios. Our Monte Carlo procedure with 1,000,000 simulated null data sets yielded a significant result for the combined test statistic, Xobs = XTDT + XSTDT = 33 (P = 0.017). A pedigree TDT test that takes family dependency into account (31) yielded similarly significant results.
Fig. 2.
Diagrams of type I (a) and type II (b) families used for the TDT analysis. The numbers in the parentheses after the genotypes are the ages of the donors when the samples were collected. For patients with genetic data, the EDSS scores are also provided. The nuclear families used for analysis are indicated by large gray ovals.
Taken together, both the TDT test for the family data and the χ2 tests for the population data suggest that the CD24v allele is a significant risk factor for the incidence of MS.
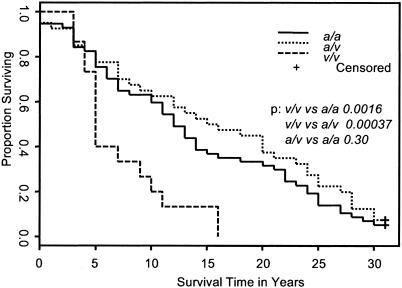
The CD24 Genotype Affects Progression of MS. The MS disease severity is usually measured according to the EDSS score. MS patients that have lost the ability to walk without aid would have reached EDSS 6.0. For the majority of the patients, their EDSS 6.0 was based on follow-up at our center. A few of the cases were based on interview. Because this is one of the most traumatic events in the patient's life, most MS patients can recall accurately the time when their disease reached EDSS 6.0. We have chosen all patients that have EDSS of 6.0 or higher, which resulted in 57, 40, and 15 patients with genotypes a/a, a/v, and v/v, respectively. We then tested whether the CD24 genotype affected the time span it took the patients to reach EDSS 6.0 from the day of the first symptom of MS. As shown in Fig. 3, 50% of the CD24v/v patients reached EDSS 6.0 in 5 years after the first symptom, whereas those with CD24a/a and CD24a/v genotypes reached EDSS 6.0 in 13 and 16 years, respectively.
Fig. 3.
CD24 genotypes and the timespan of MS patients from the year of first MS symptoms to the year they reached EDSS 6.0. Note that 50% of patients with the CD24v/v genotype reached EDSS 6.0 by 5 years as compared to 13 years for the CD24a/a and 16 years for CD24a/v patients. The P values are shown.
Furthermore, comparison of the three estimated survival curves in Fig. 3 reveals that the CD24 genotypes have a significant impact on the progression (P = 0.0008). Pairwise comparisons further show that CD24v/v patients progressed more rapidly toward EDSS 6.0 than both CD24a/v patients (P = 0.00037) and CD24a/a patients (P = 0.0016). There is no significant difference between CD24a/a and CD24a/v patients (P = 0.30).
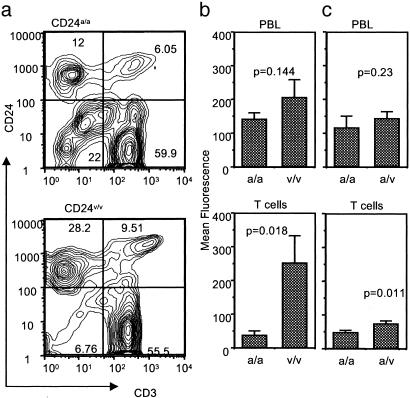
CD24v Is More Efficiently Expressed on the Cell Surface. The CD24 is a GPI anchored molecule and therefore needs to be cleaved of the C-terminal sequence before GPI attachment (32, 33). This cleavage requires specific sequence at and near the cleavage site (ω), ω+1 site, and ω+2 site (32, 33). Moreover, systematic analysis of all GPI anchored proteins with known cleavage sites suggests that the amino acid at the ω-1 and ω-2 positions may have a quantitative effect on the cleavage efficiency, and the optimal cleavage requires that the side chains in the four positions have a combined volume of 430A3 (34). As shown in Fig. 1a, CD24v and CD24a have a nonconservative replacement of A by V at the ω-1 site. Because all 4 aa in CD24a have the small side chains (A and G), replacement of A with V at ω-1 may increase the efficiency of cleavage. As a result, the CD24v protein may be expressed at a higher level than the CD24a proteins. To test this notion, we analyzed CD24 expression on the PBLs of age-, sex-, and disease-status-matched CD24a/a and CD24v/v MS patients (Table 1, Exp. 1) by two-color flow cytometry. The profiles of a representative sample in each group were presented in Fig. 4a, whereas the mean fluorescence intensities of total PBL and CD3+ T cells among the PBL were summarized in Fig. 4b. As shown in Fig. 4a, CD24 is expressed on both T cells and non-T cells, regardless of the genotypes of the MS patients. However, the percentage of positive cells and intensity of expression were higher among the PBL of CD24v/v patients. Interestingly, CD3+ T cells from the CD24a/a patients expressed 6-fold less cell-surface CD24 than did those from the CD24v/v patients. Although the same trend was found for total PBL, this was not statistically significant. In a separate experiment, we also compared six CD24a/a and six CD24a/v patients for the CD24 expression. Although the MS type was not well matched in this experiment, the MS type did not appear to influence the CD24 expression (Table 1). As shown in Table 1 (Exp. 2) and Fig. 4c, although the CD24a/v T cells expressed higher CD24 than the CD24a/a T cells, the increase was <2-fold. The small increase may explain why the CD24a/v genotype had no measurable effect on the risk and progression of MS.
Table 1. Profiles of patients and CD24 expression among MS patients with different genotypes.
| Mean fluorescence*
|
|||||||
|---|---|---|---|---|---|---|---|
| ID no. | Sex | Age, yr | EDSS | CD24 | MS type | PBL | T cells |
| Exp. 1 | |||||||
| 8a | F | 60 | 7.0 | a/a | SP | 137 | 27 |
| 11z | M | 64 | 6.5 | a/a | SP | 85 | 34 |
| 15z | F | 24 | 2.0 | a/a | RR | 148 | 22 |
| 32a | F | 62 | 2.0 | a/a | RR | 201 | 29 |
| 76z | F | 57 | 6.5 | a/a | SP | 143 | 83 |
| 25a | F | 51 | 6.0 | v/v | RR | 225 | 210 |
| 27a | F | 50 | 2.0 | v/v | RR | 351 | 545 |
| 7y | F | 47 | 2.0 | v/v | RR | 58 | 51 |
| 118z | M | 70 | 7.0 | v/v | SP | 117 | 148 |
| 122z | F | 66 | 7.0 | v/v | SP | 283 | 302 |
| Exp. 2 | |||||||
| 42z | F | 56 | 6.0 | a/a | SP | 71 | 35 |
| 43z | F | 43 | 2.0 | a/a | RR | 264 | 65 |
| 45z | F | 54 | 2.0 | a/a | RR | 56 | 20 |
| 46z | M | 61 | 7.5 | a/a | PP | 69 | 30 |
| 48z | M | 64 | 6.0 | a/a | PP | 180 | 66 |
| 12y | F | 59 | 6.5 | a/a | SP | 49 | 37 |
| 44z | F | 54 | 2.0 | a/v | RR | 204 | 92 |
| 47z | F | 33 | 2.0 | a/v | RR | 110 | 60 |
| 11y | F | 67 | 2.0 | a/v | RR | 158 | 52 |
| 21a | F | 51 | 5.0 | a/v | RR | 125 | 30 |
| 22a | M | 61 | 7.5 | a/v | SP | 185 | 92 |
| 23a | F | 59 | 2.5 | a/v | RR | 88 | 72 |
RR, remitting relapsing; SP, secondary progressive; PP, primary progressive; F, female; M, male.
Samples from RR patients were collected during the remitting phase
Fig. 4.
Higher expression of CD24 on T cells from patients with the CD24v allele. PBL was isolated from the blood of 10 MS patients who belong to either CD24a/a or CD24v/v genotypes with an approximate match in age, sex, and EDSS (see Table 1 for details). The cells were stained for CD3 and CD24 markers. (a) Contour graphs depicting expressing of CD24 and CD3 among the PBL of a representative patient in CD24a/a and CD24v/v groups. (b) The mean fluorescence of total PBL or gated CD3+ T cells. Data presented are means and SEM (n = 5). (c)Asin b, except that the expression of CD24 was compared between CD24a/a and CD24a/v patients (n = 6).
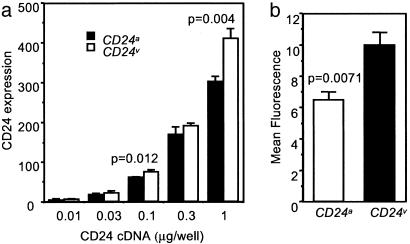
To directly address whether CD24 SNP caused variation in CD24 expression, we cloned both CD24v and CD24a cDNA and transfected the CHO cells with different concentrations of plasmids. Three days after the transfection, the cell surface expression of the CD24 gene was analyzed by flow cytometry. As shown in Fig. 5a, across a wide range of doses, the CD24v cDNA resulted in 30–40% more cell surface expression of CD24 when compared with the CD24a cDNA. To avoid variation in transfection, we also used the neomycin selection to remove untransfected cells and compared the pooled drug resistant clones for their CD24 expression. Again, CD24v cDNA transfectants expressed significantly higher cell surface CD24 (Fig. 5b).
Fig. 5.
CD24v is expressed at higher levels than CD24a allele in both transient (a) and stable (b) CHO cell transfectants. CD24v and CD24a were cloned into the PCDNA3 vector. (a) CHO cells were transfected with various amounts of CD24 cDNA. At 65 h after transfection, the transfected CHO cells were stained with saturating amounts of phycoerythrin-conjugated anti-CD24 mAbs. On the y axis, the CD24 expression shows the products of the percentage of CD24-expressing cells and mean fluorescence intensity of the positive cells. The means ± SD of triplicate samples are shown. The data are representative of three independent experiments. (b) Comparison of CD24v and CD24a expression after removing nonexpressing cells by neomycin selection. At 48 h after transfection, the CHO cells were selected with G418. The short-term drug-resistant culture (consisting of ≈500–1,000 clones) were pooled and stained with saturating amounts of phycoerythrin-conjugated anti-CD24 mAbs. Data shown were means ± SD of three independent analyses. The background fluorescence of untransfected CHO cells was subtracted. The P values from Student t tests are shown.
Discussion
We have previously reported a critical role for CD24 in the development of EAE (13), the mouse model for MS. To explore the significance of this finding in human MS, we addressed the potential contribution of the CD24 polymorphism in MS susceptibility. Our data reported here provided three lines of evidence for a significant contribution of the CD24 polymorphism to the risk and progression of MS.
First, analysis of the distribution of the CD24 genotypes among >200 MS patients and the general population of the central Ohio region indicated that the frequency of the CD24v/v genotype in MS patients is more than twice that of the general population. This result suggests the CD24v/v homozygosity raises the relative risk of MS by >2-fold. It would be of great interest to test whether this correlation can be observed in other cohorts.
Second, using the combined TDT and STDT tests, we showed that the CD24v allele is preferentially transmitted to the affected individuals compared with unaffected individuals. These data confirmed that the association at the population level most likely reflects that either CD24 or a gene linked to CD24 contributes to MS susceptibility in humans.
Third, in addition to an increased risk of MS, MS patients with the CD24v/v genotype also have a more rapid progression, as judged by the time lapse between the first MS symptom and the time when a walking aid needs to be prescribed. We have chosen EDSS 6.0 as the predetermined endpoint in experimental designs because this is a readily identifiable milestone in MS progression. We found that, among the patients that have reached EDSS 6.0, 50% of the CD24v/v patients reached that milestone in 5 years, and CD24a/a and CD24a/v patients did so in 13 and 16 years, respectively. More rapid progression in the CD24v/v patients suggests that more aggressive treatment may be warranted in this group of patients.
An important issue is how the CD24 SNP affects the risk and progression of MS. The CD24 is a GPI anchored molecule with only 32 aa in the mature protein. The SNP in CD24 resulted in a nonconservative replacement from alanine to valine at the site immediately preceding the putative cleavage site for GPI anchor (called the ω-1). Although strict conservation at this site is not necessary for the cleavage and anchor attachment, there appears to be a general requirement for the total sites of the 4 aa at positions ω+1, ω+2, ω-1, and ω-2 (34). Because the alanine and valine have a substantial difference in size, it is plausible that these two alleles may be expressed at slightly different efficiency. Our comparison revealed that the CD24v allele is expressed at 30–40% higher levels than the CD24a allele.
Indeed, the T cells in the peripheral blood of the CD24a/v patients expressed significantly higher levels of CD24 than those in the blood of the CD24a/a patients. Although resting T cells expressed very little CD24 in the mouse, its expression is rapidly induced after activation (14, 23). Because our previous work established that the CD24 gene must be functional in T cells for the T cells to be pathogenic (13), the induction of CD24 in T cells may be an important checkpoint for the pathogenesis of MS. For this reason, more efficient expression of CD24v alleles on T cells may provide a plausible explanation for the increased risk and progression of MS in the CD24v/v patients. The more efficient expression of CD24, however, is not necessarily limited to T cells, because the CD24v cDNA is more efficiently expressed even in CHO cells. Thus, the statistically insignificant difference among total PBL is most likely secondary to the vast variation in the proportion of leukocyte subsets with varying levels of CD24 (data not shown).
MS is a chronic neurological disease with strong familial influence (1–3). The most dominant factor is the HLA loci (35). Additional loci have also been suggested (8–11). Here we showed that the CD24v/v genotype is significantly increased among the MS patients and that this genotype associates with more rapid progression of the disease. This SNP exerts a strong influence in MS risk and progression. The impact of CD24 SNP, together with the requirement for CD24 in the development of EAE in the mouse, strongly suggests that CD24 is a valid target for therapy of human MS.
Acknowledgments
We thank Dr. Lynde Shaw for editorial assistance. X.B. was supported by a senior postdoctoral fellowship from the Multiple Sclerosis Society. X.L. is supported by Cancer Immunology Training Grant T32CA90223 from the National Cancer Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MS, multiple sclerosis; EAE, experimental autoimmune encephalomyelitis; SNP, single-nucleotide polymorphism; PBL, peripheral blood leukocyte; TDT, transmission disequilibrium test; EDSS, expanded disability status scale; GPI, glycosylphosphatidylinositol; CHO, Chinese hamster ovary.
References
- 1.Noseworthy, J. H. (1999) Nature 399, A40-A47. [DOI] [PubMed] [Google Scholar]
- 2.Carton, H., Vlietinck, R., Debruyne, J., De Keyser, J., D'Hooghe, M. B., Loos, R., Medaer, R., Truyen, L., Yee, I. M. & Sadovnick, A. D. (1997) J. Neurol. Neurosurg. Psychiatry 62, 329-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebers, G. C., Bulman, D. E., Sadovnick, A. D., Paty, D. W., Warren, S., Hader, W., Murray, T. J., Seland, T. P., Duquette, P., Grey, T. et al. (1986) N. Engl. J. Med. 315, 1638-1642. [DOI] [PubMed] [Google Scholar]
- 4.Kellar-Wood, H. F., Wood, N. W., Holmans, P., Clayton, D., Robertson, N. & Compston, D. A. (1995) J. Neuroimmunol. 58, 183-190. [DOI] [PubMed] [Google Scholar]
- 5.Miller, D. H., Hornabrook, R. W., Dagger, J. & Fong, R. (1989) J. Neurol. Neurosurg. Psychiatry 52, 575-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morling, N., Sandberg-Wollheim, M., Fugger, L., Georgsen, J., Hylding-Nielsen, J. J., Madsen, H. O., Rieneck, K., Ryder, L. & Svejgaard, A. (1992) Immunogenetics 35, 391-394. [DOI] [PubMed] [Google Scholar]
- 7.Olerup, O. & Hillert, J. (1991) Tissue Antigens 38, 1-15. [DOI] [PubMed] [Google Scholar]
- 8.Haines, J. L., Ter-Minassian, M., Bazyk, A., Gusella, J. F., Kim, D. J., Terwedow, H., Pericak-Vance, M. A., Rimmler, J. B., Haynes, C. S., Roses, et al. (1996) Nat. Genet. 13, 469-471. [DOI] [PubMed] [Google Scholar]
- 9.Sawcer, S., Jones, H. B., Feakes, R., Gray, J., Smaldon, N., Chataway, J., Robertson, N., Clayton, D., Goodfellow, P. N. & Compston, A. (1996) Nat. Genet. 13, 464-468. [DOI] [PubMed] [Google Scholar]
- 10.Ebers, G. C., Kukay, K., Bulman, D. E., Sadovnick, A. D., Rice, G., Anderson, C., Armstrong, H., Cousin, K., Bell, R. B., Hader, W., et al. (1996) Nat. Genet. 13, 472-476. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt, S., Barcellos, L. F., DeSombre, K., Rimmler, J. B., Lincoln, R. R., Bucher, P., Saunders, A. M., Lai, E., Martin, E. R., Vance, J. M., et al. (2002) Am. J. Hum. Genet. 70, 708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuokkanen, S., Sundvall, M., Terwilliger, J. D., Tienari, P. J., Wikstrom, J., Holmdahl, R., Pettersson, U. & Peltonen, L. (1996) Nat. Genet. 13, 477-480. [DOI] [PubMed] [Google Scholar]
- 13.Bai, X. F., Liu, J. Q., Liu, X., Guo, Y., Cox, K., Wen, J., Zheng, P. & Liu, Y. (2000) J. Clin. Invest. 105, 1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbe, M. & Altevogt, P. (1994) Eur. J. Immunol. 24, 731-737. [DOI] [PubMed] [Google Scholar]
- 15.Zhou, Q., Wu, Y., Nielsen, P. J. & Liu, Y. (1997) Eur. J. Immunol. 27, 2524-2528. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y., Jones, B., Aruffo, A., Sullivan, K. M., Linsley, P. S. & Janeway, C. A., Jr. (1992) J. Exp. Med. 175, 437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Bruijn, M. L., Peterson, P. A. & Jackson, M. R. (1996) J. Immunol. 156, 2686-2692. [PubMed] [Google Scholar]
- 18.Enk, A. H. & Katz, S. I. (1994) J. Immunol. 152, 3264-3270. [PubMed] [Google Scholar]
- 19.Liu, Y., Jones, B., Brady, W., Janeway, C. A., Jr., Linsley, P. S. & Linley, P. S. (1992) Eur. J. Immunol. 22, 2855-2859. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., Wenger, R. H., Zhao, M. & Nielsen, P. J. (1997) J. Exp. Med. 185, 251-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, Y., Zhou, Q., Zheng, P. & Liu, Y. (1998) J. Exp. Med. 187, 1151-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahne, M., Wenger, R. H., Vestweber, D. & Nielsen, P. J. (1994) J. Exp. Med. 179, 1391-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baron, J. L., Madri, J. A., Ruddle, N. H., Hashim, G. & Janeway, C. A., Jr. (1993) J. Exp. Med. 177, 57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarn, J. A., Jackson, D. G., Bell, M. V., Jones, T., Weber, E., Sheer, D., Waibel, R. & Stahel, R. A. (1995) Cytogenet. Cell Genet. 70, 119-125. [DOI] [PubMed] [Google Scholar]
- 25.McDonald, W. I., Compston, A., Edan, G., Goodkin, D., Hartung, H. P., Lublin, F. D., McFarland, H. F., Paty, D. W., Polman, C. H., Reingold, S. C., et al. (2001) Ann. Neurol. 50, 121-127. [DOI] [PubMed] [Google Scholar]
- 26.Kurtzke, J. F. (1983) Neurology 33, 1444-1452. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., Bai, X. F., Wen, J., Gao, J.-X., Liu, J., Lu, P., Wang, Y., Zheng, P. & Liu, Y. (2001) J. Exp. Med. 194, 1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agresti, A. (1990) Categorical Data Analysis (Wiley, New York).
- 29.Spielman, R. S. & Ewens, W. J. (1998) Am. J. Hum. Genet. 62, 450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleming, T. R. & Harrington, D. P. (1991) Counting Processes and Survival Analysis (Wiley, New York).
- 31.Martin, E. R., Monks, S. A., Warren, L. L. & Kaplan, N. L. (2000) Am. J. Hum. Genet. 67, 146-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Englund, P. T. (1993) Annu. Rev. Biochem. 62, 121-138. [DOI] [PubMed] [Google Scholar]
- 33.Udenfriend, S. & Kodukula, K. (1995) Annu. Rev. Biochem. 64, 563-591. [DOI] [PubMed] [Google Scholar]
- 34.Eisenhaber, B., Bork, P. & Eisenhaber, F. (1998) Protein Eng. 11, 1155-1161. [DOI] [PubMed] [Google Scholar]
- 35.Haines, J. L., Terwedow, H. A., Burgess, K., Pericak-Vance, M. A., Rimmler, J. B., Martin, E. R., Oksenberg, J. R., Lincoln, R., Zhang, D. Y., Banatao, D. R., et al. (1998) Hum. Mol. Genet. 7, 1229-1234. [DOI] [PubMed] [Google Scholar]