SUMMARY
Plant–pathogen co‐evolutionary selection processes are continuous, complex and occur across many spatial and temporal scales. Comprehensive studies of the flax–flax rust pathosystem have led to the postulation of the gene‐for‐gene model, a genetic paradigm describing recognition events between host disease resistance proteins and pathogen effector proteins. The identification of directly interacting fungal effector proteins and plant disease resistance proteins in this pathosystem has facilitated the study of both the physical nature of these interactions and the evolutionary forces that have resulted in a molecular arms race between these organisms. The flax–flax rust pathosystem has also been detailed on the scale of interacting populations, and the integration of molecular‐ and population‐scale datasets represents a unique opportunity to further our understanding of many poorly understood facets of host–pathogen dynamics. In this article, we discuss recent developments and insights in the flax–flax rust pathosystem and their implications for both long‐term co‐evolutionary dynamics in natural settings, as well as short‐term co‐evolutionary dynamics in agro‐ecosystems.
INTRODUCTION
Interaction with parasites has been postulated to be a major driver of the evolution and maintenance of diversity in both plants and animals. Infection of hosts can lead to a reduction in fitness and selection for defence or avoidance mechanisms. Conversely, pathogens are selected to circumvent the continually evolving defences mounted by their target hosts. However, despite significant advances in our understanding of these interactions at both the molecular and population levels, there are still major questions to be resolved regarding the mechanisms of host resistance and pathogen virulence, their variation in space and time, and their long‐term effect on host–pathogen co‐evolution.
The interaction between flax and flax rust has been an important model system for understanding the genetic and molecular basis of host–pathogen interactions in plant diseases, as well as for understanding the co‐evolutionary processes in natural disease systems. Flax rust, Melampsora lini, is a fungal pathogen that infects cultivated flax (Linum usitatissimum), as well as a number of related Linum species, including the native Australian flax L. marginale (Barrett et al., 2009; Lawrence et al., 2007). Rust pathogens are obligate biotrophs, which depend on living plant tissues for propagation. The invading fungal hyphae form specialized feeding structures, called haustoria, that extract nutrients from host mesophyll cells.
Working with the cultivated flax–rust system, Flor (1956) defined the gene‐for‐gene model which has proved to be widely applicable as the basic genetic paradigm of plant disease resistance. In this model, the outcome of infection is based on the interaction of dominant resistance (R) genes in the host and dominant avirulence (Avr) genes in the pathogen. This genetic interaction is now understood in terms of effector‐triggered immunity (Chisholm et al., 2006; Dodds and Rathjen, 2010; Jones and Dangl, 2006), where R proteins constitute the recognition component of the plant immune system and detect the presence of specific pathogen effector (Avr) proteins, and trigger a defence response that prevents infection. These host and pathogen genes thus confer ‘extended phenotypes’ (Dawkins, 1999), that is their effects extend to the phenotype of another organism, implying close co‐evolutionary interactions which have been the basis of much theoretical modelling (Sasaki, 2000; Thrall and Burdon, 2002). However, there are few experimental systems in which it is possible to evaluate theoretical predictions arising from different co‐evolutionary scenarios (e.g. cyclical selection vs. an escalating arms race). Recent work in the flax–rust system has now delineated the molecular basis of gene‐for‐gene resistance and has revealed new insights into the evolutionary consequences of gene‐for‐gene interactions in wild systems. In this article, we summarize the current state of knowledge of molecular‐ and population‐level interactions in the flax–rust disease system, and future directions for understanding how these organisms co‐evolve.
MOLECULAR BASIS OF GENE‐FOR‐GENE INTERACTIONS
Resistance proteins
In cultivated flax, 30 genes that confer resistance to flax rust have been mapped to five loci (K, L, M, N, P), consisting of closely linked or allelic genes (Islam and Mayo, 1990). Nineteen R genes have been cloned from flax (11 from L, three from M, three from N, two from P), all of which encode intracellular Toll interleukin 1 receptor‐nucleotide binding site‐leucine‐rich repeat (TIR‐NBS‐LRR) class proteins (Anderson et al., 1997; 2001a, 2001b; Ellis et al., 1999; 1995, 2010a). The L locus consists of a single gene with 13 allelic variants (L, L1 to L11, and LH) distinguished by their reaction to rust strains carrying different Avr genes. The N, M and P loci are more complex, containing four, up to 15, and six to eight tandemly arranged paralogues, respectively.
Most variation between alleles and paralogues at these loci occurs in the LRR domain (2001a, 2001b; 1999, 2000), and domain swap experiments have confirmed that the LRR domain is important for determining R–Avr recognition specificity. For instance, chimeric L proteins consisting of the L2 LRR and either L6 or L10 N‐termini express L2 recognition specificity (Ellis et al., 1999). In addition, the L6 and L11 proteins differ by 32‐amino‐acid polymorphisms, all in the LRR domain, and a recombinant protein with a chimeric L6/L11 LRR showed a novel recognition specificity (Dodds et al., 2006; Ellis et al., 2007). In general, LRR domains are horseshoe‐shaped molecules (Fig. 1) composed of repeating leucine‐rich units of approximately 24–30 amino acids (Kobe and Deisenhofer, 1995). Variable residues are exposed on a concave β‐sheet surface and available for participation in R–Avr interactions. Indeed, the different P and P2 specificities are a result of just six amino acid polymorphisms found in the LRR β‐sheet region (Dodds et al., 2001a). Collectively, these results indicate that the LRR domain is the major determinant of Avr recognition specificities. Congruently, in the rice–rice blast pathosystem, the LRR domain of the rice R gene Pi‐ta has been observed to interact directly with the corresponding Avr‐Pita effector protein in a yeast two‐hybrid assay (Jia et al., 2000). Likewise, domain swaps have shown that pathogen recognition specificity is controlled by the LRR region of the barley Mla resistance proteins and the Rx/Gpa2 proteins in potato (Rairdan and Moffett, 2006; Shen et al., 2003). Pull‐down experiments have shown that the LRR domain of Arabidopsis RPP1 protein associates with the Hyaloperonospora arabidopsidis ATR1 effector (Krasileva et al., 2010).
Figure 1.
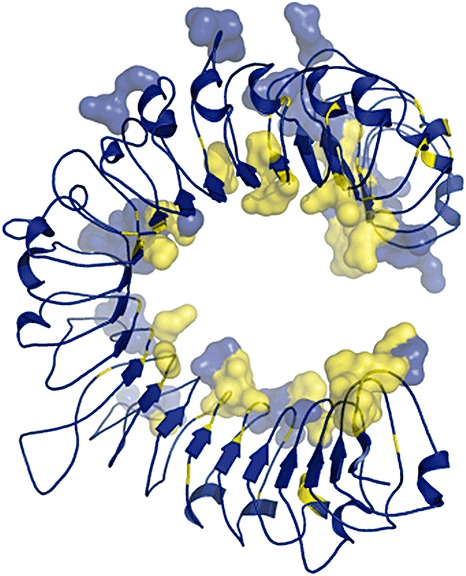
A structural model of the leucine‐rich repeat (LRR) domain of L5. Residues that may mediate interactions with AvrL567‐A have been rendered as three‐dimensional surfaces. Residues that are under significant (P > 0.95) positive selection are coloured yellow.
Although the LRR domain appears to be the primary mediator of recognition specificity, the TIR domain may also influence this function. TIR‐NBS domain swaps between L10 and L2 or L9 determined that L protein function requires co‐adapted TIR‐NBS and LRR regions, raising the possibility that these domains interact with each other (Luck et al., 2000). Indeed, positive selection has acted on the L TIR domain, suggesting that polymorphisms in this region are related to the function of the L proteins (Luck et al., 2000; M. Ravensdale, unpublished data). Hwang and Williamson (2003) have reported that intramolecular interactions between the coiled coil (CC) and LRR domains of the Mi resistance protein mediate downstream hypersensitive response (HR) signalling in tomato. Similarly, studies of the Rx resistance protein have demonstrated interactions between the CC‐NBS and LRR domains, and between the CC domain and the NBS‐LRR region (Moffett et al., 2002). Importantly, these interactions are disrupted in the presence of the cognate Rx effector ligand. In the case of the tobacco–Tobacco mosaic virus (TMV) pathosystem, the p50 fragment of the TMV replicase protein associates indirectly with the TIR domain of the N resistance protein through an intermediate protein (Burch‐Smith et al., 2007), but then appears to be recognized by binding directly to the LRR domain (Ueda et al., 2006). Collectively, these studies have led to hypothetical models of R protein function, in which the recognition of cognate effectors causes intramolecular conformational changes within the R protein, resulting in signal transduction (Rafiqi et al., 2009).
Avr effectors
Of the approximately 30 Avr specificities identified in flax rust via genetic studies, genes from four Avr families, representing nine recognition specificities, have been cloned to date: AvrL567, AvrM, AvrP123 and AvrP4 (Barrett et al., 2009; Catanzariti et al., 2006; Dodds et al., 2004). These were identified by screens for rust genes expressed during infection (AvrL567) or haustorially expressed sequence tags encoding secreted proteins (AvrM, AvrP4 and AvrP123) that co‐segregate with the Avr loci. Avirulence functions were confirmed by Agrobacterium‐mediated transient expression in flax lines expressing the corresponding R genes, which induced HR, whereas expression in flax lines without these resistance genes resulted in no HR. Recently, direct confirmation that these genes are responsible for avirulence was achieved using Agrobacterium‐mediated transformation of flax rust and RNA interference to silence AvrL567 genes; transgenic rust isolates acquired virulence on flax plants containing L5, L6 and L7 (Lawrence et al., 2010b).
All the Avr gene variants encode small secreted proteins (Table 1) that are expressed in haustoria and appear to be translocated into plant cells (Catanzariti et al., 2006; Dodds et al., 2004). They are characterized by high levels of polymorphism associated with differences in recognition specificity. For instance, there are 12 variant forms of AvrL567, seven of which are recognized by L5, L6 or L7, whereas the other five are virulence alleles (Dodds et al., 2006). Likewise, several different alleles of AvrP123 are differentially recognized by P, P1, P2 and P3, and a recombinant allele showed a novel recognition phenotype (Barrett et al., 2009; Dodds and Thrall, 2009). None of the effector families isolated from flax rust share sequence similarity with each other or with other currently known proteins, although AvrP123 contains 10 cysteine residues that conform to the consensus spacing of the kazal family of protease inhibitors (Catanzariti et al., 2006). Homologues of AvrL567, AvrM and AvrP4 occur in the poplar rust (M. larici‐populini) genome ( http://genome.jgi‐psf.org/Mellp1/Mellp1.home.html), and AvrP4 homologues occur across 22 Melampsora species (Van der Merwe et al., 2009).
Table 1.
Characteristics of cloned effector genes and their protein products from flax rust, Melampsora lini.
| Locus | Mature protein size (aa) | Number of cloned variants | Cognate R genes | Variation in rusts pathogenic on Linum marginale |
|---|---|---|---|---|
| AvrL567 | 127 | 12 | L5, L6, L7 | Not polymorphic |
| AvrM | 184–349 | 6 | M | Not polymorphic |
| AvrP123 | 88–94 | 6 | P, P1, P2, P3 | 7 alleles |
| AvrP4 | 67 | 3 | P4 | 13 alleles |
aa, amino acid.
The flax R proteins are cytoplasmic, and transient expression of Avr proteins lacking N‐terminal signal peptides results in an R gene‐dependent HR, which indicates that Avr recognition occurs inside the plant (Catanzariti et al., 2006; Dodds et al., 2004). Thus, Avr proteins must be translocated into plant cells during infection and, indeed, immunolocalization has detected the AvrM protein inside host cells containing a haustorium (Rafiqi et al., 2010) Similarly, translocation of flax rust effectors appears to be independent of the pathogen, as transient expression of AvrM in resistant flax leaves results in an HR regardless of the presence of a signal peptide (Catanzariti et al., 2006). This suggests that plant‐derived transport machinery may be exploited by these pathogens. Recently, Rafiqi et al. (2010) have demonstrated that the N‐terminus regions of AvrM and AvrL567 are sufficient to direct the translocation of secreted green fluorescent protein (GFP) fusion proteins. Likewise, translocation of effectors produced by ooymycete pathogens is mediated by a conserved N‐terminal RxLR motif (Whisson et al., 2007) and can occur independently of the pathogen (Dou et al., 2008).
Molecular basis of Avr–R interactions
The physical nature of Avr–R recognition has the potential to impact significantly on the evolution of these proteins. There are two hypothetical models that describe how pathogen effector proteins may interact with plant resistance proteins. The first is based on a direct physical interaction (receptor–ligand), and has been described in the rice—rice blast pathosystem, where Pi‐ta interacts with Avr‐Pita, in the Arabidopsis thaliana—Ralstonia solanacearum pathosystem, where RRS1 interacts with the avirulence protein PopP2, and in the tobacco–TMV pathosystem, where N interacts with the TMV replicase protein (Bernoux et al., 2008; Deslandes et al., 2003; Jia et al., 2000; Ueda et al., 2006). Alternatively, R proteins may recognize the presence of Avr proteins indirectly by detecting changes induced in other plant proteins by Avr proteins. In this scenario, R proteins guard the targets of Avr proteins. The guard hypothesis has been demonstrated in the A. thaliana—Pseudomonas syringae pathosystem, where RPM1, RPS2 and RPS5 recognize changes in RIN4 and PBS1 induced by the presence of bacterial effectors (Axtell et al., 2003; 2002, 2003; Shao et al., 2003). More recently, the guard model has been modified to include the possibility that guarded host proteins may evolve as decoys, thus functioning only as dedicated effector detectors (van der Hoorn and Kamoun, 2008). R proteins that function as guards are likely to select against the pathogenicity function of Avr proteins, whereas R proteins that bind directly to Avr effectors will impose a strong selection pressure on these proteins to evade physical detection. When Avr recognition is not related to effector function, it is possible for mutations to occur that disrupt recognition without affecting effector function. Thus, ‘guard’‐type R proteins should provide stable, long‐term resistance, whereas directly interacting R proteins should accelerate the evolution of new virulence phenotypes (Ellis and Dodds, 2003; van der Hoorn et al., 2002).
The diversification of both R and Avr allelic variants in the flax–rust system is suggestive of a direct protein–protein interaction and, indeed, yeast two‐hybrid assays have confirmed this (Catanzariti et al., 2010; Dodds et al., 2006). For example, Dodds et al. (2006) co‐expressed L5, L6 and L6L11RV (a chimeric gene derived from L6 and L11) with all 12 AvrL567 variants in the yeast two‐hybrid assay, and found a close correlation between Avr–R interactions in yeast and the induction of HR in planta. Likewise, M and AvrM interact directly in yeast, and this interaction correlates with the recognition specificities observed in planta (Catanzariti et al., 2010). Although M is approximately 80% identical to L5 and L6 at the amino acid level, AvrL567 and AvrM are unrelated. In addition, L5 and L6 are the most sequence diverged (87 polymorphisms; 59 in the LRR domain) of the L proteins and yet recognize an overlapping set of AvrL567 proteins, either as a result of convergent evolution, or because conserved sequences found in both L5 and L6 mediate these interactions. Collectively, these data suggest that the LRR domain is evolutionarily flexible and has evolved to directly recognize a diverse set of Avr effectors.
Protein sequence analysis of flax rust Avr effectors has revealed that these protein families are highly polymorphic, and appear to be under diversifying selection. For example, AvrM variants contain 14 polymorphic sites, as well as a number of deletions and truncations, and comparison with flanking sequence variation clearly indicated the effects of positive selection (Catanzariti et al., 2006). An initial study of AvrP4 revealed seven polymorphisms concentrated in the C‐terminal region of the protein that appeared to be the result of diversifying selection (Catanzariti et al., 2006), and comparison of AvrP4 homologues across 22 Melampsora species revealed significant positive selection in 15 codons located in the 3′ region (Van der Merwe et al., 2009). AvrL567 exhibits high amino acid sequence variability, with 27.5% of residues being polymorphic between variants, and DNA sequence analysis has revealed that this locus is also under significant positive diversifying selection (Dodds et al., 2006). Solution of the crystal structure of AvrL567 revealed that the side‐chains of all the polymorphic amino acids are exposed on the surface of the molecule (Wang et al., 2007). Mutational analysis confirmed the role of several of these residues in controlling recognition specificity, and it appears that these specificities are mediated by multiple amino acid contacts in a quantitative manner (Wang et al., 2007). Consequently, evolution of virulent forms of AvrL567 could occur in a stepwise manner, where single amino acid changes in avirulent forms result in partially virulent forms (i.e. weakly recognized) that would be selectively advantageous to the rust, and subsequent amino acid changes could eventually result in complete virulence.
The L locus in flax is also highly polymorphic, with 131 sites (30 in the TIR‐NBS region, 101 in the LRR domain) being under significant positive selection (M. Ravensdale, unpublished data). Wang et al. (2007) utilized the known structure of internalin A as a template for building a hypothetical structural model for the LRR domains of L5 and L6. These hypothetical L5 and L6 LRR models were then used to develop docking models for AvrL567‐A–L5 and AvrL567‐A–L6 interactions. This has resulted in a list of amino acid residues found in the LRRs of L5 and L6 that could make contact with AvrL567‐A. Superimposition of the 101 positively selected LRR residues onto the model of potential interacting residues has highlighted specific regions that may be involved in the interaction (Fig. 1). These sites can be evaluated in domain swap and mutation experiments. As Avr567 interacts with L5 and L6 in yeast, it should be possible to use the yeast two‐hybrid interaction to select for mutagenized L proteins with novel recognition specificities. A similar approach was successful in generating a variant of the potato Rx resistance protein with novel viral coat protein recognition specificities (Farnham and Baulcombe, 2006).
CO‐EVOLUTION IN A NATURAL GENE‐FOR‐GENE SYSTEM
Gene‐for‐gene resistance in L. marginale
In addition to cultivated flax, M. lini also infects the related wild flax L. marginale, a short‐lived perennial herb endemic to Australia. Both molecular and pathogenicity data indicate that the interaction between M. lini and wild flax represents a gene‐for‐gene association that is similar to, but evolutionarily differentiated from, the interaction between M. lini and cultivated flax. Of 15 pathogen isolates sampled from different L. marginale populations, only three were able to cause infection on a set of 25 L. usitatissimum rust resistance gene differential lines (Lawrence, 1989). In contrast, a set of 46 L. marginale lines, derived from across the geographical range of the native host, were all susceptible to at least several rust isolates, and six lines were susceptible to all isolates tested (Lawrence and Burdon, 1989). Furthermore, on L. marginale, isolates of M. lini display a variety of infection phenotypes, in terms of extent of sporulation and damage to the host plant, not seen on L. usitatissimum, causing, in some cases, the loss of older leaves at the bottom of the stem (Lawrence and Burdon, 1989). This differentiation between rust isolates from cultivated and wild flax is also seen in patterns of DNA variation in pathogen avirulence genes, with distinct Avr variants occurring in wild populations (Barrett et al., 2009; Dodds et al., 2006). The large variation in resistance and virulence phenotypes further suggests that L. marginale and M. lini have co‐evolved for a long time, and therefore M. lini is unlikely to be a recent introduction to Australia (Barrett et al., 2008b; Lawrence and Burdon, 1989). As in the cultivated flax system, rust resistance in L. marginale results from single dominant genes, with a minimum of 17 different R genes or alleles (Burdon, 1994).
Impact of life history on host–pathogen interactions at the population level
Host–pathogen co‐evolution is likely to be strongly influenced by life history factors, such as environmental conditions, effective population sizes (the number of individuals in a population that contribute offspring to the next generation) and pathogen dispersal mechanisms, which have been characterized extensively in the wild flax pathosystem. Linum marginale is found across Australia in various habitats, including eucalypt forests and savannah, open alpine areas covered with snow for several months a year, coastal sand dunes and along water courses in more arid inland areas (Lawrence and Burdon, 1989). Melampsora lini is also found across this extensive geographical and habitat range. Dikaryotic rust urediospores are wind dispersed and their rapid propagation can lead to local epidemics, with up to eight asexual reproduction cycles in a growing season. In colder and wetter environments (e.g. subalpine regions, referred to as the ‘Mountains’), plants flower in mid‐ to late summer before the first autumn frosts induce a large fraction of them to die, causing significant and abrupt crashes in pathogen numbers. Here, plants overwinter as underground rootstocks with or without a few green shoots protected from frost by the surrounding vegetation. These shoots may carry occasional pustules over to the next growing season, but there is also a significant probability of local loss of the pathogen. In contrast, in environments with hot and dry summers and mild winters (e.g. drier inland regions, referred to as the ‘Plains’), epidemics start earlier, last longer and are often more severe. The sexual cycle can be initiated late in the season as above‐ground shoots die back during the summer drought. This results in the production of sclerotic diploid teliospores that are resistant to environmental extremes. The decline in pathogen numbers is not as drastic as in the ‘Mountains’ region.
Several lines of evidence indicate that the pathogen has the potential to impose severe selection on the population structure of native hosts. Disease incidence varies in space and time, ranging from virtually nonexistent to epidemic levels; such patterns are consistent with varying patterns of co‐evolution across landscapes (Burdon and Thompson, 1995). Within local populations, the incidence, prevalence and disease severity start low and increase throughout the season, favoured by humid conditions that may result in epidemics with a prevalence of approximately 100% vs. approximately 20% for dry years (Jarosz and Burdon, 1992). The host survival rate is not affected by infection during the growing season, but over‐winter plant survival may be as low as 20%–30% in years of severe epidemics, in contrast with 80%–90% for a growing season with low pathogen pressure (Jarosz and Burdon, 1992). In epidemic years, disease also affects host population demography and population structure (see below). Younger plants have fewer stems and, as a result, they are less prone to infections than older (and larger) individuals, increasing their chances of survival over the winter in years of severe epidemics (Jarosz and Burdon, 1992). Following severe epidemics, host population size is often greatly reduced. Smaller populations (<100 individuals) are less likely to be infected than medium or large populations (up to 5000 individuals) in years of mild epidemics (Burdon and Jarosz, 1992). This may result from bottlenecks occurring at the end of the growing season. If too few infected hosts support overwintering pathogens, the pathogen may go extinct. Given the reduced pathogen pressure, the host population size is likely to increase, rendering the host population more prone to infection.
Patterns of resistance and virulence at multiple geographical scales
Host populations represent discrete groups that are genetically and phenotypically differentiated. For example, extensive inoculation studies have shown that, in the ‘Mountains’ region, with regard to resistance structure, populations range from being nearly monomorphic to having at least 18 resistance phenotypes (Burdon and Thompson, 1995; Jarosz and Burdon, 1991). The number of resistance phenotypes in a population shows spatial and temporal variation, indicating that host populations follow different ecological and evolutionary trajectories despite close geographical distance. This suggests that selection or drift can be stronger than gene flow. Indeed, following severe epidemics, the resistance structure of host populations can be significantly more diverse (Burdon and Thompson, 1995). In turn, among‐population variation in host resistance is a major determinant of the severity of pathogen epidemics (Thrall and Burdon, 2000). Taken together, it appears that M. lini exerts a mild to very strong selective pressure on populations of L. marginale, variable in space and time, shaping host population size, demography and genetic structure. Surprisingly, the predicted effects of selection are not consistently detected, e.g. after an epidemic in which a significant number of hosts die over the winter as a result of infection, the surviving hosts may not be more resistant to pathotypes from the previous year. Conversely, selection imposed by resistance on pathogen isolates at the local scale may be reduced by the large dispersal capacity of the pathogen. Therefore, selection may only be detected when looking at the metapopulation level, as a result of pathogen selective pressure competing with selection imposed by other environmental parameters at a local and short timescale.
Genetic analysis of M. lini isolates collected from L. marginale populations across Australia has revealed the existence of at least two distinct pathogen lineages (termed AA and AB). Lineage AB appears to have originated from hybridization between lineage AA and an extinct or as yet unidentified lineage BB (Barrett et al., 2007). The two lineages AA and AB are, for the most part, in geographical isolation from each other, with hybrids occurring mostly in areas of cool temperate climate with annual rainfall above 880 mm, whereas nonhybrids are found in hotter drier environments with under 640 mm of rainfall. No sexual reproduction was observed in the ‘Mountains’, where hybrids are prevalent, but extensive sexual reproduction was found in the ‘Plains’, where hybrids are present only at low frequencies (Barrett et al., 2007). Accordingly, AB isolates show a fixed pattern of heterozygosity (one A and one B allele at corresponding microsatellite loci). The extent to which differences in pathogen mating system are under genetic vs. environmental control is still unclear, although both field surveys and glasshouse inoculation studies indicate that telial formation (the precursor stage for sexual reproduction) is largely genetically determined (Barrett et al., 2008a). High temperatures appear to be the main environmental factor triggering telial formation in lineage AA (A. Nemri, unpublished data), and current studies have aimed to assess whether lineage AB isolates are able to proceed beyond telial formation to complete the sexual cycle. The implication is that differences in reproductive strategies and geographical distribution contribute to divergent evolutionary trajectories, and could result in the observed regional differences in virulence and diversity. Over time, this could lead to further specialization on hosts and subsequent host adaptation and, eventually, pathogen speciation.
Ecological differentiation within the host also occurs. Thus, within the ‘Mountains’ region, two morphologically discrete ecotypes (termed ‘bog’ and ‘hill’) occur at close geographical distances, but in different environments, e.g. in terms of soil moisture [A.L. Laine (University of Helsinki, Helsinki, Finland) and P.H. Thrall, unpublished data]. Populations of the ‘hill’ ecotype show distinctly more resistance than populations of the neighbouring ‘bog’ ecotype; their associated pathogen populations show correspondingly higher virulence (Thrall et al., 2001), but also have a environment‐independent lower survival rate, as shown by transplant studies (Carlsson‐Graner et al., 1999). Host differentiation in reproductive strategies is evident at larger geographical scales. Thus, the ‘Plains’ metapopulation exhibits a significant level of outcrossing, whereas plants in the ‘Mountains’ metapopulation are essentially inbred (Burdon et al., 1999). Such differences in mating system are associated with marked differences in the level and structure of resistance within and among host populations and metapopulations, which match observed differences in pathogen mating system, diversity and virulence.
Consistent with these local and regional patterns of differentiation in life history, phylogeny and variation in resistance and virulence in L. marginale and M. lini, adaptation between the pathogen and its host (a signature of co‐evolution) has also been demonstrated at multiple spatial scales. This includes continental (Burdon et al., 2002; Lawrence and Burdon, 1989), among regions (‘Plains’ vs. ‘Mountains’; (Barrett et al., 2007; Burdon et al., 1999) and within a single metapopulation [‘bog’ vs. ‘hill’ (Carlsson‐Graner et al., 1999); or local adaptation (Thrall et al., 2002)]. For example, at the continental scale, rust isolates from Victoria and southern New South Wales are significantly more virulent on hosts from these regions than are isolates from other parts of Australia. Similarly, closely adjacent inbreeding host populations in the ‘Mountains’ metapopulation were found to be more similar with regard to the relative abundance of particular resistance phenotypes than they were with more distant populations. At very local scales, the mating system has also been demonstrated recently to have an impact on the within‐population structure of resistance (A. Nemri, unpublished data). It is somewhat puzzling that, despite the broad dispersal ability of the pathogen relative to that of the host (as evidenced by the lack of distance‐dependent distribution patterns for virulence phenotypes in associated pathogen populations; (Burdon and Jarosz, 1991; Thrall and Burdon, 2003), the most virulent pathotypes do not generally dominate host populations. This contrasts with agricultural crop systems in which supervirulent pathogens rise and disperse globally over relatively short timescales (often within a few years). This discrepancy may be explained by an evolutionary trade‐off between infection strategies, which is suggested by glasshouse inoculation studies revealing a negative relationship between virulence and spore production (Thrall and Burdon, 2003). It is not known whether this negative correlation can be explained by avirulence effectors negatively contributing to pathogen fitness in their virulence form, although preliminary evidence suggests that there is no detectable fitness penalty to silencing AvrL6 in a host background lacking L6 (Lawrence et al., 2010b).
Partial infections, i.e. infections in which the pathogen cannot achieve maximum spore production, are commonly observed in glasshouse single‐spore inoculation studies. Analysis of data from multiple populations in the Linum–Melampsora system has shown that the number of partial infection phenotypes relative to the total number of resistant responses is likely to be greater when hosts are challenged with sympatric rather than allopatric pathogen isolates (Antonovics et al., 2010). Some host populations have a high prevalence of partial resistance genes compared with the average across the metapopulation, whereas some pathogen populations contain primarily isolates that are unable to cause a severe infection regardless of host origin (Antonovics et al., 2010). The occurrence of partial infection phenotypes in different host–pathogen population pairs thus depends on the host, the pathogen and the interaction. These differences constitute an important, yet so far poorly understood, aspect of the co‐evolutionary interaction between host and pathogen.
Avr gene diversity in natural populations
Melampsora lini isolates from the wild pathosystem contain homologues of the AvrL567, AvrM, AvrP123 and AvrP4 genes identified from the cultivated pathosystem. However, the patterns of selection differ between these genes. A geographically diverse set of isolates all contained the same AvrM variant, suggesting that AvrM does not contribute to the outcome of the gene‐for‐gene interaction, probably because there are no R genes in the L. marginale populations that recognize this Avr gene (Dodds and Thrall, 2009). Some variation has been observed amongst AvrL567 homologues in rust isolates from L. marginale, but the complex nature of this multicopy locus has made genetic characterization at the population level more difficult. However, isolates of M. lini from L. marginale populations show extensive variation and a signature of strong positive selection at the AvrP4 and AvrP123 loci (Barrett et al., 2009). Transient expression of these AvrP123 and AvrP4 variants in L. marginale plants can trigger HRs in some host genotypes, suggesting that there is differential recognition of these genes by R genes in host populations (Barrett et al., 2009). It is not yet known whether these recognition specificities are mediated by homologues of the P genes from L. usitatissimum. However, there is among‐population variation in the frequency of recognition of AvrP123 and AvrP4 variants, and also considerable year‐to‐year variation within populations of L. marginale (M. Ravensdale et al., unpublished data). Likewise, there is variation in the frequency of different AvrP123 and AvrP4 variants between rust populations (Barrett et al., 2009). This suggests the possibility of strong selection acting on the cognate R and Avr genes in this system. The spatial and temporal variation in the cognate R genes could explain the maintenance of the observed polymorphism at AvrP4 and AvrP123.
CONCLUDING REMARKS
The combination of in‐depth molecular‐ and population‐level information makes the flax–rust disease system a powerful model for understanding host–pathogen co‐evolution. Detailed knowledge of the molecular interaction between R and Avr proteins in cultivated flax provides a framework for understanding the selective forces underlying the population‐level variation that is observed in the wild flax–rust interaction. There still remain many challenges to the integration of these data. For instance, although we know that Avr genes identified from cultivated flax rust also operate in the wild system, we do not yet know whether the corresponding R genes in L. marginale are also homologues of the cultivated flax R genes. It will be important in the future to correlate the phenotypic analysis of wild populations with DNA‐based studies of the distribution of specific R and Avr genes. Likewise, much knowledge still needs to be gathered to enable the generalization of the findings from this system to other plant–pathogen interactions, including those occurring in agricultural crop systems. For the management of crop diseases, information on dispersal, disease dynamics and the virulence structure of rust populations is crucial. In the long term, the determination of how the selective pressure imposed by host genetic structure and other life history components shapes rust adaptation and evolution is also critical (Barrett et al., 2008b). Although considerable research effort has focused on major R genes, little is known about the epidemiological or evolutionary consequences of gene‐for‐gene interactions that result in partial infection, even though they have the potential to significantly impact disease dynamics and pathogen adaptation. The integration of the molecular understanding of gene‐for‐gene interactions with population genetics in the flax–rust system is now providing a powerful approach for measuring the effects of gene‐for‐gene co‐evolution. However, such studies also need to account for other selective factors in the environment, i.e. life history, with which they probably interact. A specific experimental challenge will be to find the spatial and temporal scales at which one can verify predictions emerging from the arms race model in the context of co‐evolving metapopulations of hosts and pathogens. This will be necessary not only to address issues such as the maintenance of host and pathogen diversity, but also to explain the modality of the arms race and the maintenance of sexual reproduction in the pathogen and outcrossing in the host with respect to the Red Queen hypothesis (Van Valen, 1973).
GLOSSARY BOX
Definitions
Avr proteins: pathogen effectors that are recognized by host R proteins.
Co‐evolutionary arms race in host–parasite interactions: an asymmetrical genetic interaction in which the fitness of the host is negatively correlated with the fitness of the parasite. To try to increase its fitness, the host evolves defence mechanisms, such as resistance, that have to be circumvented by the pathogen by evolving virulence mechanisms, such as effector‐triggered immunity. This results in the constant accumulation of defence mechanisms on the host side and virulence mechanisms on the pathogen side. At the gene level, it is characterized by the rapid and continuous fixation of novel mutations with advantageous effect.
Diversifying selection: characterized by alleles at both extremes of a phenotype spectrum being selected at the same time, whereas individuals with alleles encoding intermediate phenotypes would be selected against.
Effectors: pathogen proteins that are produced to interfere with host processes and allow disease establishment.
Effector‐triggered immunity: an immune response triggered by R–Avr recognition.
Metapopulation: a group of populations located at a geographical distance that permits the genetic exchange of propagules (pollen, seeds, spores) between them.
Positive selection: a type of directional selection in which one allele is favoured and rises from rare to predominant in a population. Genes under positive selection typically have a high ratio of nonsynonymous mutations (Ka/Ks).
Virulence (as used in plant pathology) describes the capability of causing infection, and aggressiveness refers to the reproductive output of the pathogen. Pathogen fitness is referred to as aggressivity.
ACKNOWLEDGEMENTS
The support of the National Institutes of Health (NIH grant 5RO1 GM074265‐01A2) is gratefully acknowledged. This Review was developed from a presentation that was to be given at the British Society for Plant Pathology's 2009 presidential meeting.
REFERENCES
- Anderson, P.A. , Lawrence, G.J. , Morrish, B.C. , Ayliffe, M.A. , Finnegan, E.J. and Ellis, J.G. (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine‐rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics, J. , Thrall, P.H. , Laine, A.L. and Burdon, J.J. (2010) Partial resistance in the Linum‐Melamspora host‐pathogen system: does partial resistance make the Red Queen run slower? Evolution, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J. , Chisholm, S.T. , Dahlbeck, D. and Staskawicz, B.J. (2003) Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49, 1537–1546. [DOI] [PubMed] [Google Scholar]
- Barrett, L.G. , Thrall, P.H. and Burdon, J.J. (2007) Evolutionary diversification through hybridization in a wild host–pathogen interaction. Evolution, 61, 1613–1621. [DOI] [PubMed] [Google Scholar]
- Barrett, L.G. , Thrall, P.H. , Burdon, E.J. , Nicotra, A.B. and Linde, C.C. (2008a) Population structure and diversity in sexual and asexual populations of the pathogenic fungus Melampsora lini . Mol. Ecol. 17, 3401–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, L.G. , Thrall, P.H. , Burdon, J.J. and Linde, C.C. (2008b) Life history determines genetic structure and evolutionary potential of host–parasite interactions. Trends Ecol. Evol. 23, 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, L.G. , Thrall, P.H. , Dodds, P.N. , Van Der Merwe, M. , Linde, C.C. , Lawrence, G.J. and Burdon, J.J. (2009) Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini . Mol. Biol. Evol. 26, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux, M. , Timmers, T. , Jauneau, A. , Briere, C. , De Wit, P. , Marco, Y. and Deslandes, L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1‐R‐mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum Popp2 effector. Plant Cell, 20, 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Schiff, M. , Caplan, J.L. , Tsao, J. , Czymmek, K. and Dinesh‐Kumar, S.P. (2007) A novel role for the TIR domain in association with pathogen‐derived elicitors. PLoS Biol. 5, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon, J.J. (1994) The distribution and origin of genes for race‐specific resistance to Melampsora lini in Linum marginale . Evolution, 48, 1564–1575. [DOI] [PubMed] [Google Scholar]
- Burdon, J.J. and Jarosz, A.M. (1991) Host–pathogen interactions in natural populations of Linum marginale and Melampsora lini. I. Patterns of resistance and racial variation in a large host population. Evolution, 45, 205–217. [DOI] [PubMed] [Google Scholar]
- Burdon, J.J. and Jarosz, A.M. (1992) Temporal variation in the racial structure of flax rust (Melampsora lini) populations growing on natural stands of wild flax (Linum marginale): local versus metapopulation dynamics. Plant Pathol. 41, 165–179. [Google Scholar]
- Burdon, J.J. and Thompson, J.N. (1995) Changed patterns of resistance in a population of Linum marginale attacked by the rust pathogen Melampsora lini . J. Ecol. 83, 199–206. [Google Scholar]
- Burdon, J.J. , Thrall, P.H. and Brown, A.H.D. (1999) Resistance and virulence structure in two Linum marginale–Melampsora lini host–pathogen metapopulations with different mating systems. Evolution, 53, 704–716. [DOI] [PubMed] [Google Scholar]
- Burdon, J.J. , Thrall, P.H. and Lawrence, G.J. (2002) Coevolutionary patterns in the Linum marginale–Melampsora lini association at a continental scale. Can. J. Bot.-Rev. Can. Bot. 80, 288–296. [Google Scholar]
- Carlsson‐Graner, U. , Burdon, J.J. and Thrall, P.H. (1999) Host resistance and pathogen virulence across a plant hybrid zone. Oecologia, 121, 339–347. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.‐M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Ve, T. , Kobe, B. , Ellis, J.G. and Staskawicz, B.J. (2010) The Avrm effector from flax rust has a structured C‐terminal domain and interacts directly with the M resistance protein. Mol. Plant–Microbe Interact. 23, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Dawkins, R. (1999) The Extended Phenotype: The Long Reach of the Gene. Oxford: Oxford University Press. [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, L. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dodds, P. and Thrall, P. (2009) Recognition events and host–pathogen co‐evolution in gene‐for‐gene resistance to flax rust. Funct. Plant Biol. 36, 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Teh, T. , Wang, C.‐I.A. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. and Ellis, J.G. (2001a) Six amino acid changes confined to the leucine‐rich repeat beta‐strand/beta‐turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell, 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. and Ellis, J.G. (2001b) Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27, 439–453. [DOI] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008) RxLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. and Dodds, P. (2003) Plant pathology: monitoring a pathogen‐targeted host protein. Curr. Biol. 13, R400–R402. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. , Luck, J.E. and Dodds, P.N. (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene‐for‐gene specificity. Plant Cell, 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. , Dodds, P. and Pryor, T. (2000) The generation of plant disease resistance gene specificities. Trends Plant Sci. 5, 373–379. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. and Dodds, P.N. (2007) Further analysis of gene‐for‐gene disease resistance specificity in flax. Mol. Plant Pathol. 8, 103–109. [DOI] [PubMed] [Google Scholar]
- Farnham, G. and Baulcombe, D.C. (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease‐resistance gene from potato. Proc. Natl. Acad. Sci. USA, 103, 18 828–18 833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1956) The complementary genic systems in flax and flax rust. Adv. Genet. 8, 29–54. [Google Scholar]
- Van Der Hoorn, R.A.L. and Kamoun, S. (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell, 20, 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hoorn, R.A.L. , Wit, P.J.G.M.D. and Joosten, M.H.A.J. (2002) Balancing selection favors guarding resistance proteins. Trends Plant Sci. 7, 67–71. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F. and Williamson, V.M. (2003) Leucine‐rich repeat‐mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J. 34, 585–593. [DOI] [PubMed] [Google Scholar]
- Islam, M.R. and Mayo, G.M.E. (1990) A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104, 89–100. [Google Scholar]
- Jarosz, A.M. and Burdon, J.J. (1991) Host–pathogen interactions in natural populations of Linum marginale and Melampsora lini. II. Local and regional variation in patterns of resistance and racial structure. Evolution, 45, 1618–1627. [DOI] [PubMed] [Google Scholar]
- Jarosz, A.M. and Burdon, J.J. (1992) Host–pathogen interactions in natural populations of Linum marginale and Melampsora lini. III. Influence of pathogen epidemics on host survivorship and flower production. Oecologia, 89, 53–61. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , McAdams, S.A. , Bryan, G.T. , Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kobe, B. and Deisenhofer, J. (1995) A structural basis of the interactions between leucine‐rich repeats and protein ligands. Nature, 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Krasileva, K.V. , Dahlbeck, D. and Staskawicz, B.J. (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine‐rich repeat domain with the cognate oomycete effector. Plant Cell, DOI: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J. (1989) Flax rust from Linum marginale—pathogenicity reactions on the Linum usitatissimum set of differential varieties. Can. J. Bot.-Rev. Can. Bot. 67, 3187–3191. [Google Scholar]
- Lawrence, G.J. and Burdon, J.J. (1989) Flax rust from Linum marginale—variation in a natural host–pathogen interaction. Can. J. Bot.-Rev. Can. Bot. 67, 3192–3198. [Google Scholar]
- Lawrence, G.J. , Finnegan, E.J. , Ayliffe, M.A. and Ellis, J.G. (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial‐resistance gene Rps2 and the tobacco viral resistance gene‐N . Plant Cell, 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J. , Dodds, P.N. and Ellis, J.G. (2007) Rust of flax and linseed caused by Melampsora lini . Mol. Plant Pathol. 8, 349–364. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J. , Anderson, P.A. , Dodds, P.N. and Ellis, J.G. (2010a) Relationships between rust resistance genes at the M locus in flax. Mol. Plant Pathol. 11, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J. , Dodds, P.N. and Ellis, J.G. (2010b) Transformation of the flax rust fungus, Melampsora lini: selection via silencing of an avirulence gene. Plant J. 61, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J.E. , Lawrence, G.J. , Dodds, P.N. , Shepherd, K.W. and Ellis, J.G. (2000) Regions outside of the leucine‐rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell, 12, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Holt, B.F. , Wiig, A. and Dangl, J.L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for Rpm1‐mediated resistance in Arabidopsis . Cell, 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mackey, D. , Belkhadir, Y. , Alonso, J.M. , Ecker, J.R. and Dangl, J.L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates Rps2‐mediated resistance. Cell, 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Moffett, P. , Farnham, G. , Peart, J. and Baulcombe, D.C. (2002) Interaction between domains of a plant NBS‐LRR protein in disease resistance‐related cell death. EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi, M. , Bernoux, M. , Ellis, J.G. and Dodds, P.N. (2009) In the trenches of plant pathogen recognition: role of NB‐LRR proteins. Semin. Cell Dev. Biol. 20, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Rafiqi, M. , Gan, P.H.P. , Ravensdale, M. , Lawrence, G.J. , Ellis, J.G. , Jones, D.A. , Hardham, A.R. and Dodds, P.N. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell, 22, 2017–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J. and Moffett, P. (2006) Distinct domains in the arc region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell, 18, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A. (2000) Host–parasite coevolution in a multilocus gene‐for‐gene system. Proc. R. Soc. Lond. B Biol. Sci. 267, 2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F. , Golstein, C. , Ade, J. , Stoutemyer, M. , Dixon, J.E. and Innes, R.W. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science, 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H. , Zhou, F. , Bieri, S. , Haizel, T. , Shirasu, K. and Schulze‐Lefert, P. (2003) Recognition specificity and Rar1/Sgt1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell, 15, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall, P.H. and Burdon, J.J. (2000) Effect of resistance variation in a natural plant host–pathogen metapopulation on disease dynamics. Plant Pathol. 49, 767–773. [Google Scholar]
- Thrall, P.H. and Burdon, J.J. (2002) Evolution of gene‐for‐gene systems in metapopulations: the effect of spatial scale of host and pathogen dispersal. Plant Pathol. 51, 169–184. [Google Scholar]
- Thrall, P.H. and Burdon, J.J. (2003) Evolution of virulence in a plant host–pathogen metapopulation. Science, 299, 1735–1737. [DOI] [PubMed] [Google Scholar]
- Thrall, P.H. , Burdon, J.J. and Young, A. (2001) Variation in resistance and virulence among demes of a single host–pathogen metapopulation. J. Ecol. 89, 736–748. [Google Scholar]
- Thrall, P.H. , Burdon, J.J. and Bever, J.D. (2002) Local adaptation in the Linum marginale–Melampsora lini host–pathogen interaction. Evolution, 56, 1340–1351. [DOI] [PubMed] [Google Scholar]
- Ueda, H. , Yamaguchi, Y. and Sano, H. (2006) Direct interaction between the tobacco mosaic virus helicase domain and the ATP‐bound resistance protein, N Factor, during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61, 31–45. [DOI] [PubMed] [Google Scholar]
- Van der Merwe, M.M. , Kinnear, M.W. , Barrett, L.G. , Dodds, P.N. , Ericson, L. , Thrall, P.H. and Burdon, J.J. (2009) Positive selection in AvrP4 avirulence gene homologues across the genus Melampsora . Proc. R. Soc. Lond. B Biol. Sci. 276, 2913–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen, L. (1973) A new evolutionary law. Evol. Theory, 1, 1–30. [Google Scholar]
- Wang, C.‐I.A. , Guncar, G. , Forwood, J.K. , Teh, T. , Catanzariti, A.‐M. , Lawrence, G.J. , Loughlin, F.E. , Mackay, J.P. , Schirra, H.J. , Anderson, P.A. , Ellis, J.G. , Dodds, P.N. and Kobe, B. (2007) Crystal structures of flax rust avirulence proteins AvrL567‐A and ‐D reveal details of the structural basis for flax disease resistance specificity. Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , Van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]


