Abstract
Glucuronidation is an important pathway in the metabolism of nicotine, with previous studies suggesting that ~22% of urinary nicotine metabolites are in the form of glucuronidated compounds. Recent in vitro studies have suggested that the UGTs 2B10 and 2B17 play major roles in nicotine glucuronidation with polymorphisms in both enzymes shown to significantly alter the levels of nicotine-, cotinine-, and trans-3′-hydroxy-cotinine (3HC)-glucuronides in human liver microsomes in vitro. In the present study, the relationship between the levels of urinary nicotine metabolites and functional polymorphisms in UGTs 2B10 and 2B17 were analyzed in urine specimens from 104 Caucasian smokers. Based on their percentage of total urinary nicotine metabolites, the levels of nicotine-glucuronide and cotinine-glucuronide were 42% (p<0.0005) and 48% (p<0.0001), respectively, lower in the urine from smokers exhibiting the UGT2B10 (*1/*2) genotype and 95% (p<0.05) and 98% (p<0.05), respectively, lower in the urine from smokers with the UGT2B10 (*2/*2) genotype as compared to the urinary levels in smokers having the wild-type UGT2B10 (*1/*1) genotype. The levels of 3HC-glucuronide was 42% (p<0.001) lower in the urine from smokers exhibiting the homozygous UGT2B17 (*2/*2) deletion genotype as compared to the levels in urine from wild-type UGT2B17 subjects. These data are consistent with previous in vitro studies and demonstrate that UGTs 2B10 and 2B17 play important roles in the glucuronidation of nicotine, cotinine and 3HC and suggest that the UGT2B10 codon 67 SNP and the UGT2B17 deletion significantly reduce overall glucuronidation rates of nicotine and its major metabolites in smokers.
Introduction
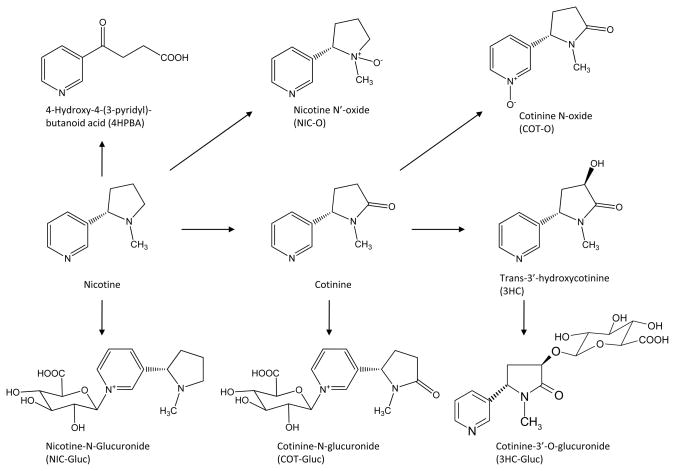
Tobacco smoking causes 500,000 deaths annually in the United States, and nicotine is the single most important pharmacological agent responsible for tobacco addiction (1). Analysis of urinary nicotine metabolite profiles indicate that about 70–80% of nicotine is metabolized to cotinine and cotinine is further metabolized to trans-3′-hydroxycotinine (3HC) and other compounds (2) (see Figure 1). Hepatic metabolism of nicotine to cotinine and then to 3HC is catalyzed primarily by CYP2A6 (3). Nicotine, cotinine and 3HC undergo further phaseII detoxification reactions by conjugation with glucuronic acid via catalysis by the UDP-glucuronosyltransferase (UGT) family of enzymes. Up to 31% of nicotine urinary metabolites are in the form of phase II glucuronidated compounds, with nicotine-N-glucuronide (nicotine-Gluc), cotinine-N-glucuronide (cotinine-Gluc), and cotinine-3′-O-glucuronide (3HC-Gluc) comprising the majority of these conjugates (3). Both cotinine and nicotine are glucuronidated on the nitrogen of the pyridine ring, and N-glucuronidation of both compounds is observed in human liver microsomes (HLM) and in the urine of smokers (4–7). Approximately 90% of the systemic dose of nicotine is excreted through the urine, and while both O- and N-glucuronidation of 3HC was observed in HLM, only its O-glucuronide, 3HC-Gluc, was detectedin the urine of smokers (8).
Figure 1.
Major nicotine metabolites in the urine of smokers.
A high correlation exists between the in vivo urinary ratio of nicotine-Gluc/(nicotine + nicotine-Gluc) and the ratio of cotinine-Gluc/(cotinine + cotinine-Gluc) in smokers. By contrast, the in vivo urinary nicotine-Gluc/(nicotine + nicotine-Gluc) ratio is only moderately correlated with the ratio of 3HC-Gluc/(3HC+ 3HC-Gluc) (2). This suggests that the glucuronidation of nicotine/cotinine versus 3HC may be via different enzyme pathways. Previous studies have also demonstrated a high correlation between the glucuronidation of nicotine and cotinine in HLM in vitro and that the hepatic UGT2B10 was the major enzyme responsible for their glucuronidation; no correlation exists between either nicotine-Gluc or cotinine-Gluc formation and 3HC-Gluc formation in HLM in vitro (9, 10). While earlier studies identified UGTs 2B7 and 1A9 to be important in the glucuronidation of 3H-cotinine, more recent studies have demonstrated that UGT2B17 exhibits the highest activity against 3H-cotinine in vitro (Chen and Lazarus, unpublished results).
A functional polymorphism (rs61750900) at codon 67 (Asp>Tyr) of the UGT2B10 gene with an allelic prevalence of ~10% in Caucasians has been reported (9). The UGT2B10Tyr variant was shown to be associated with significantly reduced N-glucuronidation of nicotine and cotinine in HLM and UGT2B10-overexpressing cell lines in vitro (9, 11). The UGT2B10Tyr variant was also shown to be associated with significantly reduced N-glucuronidation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), the major metabolite of the nicotine-derived tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), in HLM and in UGT2B10-overexpressing cell lines in vitro (11). Similarly, a polymorphic whole-gene deletion of the UGT2B17 gene, with an allelic prevalence of ~30% in Caucasians, has been shown to be associated with a significant decrease in the O-glucuronidation of 3HC in HLM in vitro (Chen et al. submitted). In similar studies, this polymorphism was also shown to be associated with a significant decrease in the O-glucuronidation of NNAL (12). Together, these data suggest that both polymorphisms could significantly alter how nicotine is metabolized in vivo.
The goal of the present study was to assess the levels of nicotine and a panel of nicotine metabolites (cotinine, 3HC, nicotine-Gluc, cotinine-Gluc, 3HC-Gluc, nicotine N′-oxide, cotinine N-oxide, and 4-hydroxy-4-(3-pyridyl)-butanoic acid [4HPBA]) in the urine of smokers and to determine whether the relative levels of urinary nicotine-Gluc, cotinine-Gluc, and 3HC-Gluc are associated with functional polymorphisms in the UGT2B10 and UGT2B17 genes in vivo.
Materials and Methods
Chemicals
Nicotine and creatinine were purchased from Sigma-Aldrich (St. Louis, MO). Cotinine, 3HC, nicotine-Gluc, cotinine-Gluc, 3HC-Gluc, nicotine N′-oxide, cotinine N-oxide, and deuterium-labeled internal standards including nicotine-methyl-D3, cotinine-methyl-D3, trans-3′-hydroxycotinine-methyl-D3, nicotine(methyl-D3)-glucuronide, cotinine(methyl-D3)-glucuronide, cotinine(methyl-D3)-3′-O-glucuronide, nicotine N′-oxide-methyl-D3, cotinine N-oxide-methyl-D3 and creatinine-D3 were purchased from Toronto Research Chemicals Inc (North York, Canada). All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA).
Subjects and Samples
Spot urine specimens and matching genomic DNA from buccal cells were from 107 subjects that were a subset of the group of healthy subjects that were recruited as controls as part of a case-control study conducted at the H. Lee Moffitt Cancer Center (Tampa, FL) from 2000 to 2003 as previously described (13). All of the 107 subjects in the present study were Caucasian and were self-reported current smokers. The age of the subjects ranged from 30 to 74 y with an average of 56.8 y, and 43% (n=46) were female. All urine and DNA samples were stored at −80°C.
Sample Preparation
A 10 μl aliquot of each urine sample was spiked with 5 μl of a mixture of deuterium-labeled internal standards, which included nicotine-methyl-D3, nicotine-oxide-methyl-D3, cotinine-methyl-D3, cotinine-N-oxide-methyl-D3, trans-3′-hydroxycotinine-methyl-D3, nicotine(methyl-D3)-glucuronide, cotinine(methyl-D3)-glucuronide, trans-3′-hydroxycotinine(methyl-D3)-glucuronide and creatinine-D3, each at a concentration of 10 ppm. Twenty-five μl of 40 mM NH4AC (pH 6.7) and 160 μl of acetonitrile/methanol (75%/25% [v/v]) were added to buffer the pH and to precipitate out proteins. After vortexing and subsequent centrifugation at 12,000 g for 10 min at 4°C, 100 μl of supernatant was transferred to a sample vial for analysis by ultra-pressure liquid chromatography (UPLC)-mass spectrometry (MS) analysis.
UPLC-MS-MS Conditions
Urine samples prepared as described above were analyzed using an Acquity LC-MS-MS system (Waters Corporation, Milford, MA, USA), consisting of an Acquity UPLC pump, an auto sampler, an ACQUITY UPLC BEH HILIC (2.1 mm × 100 mm, 1.7 μm particle size; Waters Corp.) column at 30°C, and an Acquity TQ tandem mass spectrometer (Waters). UPLC was performed at a flow rate of 0.4 ml/min using the following conditions: 1.5 min in 20% solvent A, a linear gradient for 1 min to 100% solvent A, and 3 min in 100% solvent A, where solvent A is 5 mM NH4AC (pH 6.7) and 50% acetonitrile (v/v), and solvent B is 5 mM NH4AC (pH 6.7) and 90% acetonitrile (v/v). The injection volume of each prepared urine sample was 5 μL, which represented 0.25 μL of urine collected from each subject. To verify the validity of values obtained from the analysis of one sample per subject, 6 sample preparations and analyses were carried out on urine specimens from 5 individual subjects. The inter-assay coefficient of variation (CV) was less than 10% for all 5 subjects.
The Waters Acquity TQ tandem mass spectrometer was equipped with an electrospray ionization (ESI) probe operated in the positive-ion mode, with capillary voltage at 0.64 kV. Nitrogen was used as both the cone and desolvation gases with flow rates maintained at 20 and 760 L/h, respectively. Ultra-pure argon was used as the collision gas with a flow rate of 0.1 L/h for collision-induced dissociation. The source and desolvation gas temperatures were 140 and 450°C, respectively. For the assay of nicotine, nicotine N′-oxide, , nicotine-Gluc, cotinine, cotinine-Gluc, cotinine N-oxide, 3HC, 3HC-Gluc, 4HPBA and creatinine in urine samples, the mass spectrometer was operated in the multiple reaction monitoring mode (MRM) and the concentrations of all analytes were determined simultaneously. The dwell time for each ion was 100 ms with 5 ms of inter-scan-delay. The ion related parameters for the 16 transitions monitored are listed in Table 1.
Table 1.
MRM transitions and ion optics parameters for the test compounds
| Compound | ES+ MS transition (M/z)+ | Cone Voltage (Volt) | Collision Energy (Volt) |
|---|---|---|---|
| Nicotine | 163.1>106 | 35 | 15 |
| Nicotine-methyl-D3 | 166.1>106 | 35 | 15 |
| Cotinine | 177.1>98 | 40 | 20 |
| Cotinine-methyl-D3 | 180.1>101 | 40 | 20 |
| 3HC | 193.1>80 | 40 | 20 |
| 3HC-methyl-D3 | 196.1>80 | 40 | 20 |
| Nicotine-Gluc | 339.1>163.1 | 30 | 15 |
| Nicotine-methyl-D3-Gluc | 342.1>166.1 | 30 | 15 |
| Cotinine-Gluc | 353.1>177.1 | 30 | 15 |
| Cotinine-methyl-D3-Gluc | 356.1>180.1 | 30 | 15 |
| 3HC-Gluc | 369.1>193.1 | 30 | 15 |
| 3HC-(methyl-D3)-Gluc | 372.1>196.1 | 30 | 15 |
| Creatinine | 114.1>44 | 30 | 15 |
| D3-creatinine | 117.1>47 | 30 | 15 |
| Nicotine N′-Oxide | 179.1>117.1 | 35 | 30 |
| Nicotine-(methyl-D3)-N′-Oxide | 182>117.1 | 35 | 30 |
| Cotinine N-Oxide | 193.1>96 | 40 | 25 |
| Cotinine-(methyl-D3)-N-Oxide | 196.1>96 | 40 | 25 |
| 4HPBA | 182>109 | 35 | 25 |
Quantification
Standard curves were constructed by plotting the ratio of analyte peak area to peak area of the corresponding internal standard (described above) versus analyte concentration for at least 8 analyte concentrations. For the nicotine and nicotine metabolites standards, a 1,000 ppm stock solution was made in water. The stock solution was serially diluted in water and then mixed with an equal volume of urine from a nonsmoker. Standards at concentrations ranging from 0.05 ppm to 50 ppm were used to establish standard curves. For creatinine, the stock solution (100,000 ppm) was serially diluted in water to concentrations ranging from 10 to 10,000 ppm. Urinary analyte concentrations were determined by measuring the peak area ratios of analyte to internal standard and then calculating analyte concentration from the appropriate standard curve using Waters’ MassLynx software. Standard curves were included in each batch of analyses, as were aliquots of urine from both a smoker and a non-smoker as quality controls. The quantification limits (signal/noise>10) for each compound were approximately 0.01 ppm for cotinine-N-oxide, nicotine-Gluc and nicotine-N-oxide, 0.05 ppm for cotinine and cotinine-Gluc, 0.1 ppm for 3HC and 3HC-Gluc, and 0.2 ppm for nicotine. The total nicotine equivalents (Total-NIC-Eq) present in each sample were determined as the sum of the micro-molar concentrations of nicotine, nicotine-Gluc, nicotine N′-oxide, cotinine, cotinine-Gluc, cotinine N-oxide, 3HC, 3HC-Gluc, and 4HPBA. The percentage of each analyte in the Total-NIC-Eq was calculated based on the equation: C (μM)/[Total-NIC-Eq (μM)]*100, where ‘C’ is the urinary metabolite concentration. Creatinine concentration in each sample was measured as a control for urine secretion and was also used for normalization of concentrations of nicotine metabolites from each sample.
Genotyping of UGTs 2B10 and 2B17
Genomic DNA was purified from oral buccal cell swabs collected from the same subjects who provided urine specimens as previously described (13). All subjects were recruited as healthy controls as part of a case-control study conducted at the H. Lee Moffitt Cancer Center (Tampa, FL) from 2000 to 2003. Subjects were randomly-selected from community residents attending the Lifetime Cancer Screening facility of the Moffitt Cancer Center who underwent prostate-specific antigen testing, skin examinations, endoscopy, or mammography. A list of control IDs were matched against the hospital patient database to identify subjects, if any, who might have developed cancer. All control subjects with a new cancer diagnosis were excluded from this study. Ninety-seven percent of subjects who were asked to participate signed a consent form approved by the institutional review board and all were interviewed using a structured questionnaire.
The UGT2B10 codon 67 polymorphism was determined by real-time PCR using a custom designed TaqMan Genotype assay (Applied Biosystems, Foster City, CA) on the ABI 7900 HT (Applied Biosystems). Real-time results were confirmed for 50% of samples, including representative samples of each of the three UGT2B10 genotypes, by PCR-restriction fragment length polymorphism (RFLP) analysis using the HinfI restriction enzyme as described previously (9). Using the same two primers used for PCR amplification for RFLP, direct sequencing was performed on two subjects for each of the three potential UGT2B10 genotypes (Asp/Asp = [*1/*1], Asp/Tyr = [*1/*2], Tyr/Tyr = [*2/*2]) and genotypes were confirmed in all cases. The UGT2B17 gene deletion polymorphism genotype were determined by real-time PCR using a previously described Taqman assay (13).
Statistical Analysis
All statistical analyses were performed using R (14). Linear regression analysis was used to measure the linear relationship between nicotine-Gluc, cotinine-Gluc and/or 3HC-Gluc measured either as a percentage of Total-NIC-Eq or as creatinine-adjusted levels. The Students t-test and the regression trend test were used to compare levels of urinary 3HC-Gluc in smokers stratified by UGT2B17 deletion genotypes, whether measured as a percentage of Total-NIC-Eq, as creatinine-normalized levels, or as the ratio of 3HC-Gluc/(3HC+3HC-Gluc) ratio. As the UGT2B10 (*2/*2) genotype had limited sample size (n=2), nonparametric rank-based statistical tests were used to reduce the assumptions made on the data. The Mann-Whitney U test and the Jonckheere-Terpstra trend test (15) were used to compare the levels of urinary nicotine metabolites in smokers stratified by UGT2B10 codon 67 genotypes, whether measured as a percentage of Total-NIC-Eq, or as creatinine-normalized levels.
Results
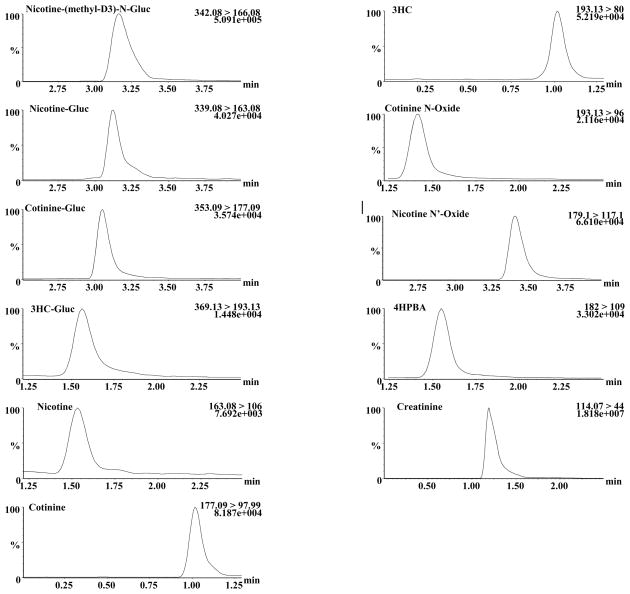
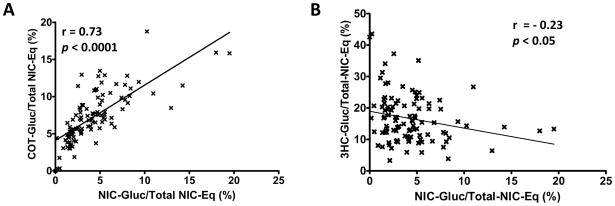
The levels of nicotine and its eight major metabolites - nicotine-Gluc, cotinine, cotinine-Gluc, nicotine-N′-oxide, cotinine-N-oxide, 3HC, 3HC-Gluc and 4HPBA - were determined simultaneously in the urine of 107 smokers. Following UPLC-MS-MS analysis, three subjects were excluded from further evaluation because the Total-NIC-Eq value for each was less than 5 μM and individual nicotine metabolites from those samples could not be quantified reliably. Chromatograms obtained from a representative analysis of the urine of a single subject are shown in Figure 2. The levels of each metabolite are shown in Table 2 and are expressed as both creatinine-adjusted concentrations as well as a percentage of Total-NIC-Eq. Similar to that observed in previous studies (16), 3HC was the major nicotine metabolite based on metabolite levels as a percentage of Total-NIC-Eq in this population. The ranking of the mean levels of nicotine metabolites as a percentage of Total-NIC-Eq was 3HC > 3HC-Gluc > cotinine > nicotine > 4HPBA > cotinine-Gluc > nicotine-N′-oxide > nicotine-Gluc > cotinine-N-oxide. Identical results were obtained when comparing the levels of creatinine-adjusted metabolites (Table 2). Of the glucuronides, the mean Total-NIC-Eq-adjusted 3HC-Gluc levels were 2.3- and 3.9-fold higher than cotinine-Gluc and nicotine-Gluc, respectively, with nicotine-Gluc comprising an average 4.3% of Total-NIC-Eq. The levels of 3HC-Gluc and cotinine-Gluc comprised 16.7% and 7.3%, respectively of the Total-NIC-Eq. Two subjects exhibited urinary nicotine-Gluc levels of ≥18% and six exhibited urinary nicotine-Gluc levels of ≥10% of Total-NIC-Eq (results not shown). Urinary nicotine-Gluc + cotinine-Gluc comprised ≥15% of Total-NIC-Eq in 27 subjects, with two subjects exhibiting urinary nicotine-Gluc + cotinine-Gluc levels that were >33% of Total-NIC-Eq (results not shown). To determine whether any correlations existed between the amount of glucuronidated product formed for nicotine or its major metabolites, the levels of nicotine-Gluc measured as a percentage of Total-NIC-Eq were compared to the percentages of Total-NIC-Eq for cotinine-Gluc and 3HC-Gluc (Figure 3). There was a significant (p<0.0001, r= 0.73) correlation between the levels of urinary nicotine-Gluc and cotinine-Gluc in smokers (panel A). A weak negative association was observed between the percentages of Total-NIC-Eq for urinary nicotine-Gluc and 3HC-Gluc (p<0.05, r=−0.23; panel B) or for urinary cotinine-Gluc and 3HC-Gluc (p<0.05, r=−0.23; results not shown).
Figure 2. UPLC-MS-MS analysis of nicotine and its metabolites in the urine of smokers.
Shown are representative chromatograms for each analyte and a representative deuterated standard [nicotine (methyl-D3)- Gluc)] that was identified simultaneously from the MRM analysis of a single urine sample as described in the Materials and Methods. For each compound, the ratio of analyte peak area to that of the deuterated standard was used for quantification as described. The m/z transition monitored for each analyte is detailed on each graph.
Table 2.
Urinary nicotine metabolite profile in 104 smokers.
| % of Total-NIC-Eqa |
Creatinine-adjusted levels (nmol/mg creatinine)b |
|||
|---|---|---|---|---|
| Mean ± S.D. (95% CI)c | Range | Mean ± S.D. (95% CI) | Range | |
| 3HC | 33.6 ± 8.7 (32.0–35.4) | 5.9–53.4 | 42.4 ± 25.2 (37.6–47.3) | 0.78–145 |
| 3HC-Gluc | 16.7 ± 7.8 (15.1–18.2) | 3.3–43.6 | 21.8 ± 18.5 (18.2–25.3) | 0.44–124 |
| Cotinine | 11.0 ± 6.6 (9.8–12.2) | 2.9–32.0 | 14.2 ± 11.9 (11.9–16.5) | 0.97–70 |
| Nicotine | 10.6 ± 7.3 (9.2–12.0) | 0.58–31.7 | 12.9 ± 11.5 (11.0–15.2) | 0.42–64 |
| 4HPBA | 8.9 ± 3.7 (8.1–9.6) | 3.1–32.3 | 10.6 ± 6.0 (9.4–11.8) | 0.95–37 |
| Cotinine-Gluc | 7.3 ± 3.4 (6.6–8.0) | 0–18.8 | 8.7 ± 5.3 (7.6–9.7) | 0.00–26 |
| Nicotine-N′-Oxide | 4.8 ± 2.5 (4.3–5.3) | 1.4–13.4 | 5.6 ± 3.2 (4.9–6.2) | 0.36–18 |
| Nicotine-Gluc | 4.3 ± 3.4 (3.6–4.9) | 0–19.5 | 5.0 ± 4.6 (4.2–5.8) | 0.00–31 |
| Cotinine-N-Oxide | 2.8 ± 0.77 (2.7–3.0) | 1.2–5.0 | 3.4 ± 1.6 (7.6–9.7) | 0.58–8.1 |
Ratio of individual nicotine metabolite (μM) measured as a percentage of total urinary nicotine metabolites [Total-NIC-Eq (μM)].
Ratio of individual nicotine metabolite (μM) measured as a percentage of urinary creatinine (mg).
S.D. = standard deviation; CI = confidence interval.
Figure 3. Correlation between percentages of Total-NIC-Eq for nicotine-, cotinine-and 3HC-glucuronide in the urine of smokers.
Nicotine and cotinine glucuronides were determined for 104 smokers as described in the Materials and Methods. Panel A, percentage of Total-NIC-Eq for nicotine-glucuronide and cotinine-glucuronide; panel B, percentage of Total-NIC-Eq for nicotine-glucuronide and 3HC-glucuronide.
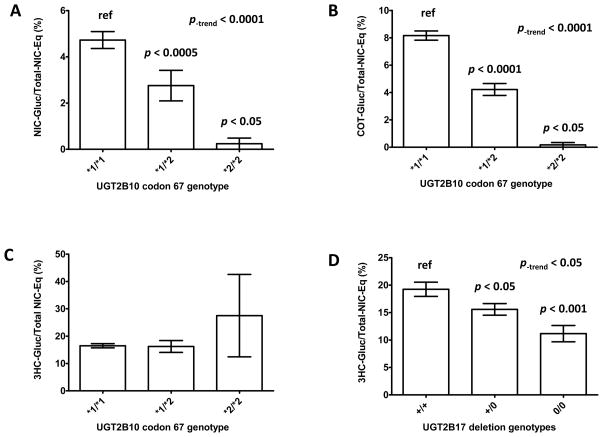
Informative genotyping data was obtained for all of the 104 subjects tested for the UGT2B10 codon 67 polymorphism. The UGT2B10 codon 67 (Asp>Tyr) polymorphism genotype distribution was as follows: 80% (n=83) exhibited the homozygous wild-type UGT2B10 (*1/*1) genotype, 18% (n=19) exhibited the heterozygous UGT2B10 (*1/*2) genotype and 2% (n=2) exhibited the homozygous UGT2B10 (*2/*2) genotype. This genotype distribution was consistent with Hardy-Weinberg equilibrium (p = 0.5) and the UGT2B10 allelic frequency (11%) was consistent with that observed in previous studies (9). The average percentage of Total-NIC-Eq determined for nicotine-Gluc in the urine from smokers with the wild-type UGT2B10 (*1/*1) genotype was 4.7% while that for smokers of each of the variant genotypes was significantly lower, at 2.8% for smokers with the UGT2B10 (*1/*2) genotype (p<0.05) and 0.24% (p<0.001) for smokers with the UGT2B10 (*2/*2) genotype (Figure 4, panel A). A similar pattern was observed for the percentage of Total-NIC-Eq determined for cotinine-Gluc, with significant 1.9- (p<0.0001) and 48-fold (p<0.0001) lower levels in the urine from smokers with the UGT2B10 (*1/*2) (4.2%) and UGT2B10 (*2/*2) (0.17%) genotypes, respectively, as compared to the level (8.2%; Figure 4, panel B) in the urine of smokers with the wild-type UGT2B10 (*1/*1) genotype. There was a significant trend towards a decreased percentage of Total-NIC-Eq for nicotine-Gluc (p<0.001) and cotinine-Gluc (p<0.0001) levels in the urine of smokers with an increasing number of the variant UGT2B10*2 allele. A similarly significant pattern was observed when examining the creatinine-adjusted levels of nicotine-Gluc or cotinine-Gluc (results not shown). No correlation was observed between the percentage of Total-NIC-Eq for 3HC-Gluc and UGT2B10 codon 67 genotype (Figure 4, panel C). Additionally, no correlation was observed between UGT2B10 codon 67 genotypes and the percentage of Total-NIC-Eq for any of the other nicotine metabolites analyzed in this study (data not shown).
Figure 4. Effect of UGT2B10 and UGT2B17 polymorphisms on nicotine-Gluc, cotinine-Gluc, and 3HC-Gluc levels in urine samples from smokers.
Shown are the percentage of Total-NIC-Eq for urinary nicotine and nicotine metabolite glucuronides for subjects stratified by UGT2B10 codon 67 or UGT2B17 deletion genotypes. Panel A, UGT2B10 genotype vs. urinary nicotine-Gluc; panel B, UGT2B10 genotype vs. urinary cotinine-Gluc; panel C, UGT2B10 genotype vs. urinary 3HC-Gluc; panel D, UGT2B17 genotype versus urinary 3HC-Gluc. The Mann-Whitney U test was used to compare urine from subjects with the UGT2B10 (*2/*2) or (*1/*2) genotypes to specimens from subjects with the wild-type UGT2B10 (*1/*1) genotype, and the Jonckheere-Terpstra trend test was used to examine the overall effect of UGT genotypes on nicotine-, cotinine-, and 3HC-glucuronide levels determined as a percentage of Total-NIC-Eq. The Students t-test was used to compare urine from subjects with the UGT2B17 (0/0) or (0/+) genotypes to specimens from subjects with the wild-type UGT2B17 (+/+) genotype, and the trend test was performed to examine the overall effect of UGT genotypes on nicotine-, cotinine-, and 3HC-glucuronide levels determined as a percentage of Total-NIC-Eq. Data are shown as the mean ± standard error.
Of the 88 samples for whom informative UGT2B17 deletion genotype data were obtained, the UGT2B17 copy number distribution was as follows: 53% (n=47) exhibited both copies of the wild-type UGT2B17*1 gene, 39% (n=34) exhibited one copy of the UGT2B17*2 deletion allele, and 8% (n=7) exhibited the UGT2B17 (*2/*2) homozygous deletion genotype. This genotype distribution was consistent with Hardy-Weinberg equilibrium (p = 0.5) and the UGT2B17*2 deletion allelic frequency (27%) was consistent with that observed in previous studies (12, 17, 18). The average percentage of Total-NIC-Eq for 3HC-Gluc in the urine of subjects exhibiting the wild-type UGT2B17 (*1/*1) genotype was 19.2%, but was significantly lower for subjects heterozygous (15.6%; p<0.05) or homozygous (11.2%; p<0.001) for the UGT2B17 deletion (Figure 4, panel D). There was a significant trend towards a decreased percentage of Total-NIC-Eq for urinary 3HC-Gluc (p<0.05) in subjects with increasing copies of the UGT2B17*2 deletion allele. A similarly significant trend was observed when examining the levels of 3HC-Gluc as a ratio with (i) 3HC + 3HC-Gluc (p<0.001), or (ii) creatinine (p<0.05), as the denominator (results not shown).
Discussion
The present study is the first to directly examine the role of functional variants in both UGTs 2B10 and 2B17 in nicotine metabolism in smokers. Previous studies have demonstrated that while UGTs 1A4 and 2B10 exhibit glucuronidating activity against both nicotine and cotinine in vitro, UGT2B10 exhibits a 3–37-fold lower Km than UGT1A4 against these two compounds and a functional polymorphism at codon 67 of the UGT2B10 gene results in a 5- and 16-fold decrease in liver activity against nicotine and cotinine, respectively (9, 19, 20). In the present study, it was demonstrated that the percentage of urinary nicotine-Gluc and cotinine-Gluc in smokers homozygous for the UGT2B10*2 variant were decreased by 95% and 98%, respectively, as compared to smokers exhibiting the wild-type UGT2B10 (*1/*1) genotype. This UGT2B10*2 allele-associated decrease in nicotine and cotinine glucuronidation was consistent with that observed in recent studies for urine specimens from subjects heterozygous for a tagSNP linked with the UGT2B10 codon 67 SNP (21). This pattern is also consistent with previous in vitro studies in HLM and UGT-over-expressing cell lines indicating that UGT2B10 is the major enzyme active in the glucuronidation of these compounds (9). In addition, this confirms the functionality of the UGT2B1067Tyr variant against nicotine and cotinine, an effect that is consistent with the drastic change in amino acid sequence from the acidic aspartic acid encoded by the UGT2B10*1 allele to the bulky, phenolic tyrosine residue encoded by UGT2B10*2.
Two previous studies (8, 22) have suggested that UGTs 1A9 and 2B7 are primarily responsible for 3HC-Gluc formation. For both of these studies, several UGTs including UGT2B17 were not screened for activity against 3HC. Recent data indicates that UGT2B17 exhibits a Km that is lower than both UGTs 1A9 and 2B7 against 3HC and that the homozygous deletion of the UGT2B17 gene results in a ~2-fold decrease in liver microsomal activity against 3HC in vitro (Chen and Lazarus, unpublished results). In the present study, it was demonstrated that there was a 2.8-fold decrease in the levels of urinary 3HC-Gluc in smokers homozygous for the UGT2B17 deletion variant as compared to urine specimens from smokers with the wild-type UGT2B17 (*1/*1) genotype. This is consistent with an important role for UGT2B17 in 3HC glucuronidation. Based on the difference in urinary 3HC levels in UGT2B17 (*1/*1) versus UGT2B17 (*2/*2) smokers and the fact that the UGT2B17 deletion is in effect a functional knock-out of the UGT2B17 gene, UGT2B17 accounts for approximately 42% of all glucuronidated 3HC in humans, with UGTs 2B7 and 1A9 likely comprising the other UGTs active in this process (8, 22).
This is the first study to simultaneously and directly determine the levels of nicotine and eight of its major metabolites in the urine of smokers. While the means of the percentage of Total-NIC-Eq for most of the nicotine metabolites examined in this study were similar to that described in previous reports, the relative levels of cotinine-Gluc and 3HC-Gluc were, on average, 1.7-fold lower and 2-fold higher, respectively, than the levels reported in other studies for these two metabolites in the urine of smokers (16, 23, 24). While differences in the methodologies used for nicotine metabolite detection and quantification may contribute to the differences observed between studies, sample size variation could also play an important role. The current study included 104 subjects while only up to 12 subjects were included in each of the two previous studies (16, 23). In addition, the method of urine sample collection was different between studies. Spot urine samples were analyzed in the present study, while 24-hour urine specimens were utilized in previous studies. Due to the relatively short elimination half-life [~2 h (25)] of nicotine, the levels of nicotine metabolites in spot urine samples may vary in relation to when the last cigarette was smoked; 24-hour urine has the advantage of providing steady state urinary nicotine excretion data. However, virtually identical trends were observed with respect to UGT2B10 genotype and the formation of both nicotine-glucuronide as well as cotinine-glucuronide, suggesting that the use of spot urine samples was not confounding the results observed in this study.
Interestingly, while the levels of glucuronide conjugates of nicotine and cotinine were low in most subjects analyzed in this study (mean % of Total-NIC-Eq of 4.3 and 7.3, respectively), higher relative levels of nicotine-Gluc and cotinine-gluc were observed in some subjects, with the nicotine-Gluc reaching as high as 20% of total urinary nicotine metabolites, and total nicotine-Gluc + cotinine-Gluc levels reaching as high as one-third of total urinary nicotine metabolites. Therefore, the glucuronidation pathway may play an even more important role in the overall metabolism of nicotine and cotinine in a subset of the population than previously identified.
Alteration in the rate of clearance or any shift in the distribution of dose between nicotine and its metabolites and or conjugates could have an effect on addiction and possibly carcinogen exposure resulting from altered smoking dose. The glucuronidation pathway may be particularly important in subjects for whom nicotine metabolism by hydroxylation/oxidation by CYP450 enzymes is low. CYP2A6 is the major hepatic enzyme involved in both the metabolism of nicotine to its non-addictive metabolite cotinine, and in the metabolism of cotinine to 3HC (3). It has been demonstrated that approximately 20% of Caucasians and African-Americans as well as 70% of Asians exhibit a CYP2A6 enzyme phenotype that results in reduction of CYP2A6 enzyme activity by at least 25% and thus results in poor metabolism of nicotine and cotinine (26). In Asian smokers homozygous for the CYP2A6*4 deletion allele and who therefore have no active CYP2A6 enzyme, the urinary levels of nicotine and nicotine-Gluc were shown to be as high as 45% and 55%, respectively, of total urinary nicotine metabolites (27). In subjects with a CYP2A6 enzyme phenotype that results in decreased metabolism of nicotine and cotinine, the percentage of the parent compounds would also likely be increased to even higher levels if that individual also had one or more UGT2B10*2 variant alleles that resulted in less efficient glucuronidation of these compounds. Therefore, the UGT2B10 codon 67 polymorphism could be a very important modifier of nicotine metabolism in subjects with CYP2A6 low-activity phenotypes and could therefore play an important role in inter-individual variations in nicotine addiction in this population.
The nicotine metabolite ratio of 3HC:cotinine has been used as an indicator of CYP2A6 activity (28, 29) and as a variable in clinical studies of smoking cessation (30–35). Since the UGT2B17 deletion affects overall clearance of 3HC, this polymorphism could significantly affect the efficacy of this ratio as a determinant of CYP2A6 activity and variable in nicotine addiction. Further large-scale studies examining urinary nicotine metabolites in smokers of different combined CYP2A6 and UGT2B17 genotypes will be required to better assess these possibilities.
In conclusion, this study demonstrates that UGT2B10 is the enzyme responsible for the majority of the glucuronidation of nicotine and cotinine while UGT2B17 is an important enzyme responsible for the glucuronidation of 3HC in vivo. Additionally, this study has established that, (i) the UGT2B10 codon 67 (Asp>Tyr) polymorphism is associated with large reductions in the levels of nicotine-Gluc and cotinine-Gluc, and (ii) the UGT2B17 deletion is associated with significant decreases in the levels of 3HC-Gluc, in the urine of smokers. Population-based studies will be required to better assess their potential contribution in overall smoking behavior and nicotine dependency.
Acknowledgments
We are highly appreciative of Diane McCloskey for critical reading of the manuscript. We thank the Tissue Procurement Facility at the H. Lee Moffitt Cancer Center for tissue procurement for these studies. The authors also thank the Functional Genomics Core Facility at the Penn State University College of Medicine for DNA genotyping and sequencing services. These studies were supported by a Dean’s Feasibility Grant from Penn State College of Medicine (Chen), two formula grants under the Pennsylvania Department of Health’s Health Research Formula Funding Program (SAP4100038715 [Lazarus] and SAP4100038714 [Whitehead]; State of PA, Act 2001-77 – part of the PA Tobacco Settlement Legislation), and Public Health Service grants P01-CA68384 (Lazarus) and R01-DE13158 (Lazarus) from the National Institutes of Health.
References
- 1.Hatsukami DK, Stead LF, Gupta PC. Tobacco addiction. Lancet. 2008;371:2027–38. doi: 10.1016/S0140-6736(08)60871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56:483–93. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 3.Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell WS, Greene JM, Byrd GD, et al. Characterization of the glucuronide conjugate of cotinine: a previously unidentified major metabolite of nicotine in smokers’ urine. Chem Res Toxicol. 1992;5:280–5. doi: 10.1021/tx00026a021. [DOI] [PubMed] [Google Scholar]
- 5.Byrd GD, Uhrig MS, deBethizy JD, et al. Direct determination of cotinine-N-glucuronide in urine using thermospray liquid chromatography/mass spectrometry. Biol Mass Spectrom. 1994;23:103–7. doi: 10.1002/bms.1200230210. [DOI] [PubMed] [Google Scholar]
- 6.Byrd GD, Caldwell WS, Bhatti BS, Ravard A, Crooks PA. Determination of nicotine N-1-glucuronide, a quaternary N-glucuronide conjugate, in human biological samples. Drug Metabol Drug Interact. 2000;16:281–97. doi: 10.1515/dmdi.2000.16.4.281. [DOI] [PubMed] [Google Scholar]
- 7.Ghosheh O, Vashishtha SC, Hawes EM. Formation of the quaternary ammonium-linked glucuronide of nicotine in human liver microsomes: identification and stereoselectivity in the kinetics. Drug Metab Dispos. 2001;29:1525–8. [PubMed] [Google Scholar]
- 8.Kuehl GE, Murphy SE. N-glucuronidation of trans-3′-hydroxycotinine by human liver microsomes. Chem Res Toxicol. 2003;16:1502–6. doi: 10.1021/tx034173o. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, Lazarus P. Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp>Tyr) polymorphism. Cancer Res. 2007;67:9024–9. doi: 10.1158/0008-5472.CAN-07-2245. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura M, Naito S. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab Pharmacokinet. 2006;21:357–74. doi: 10.2133/dmpk.21.357. [DOI] [PubMed] [Google Scholar]
- 11.Chen G, Dellinger RW, Gallagher CJ, Sun D, Lazarus P. Identification of a prevalent functional missense polymorphism in the UGT2B10 gene and its association with UGT2B10 inactivation against tobacco-specific nitrosamines. Pharmacogenet Genomics. 2008;18:181–91. doi: 10.1097/FPC.0b013e3282f4dbdd. [DOI] [PubMed] [Google Scholar]
- 12.Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15:769–78. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher CJ, Muscat JE, Hicks AN, et al. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:823–8. doi: 10.1158/1055-9965.EPI-06-0823. [DOI] [PubMed] [Google Scholar]
- 14.R-project.org [homepage on the Internet]. R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [update 2009; cited 2010 Mar 26]. Available from: http://www.R-project.org. [Google Scholar]
- 15.Broberg P. SAGx: Statistical Analysis of the GeneChip [homepage on the Internet] R package version 1.20.0. [Cited 2010 Mar 26]. Available from: http://home.swipnet.se/pibroberg/expression_hemsida1.html.
- 16.Benowitz NL, Jacob P, 3rd, Fong I, Gupta S. Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. J Pharmacol Exp Ther. 1994;268:296–303. [PubMed] [Google Scholar]
- 17.Murata M, Warren EH, Riddell SR. A human minor histocompatibility antigen resulting from differential expression due to a gene deletion. J Exp Med. 2003;197:1279–89. doi: 10.1084/jem.20030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson W, 3rd, Pardo-Manuel de Villena F, Lyn-Cook BD, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–14. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Kuehl GE, Murphy SE. N-glucuronidation of nicotine and cotinine by human liver microsomes and heterologously expressed UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31:1361–8. doi: 10.1124/dmd.31.11.1361. [DOI] [PubMed] [Google Scholar]
- 20.Kaivosaari S, Toivonen P, Hesse LM, Koskinen M, Court MH, Finel M. Nicotine Glucuronidation and the Human UDP-Glucuronosyltransferase UGT2B10. Mol Pharmacol. 2007;72:761–8. doi: 10.1124/mol.107.037093. [DOI] [PubMed] [Google Scholar]
- 21.Zinggeler Berg J, Mason J, Boettcher AJ, Hatsukami DK, Murphy SE. Nicotine metabolism in African Americans and European Americans: variation in glucuronidation by ethnicity and UGT2B10 haplotype. J Pharmacol Exp Ther. 2009 doi: 10.1124/jpet.109.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka H, Nakajima M, Katoh M, et al. Trans-3′-hydroxycotinine O- and N-glucuronidations in human liver microsomes. Drug Metab Dispos. 2005;33:23–30. doi: 10.1124/dmd.104.001701. [DOI] [PubMed] [Google Scholar]
- 23.Hecht SS, Carmella SG, Murphy SE. Effects of watercress consumption on urinary metabolites of nicotine in smokers. Cancer Epidemiol Biomarkers Prev. 1999;8:907–13. [PubMed] [Google Scholar]
- 24.Hecht SS, Hatsukami DK, Bonilla LE, Hochalter JB. Quantitation of 4-oxo-4-(3-pyridyl)butanoic acid and enantiomers of 4-hydroxy-4-(3-pyridyl)butanoic acid in human urine: A substantial pathway of nicotine metabolism. Chem Res Toxicol. 1999;12:172–9. doi: 10.1021/tx980214i. [DOI] [PubMed] [Google Scholar]
- 25.Benowitz NL, Jacob P., 3rd Nicotine and cotinine elimination pharmacokinetics in smokers and nonsmokers. Clin Pharmacol Ther. 1993;53:316–23. doi: 10.1038/clpt.1993.27. [DOI] [PubMed] [Google Scholar]
- 26.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther. 2005;77:145–58. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima M, Yokoi T. Interindividual variability in nicotine metabolism: C-oxidation and glucuronidation. Drug Metab Pharmacokinet. 2005;20:227–35. doi: 10.2133/dmpk.20.227. [DOI] [PubMed] [Google Scholar]
- 28.Benowitz NL, Hukkanen J, Jacob P., 3rd Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009:29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dempsey D, Tutka P, Jacob P, 3rd, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Ho MK, Mwenifumbo JC, Zhao B, Gillam EM, Tyndale RF. A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenet Genomics. 2008;18:67–75. doi: 10.1097/FPC.0b013e3282f3606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerman C, Tyndale R, Patterson F, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79:600–8. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Patterson F, Schnoll RA, Wileyto EP, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84:320–5. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 33.Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J Neurogenet. 2009;23:252–61. doi: 10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav. 2009;92:6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho MK, Mwenifumbo JC, Al Koudsi N, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther. 2009;85:635–43. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]