Abstract
Recently, agonist antibodies to glucocorticoid-induced tumor necrosis factor receptor (GITR) (tumor necrosis factor receptor superfamily 18) have been shown to neutralize the suppressive activity of CD4+CD25+ regulatory T cells. It was anticipated that this would be the role of the physiological ligand. We have identified and expressed the gene for mouse GITR ligand and have confirmed that its interaction with GITR reverses suppression by CD4+CD25+ T cells. It also, however, provides a costimulatory signal for the antigen-driven proliferation of naïve T cells and polarized T helper 1 and T helper 2 clones. RT-PCR and mAb staining revealed mouse GITR ligand expression in dendritic cells, macrophages, and B cells. Expression was controlled by the transcription factor NF-1 and potentially by alternative splicing of mRNA destabilization sequences.
Mouse glucocorticoid-induced tumor necrosis factor receptor (mGITR) was originally identified in dexamethasone-treated T cell hybridoma cells (1) and encodes a 228-aa cysteine-rich protein that is defined as tumor necrosis factor receptor (TNFR) superfamily 18 (Tnfrsf 18). The human counterpart and its ligand were characterized soon after (2, 3). The interaction of human GITR (hGITR) with its ligand activates NF-κB via a TNFR-associated factor 2-mediated pathway (1-3). Overexpression of hGITR mRNA was shown to rescue T cells from T cell receptor (TCR)-mediated cell death (1-3), although an agonist anti-mGITR antibody did not reproduce this effect (4).
The identification of GITR on CD4+CD25+ regulatory T cells and the ability of agonist antibodies to reverse regulation both in vitro and in vivo suggested that mouse GITR ligand (mGITRL) could be a physiological modulator of T cell regulation (4, 5). However, the same agonist anti-GITR antibodies did not appear to have an effect on CD4+CD25- T cells even though these cells express GITR weakly (4, 5). This finding implied no role for the ligand in the function of these cells.
To establish whether the effects of agonist antibodies truly reflect the action of the physiological mGITRL, we have cloned and expressed the mGITRL and studied its effect on T cell proliferative responses in vitro.
The mGITRL gene was identified in the genome database by using sequence information from the published human GITR ligand (2, 3). Transcripts of this gene were abundantly expressed in murine dendritic cells (DCs) and macrophages. Expression of mGITRL is transiently up-regulated, and then rapidly down-modulated, after lipopolysaccharide (LPS) stimulation, this effect being mediated through the transcription factor NF-1. Using the recombinant protein and transfectants of mGITRL, we show that mGITR signaling reverses the suppressive effects of CD4+CD25+ regulatory T cells. In addition, mGITR signaling can enhance the proliferation of antigen-stimulated T helper 1 (Th1), T helper 2 (Th2), and naïve CD4+ cells from monospecific TCR transgenic mice (6) when exposed to low doses of antigen. Our findings suggest that mGITR functions as a general costimulatory receptor for TCR-stimulated T cells.
Materials and Methods
cDNA Cloning and Mapping of cDNA Ends. The mGITRL gene was identified by using the Celera database. Mapping of 5′ and 3′ ends of the cDNA was performed by RACE (7) with minor modifications (8). 5′ RACE primer 1 (TGAGTGAAGTATAGATCAGTG), 5′ RACE primer 2 (GCATCAGTAACAGAG CCACTATG), 3′ RACE primer 1 (GATGGGAAGCTGAAGATACTG), and 3′ RACE primer 2 (GAACTGCATGCTGGAGATAAC) were used for this assay. For 3′ RACE, cDNA was prepared by using a GeneRacer kit (Invitrogen). The mGITRL cDNAs containing the 3′ ends were amplified by using 3′ RACE primers and UAP (Invitrogen).
Cell Culture and Transfection. Preparation of bone marrow-derived (bm) DC (9), a B cell-enriched fraction (9), and peritoneal cells (10) have been described. Briefly, a B cell-enriched fraction was prepared from T cell-depleted CBA/Ca mice by passing through splenocytes over a Sephadex G-10 column. To prepare bmDC and bm macrophage, bone marrow cells from CBA/Ca mice were cultured for 7 days with granulocyte/macrophage colony-stimulating factor (5 ng/ml). bmDC were separated by gentle aspiration from bm macrophage, which tightly bound to cell culture dishes. If required, these cells were stimulated with LPS (10 μg/ml). To prepare IL-10/DC, IL-10 (final 20 ng/ml) was added to bmDC culture at day 6 and harvested at day 9. Peritoneal cells were isolated from >6 week-old CBA/Ca mice by peritoneal lavage by using ice-cold DMEM containing 0.38% sodium citrate. To isolate primary macrophages, cells were cultured with LPS (10 μg/ml) in bacterial Petri dishes. After 6 or 24 h, binding macrophages on the plastic surface were isolated.
mGITRL transfectants were generated by using NB2 6TG, HEK293 cells, and a mGITRL expression plasmid in pMTF vector (9). mGITR transfectants were also generated by using a mGITR expression plasmid (in pMTF) and Jurkat (JE6.1) cells. Stable transfectants were selected by G418 (1 mg/ml).
Preparation of Recombinant mGITRL and Anti-mGITRL Antibody. cDNAs encoding extracellular domains of mGITRL (Fig. 1A, amino acid positions 43-173) and human CD40 ligand (hCD40L) (amino acid positions 47-261) were amplified by using PCR primers (mGITRL sense, TCGGATCCTCACTCAAGCCAACTGC; mGITRL antisense, AAGAATTCAATCTCTAAGAGATGAATGG; hCD40L sense, GTGGGATCCCATAGAAGGTTGGACAAGATAG; hCD40L antisense, GTGGAATTCATCAGAGTTTGAGTAAGCCAAAGG). The amplified fragments were cloned into BamHI and EcoRI sites of pRSET vector (Invitrogen). The resulting plasmids were transferred into Escherichia coli BL21(DE3)pLysS to produce recombinant proteins by isopropyl β-d-thiogalactoside induction. A 6× His tag recombinant protein was purified by using Ni-NTA agarose (Qiagen, Chatsworth, CA), and the eluted protein was dialyzed against PBS.
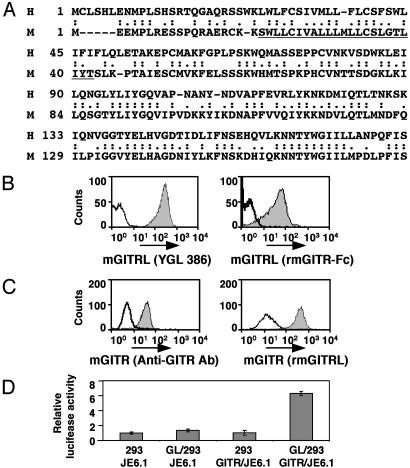
Fig. 1.
Identification of mGITRL. (A) Amino acid sequences of human (H) and mouse (M) GITRL are aligned. The predicted transmembrane domain is underlined. (B) Binding of GITR and the putative mGITRL was analyzed by using recombinant GITR-Fc and transfectants expressing this ligand. The putative mGITRL transfectants (filled with gray) and nontransfectants (solid line) were stained with mAb (YGL 386) or recombinant mGITR-Fc (rmGITR-Fc). (C) Binding of mGITR and the mGITRL was analyzed by using mGITR transfectants and recombinant protein of this ligand. mGITR transfectants (filled with gray) and nontransfectants (solid line) were stained with anti-GITR antibody (Anti-GITR Ab) or the recombinant putative mGITRL (rmGITRL). Binding of the recombinant protein was detected with anti-His tag antibody. (D) Signaling through mGITR with the mGITRL was analyzed by a luciferase assay using the NF-κB reporter plasmid. mGITR transfectant (GITR/JE6.1) and nontransfectant (JE6.1) were electroporated with the NF-κB reporter plasmid. Five hours postelectroporation, these cells were harvested and mixed with either growth-arrested (by mitomycin C treatment) HEK293/mGITRL transfectants (GL/293) or HEK293 (293). Mixed combinations are indicated under the graph. Luciferase activities generated these cells were compared with that in JE6.1 with HEK293.
To produce anti-mGITRL mAb, DA rats were immunized by using this purified recombinant protein. An anti-mGITRL mAb, YGL386 (IgG1), was obtained from the fusion of the immunized rat spleen with a myeloma line, Y3/Ag1.2.3. This antibody was purified by using a protein G column (Amersham Pharmacia Bioscience) and biotinylated. Rat anti-canine CD8 antibody (IgG1) was used as an isotype control antibody.
For flow cytometric analysis using bmDC and transfectants, biotinylated anti-mGITR (R & D Systems, BAF524), anti-mGITRL mAb (YGL386), and recombinant mGITR-Fc (Alexis Biochemicals, Lausen, Switzerland) were used. Allophycocyanin-conjugated streptavidin was used as a secondary reagent. bmDC were costained with FITC-conjugated anti-CD11c antibody (Becton Dickinson Pharmingen). To stain spleen and peritoneal cells, Alexa 488-conjugated YGL386, allophycocyanin-conjugated anti-CD3 (Becton Dickinson), phycoerythrin (PE)-conjugated anti-F4/80 (Becton Dickinson Pharmingen), and PE-conjugated anti-B220 (Becton Dickinson Pharmingen) antibodies were used.
RT-PCR. To detect mGITRL mRNA, RT-PCR was performed as described (11). To compare expression levels and minimize PCR artifacts, the number of PCR cycles was kept low [17 cycles for hypoxanthine phosphoribosyltransferase (HPRT), 25 cycles for mGITRL], and PCR products from mGITRL mRNA were detected by Southern blot hybridization using a cDNA probe. Cycle numbers of PCR were determined by preliminary experiments, and under these conditions, PCR was not saturated. The PCR primers used were: mGITRL sense, AGCCTCATGGAGGAAATG; mGITRL antisense, ATATGTGCCACTCTGCAGTATC; HPRT sense, ACAGCCCCAAAATGGTTAAGG; and HPRT antisense, TCTGGGGACGCAGCAACTGAC.
Luciferase Reporter Assay. To examine NF-κB activity in mGITR transfectants, a luciferase assay was performed as described in Results. pNF-κB luc (Stratagene) was used as a NF-κB reporter plasmid. Ten mGITRL promoter fragments (5′ ends are indicated in Fig. 5B) were cloned into the pGL3-basic Vector (Promega). RAW 264 cells (1.5 × 107 cells) were transfected with the resulting luciferase reporter plasmids (10 μg) by Gene Pulser (BioRad). If required, cells were stimulated with LPS (10 μg/ml) 5 h postelectroporation. After 48 h culture, cells were harvested, and promoter activities were analyzed by using the Dual-Luciferase Reporter Assay System (Promega). These assays were repeated at least three times, and firefly luciferase activities (mGITRL promoter activities) were normalized to Renilla luciferase (internal control) activities.
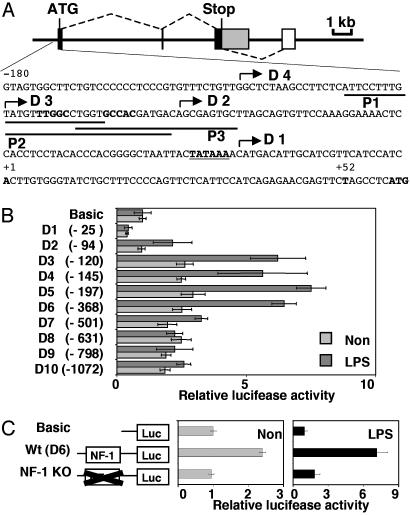
Fig. 5.
Gene structure and promoter activity of mGITRL. (A) Coding exons are indicated by black boxes, and a 3′ noncoding region is indicated by a gray box. An alternative 3′ noncoding exon is indicated by a white box. Splice joints are indicated by dotted lines. Partial promoter and 5′ noncoding sequences are shown under mGITRL gene structure. A major transcription start site (+1), the 3′ end (+52) of the promoter fragments in the luciferase reporter plasmids (in B), and the first ATG are indicated in bold. The TATA box sequence is indicated in bold and underlined. 5′ Ends of the promoter fragments in the luciferase reporter plasmids (in B, D1-D4) are indicated by arrows, and the locations of probes P1, P2, and P3 for EMSA (in Fig. 6A) are indicated by solid lines. (B and C) mGITRL promoter activity was analyzed by luciferase assays. Luciferase activity generated using the reporter plasmids were compared with that generated using the negative control plasmid (no insert) pGL3-Basic Vector (Basic) in nonstimulated and LPS-stimulated RAW 264 cells. These assays were repeated at least three times. (B) The luciferase reporter plasmids were constructed by using the mGITRL promoter fragments. The 5′ end of each promoter fragment is indicated in parentheses. (C) The NF-1 site in the luciferase reporter D6 (in B) was mutated, and the structure of these plasmids used in the luciferase assay are illustrated. The mutated NF-1 site (TTGGCCTGGTGCCAC to TGGCCTGGGAATTC) is indicated (X).
Preparation of Nuclear Extracts, Electrophoretic Mobility-Shift Assay (EMSA), and Immunoblotting. EMSA was performed as described (8). Five micrograms of nuclear extract was used for this assay. For the competition assay, a 100-fold excess of unlabeled competitor was added to EMSA reaction mixture. To perform the super shift assay, nuclear extracts in EMSA reaction buffer was incubated with anti-NF-1 antibody (Santa Cruz Biotechnology, H-300) for 15 min, at which time probes were then added.
Ten-microgram nuclear extracts and anti-NF-1 antibody (Santa Cruz Biotechnology, H-300) were used for immunoblotting as described (8).
Proliferation Assays. Th1 (R2.2) and Th2 (R2.4) clones, established from the spleen of a female A1(M)RAG-1-/- mouse, have been described (6) and were used 14 days after antigen stimulation. Naïve CD4+ cells were purified from the spleens of female A1(M)RAG-1-/- mice using the CD4 isolation kit (Miltenyi Biotech, Auburn, CA). Total CD4+ T cells were purified from naïve female CBA/Ca mice as described (12). CD4+CD25- and CD4+CD25+ cells were separated by cell sorter (MoFlo, Dako Cytomation, Glostrup, Denmark) using FITC-conjugated anti-CD4 and phycoerythrin-conjugated anti-CD25 (both Becton Dickinson Pharmingen) antibodies. Where appropriate, cells were activated by the H-Y peptide (REEALHQFRSGRKPI; 1-100 nM), plate-bound or soluble anti-CD3 antibody (145-2C11), and soluble anti-CD28 antibody (37.51).
For proliferation assays, 1 × 104 clones (R2.2 and R2.4) or 5 × 104 naïve CD4+ cells from A1(M)RAG-1-/- mice were used. These cells were cultured with 1 × 105 mitomycin C-treated, T cell-depleted female spleen cells and H-Y peptide (0-100 nM). If required, recombinant mGITRL (10 μg/ml), recombinant hCD40L (10 μg/ml), mGITRL/NB2 transfectants, or NB2 6TG cells were added to the culture. Suppression assays of CD4+CD25- by CD4+CD25+ cells were activated by anti-CD3 antibody, 5% final concentration of culture supernatant, and performed as described (5); proliferation was measured at 48 h by 3H-thymidine incorporation. For the suppression assay with naïve CD4+ cells from A1(M)RAG-1-/- mice, CD4+CD25+ cells were preactivated overnight with plate-bound anti-CD3 antibody (145-2C11, 10 μg/ml) and soluble anti-CD28 antibody (37.51, 1 μg/ml), and then treated with mytomycin C. Cultures were established with equal numbers (5 × 104) CD4+ A1(M)RAG-1-/-, CD4+CD25+, and T cell-depleted, mytomycin C-treated, female CBA spleen. Peptide was added at 1-100 nM. After 72 h culture, 0.5 μCi 3H-thymidine was added to all cultures and terminated 18 h later.
Results
Identification of mGITRL and Its Gene. mGITRL was found by searching the Celera database by using an amino acid sequence of human GITR ligand. Two homologous peptide sequences were identified. These two sequences did not overlap, and as encoding regions mapped in the same orientation within 9.1 kb, this finding suggested that these two peptides were encoded within one gene (Fig. 7, which is published as supporting information on the PNAS web site). The putative gene consists of at least two exons and maps to a region between OX40 ligand and Fas ligand genes on chromosome 1. This position is equivalent to human GITR ligand gene (chromosome 1q23). RT-PCR and RACE were performed to confirm this finding and determine the full nucleotide sequence of the transcript from the putative gene. Amplified cDNA containing these two exon sequences were obtained from the macrophage cell line RAW 264 and bmDC. This cDNA encodes a 173-aa protein with a type 2 transmembrane topology similar to other TNF family members and shows 51% identity with that of human GITR ligand (Fig. 1 A).
To demonstrate that the identified gene product is indeed mGITRL, we examined the ability of the identified gene product to bind to mGITR. A mAb (YGL 386) generated against this gene product was able to bind the surface of transfected mammalian cells expressing the putative mGITRL but not to a control parent cell (Fig. 1B, YGL 386), in a manner similar to a recombinant mGITR Fc immunofusion protein (Fig. 1B, GITR Fc). Furthermore, the recombinant protein from the identified gene was shown to bind a mGITR transfectant but not a control parent cell (Fig. 1C). These results clearly indicate that the identified molecule is mGITRL.
This mGITRL was then shown to be capable of signaling through mGITR. mGITR transfectant and control cells were electroporated with a NF-κB/luciferase reporter plasmid. These cells were cultured with either growth-arrested transfectants expressing mGITRL or control parent cells. The luciferase activity (48-h incubation) in GITR-expressing cells cultured with mGITRL-expressing cells was 6-fold greater than the controls (Fig. 1D), indicating that NF-κB is activated via signaling through mGITR.
Ligand Engagement of mGITR Provides a Costimulatory Signal for T Cell Proliferation. Agonist anti-mGITR antibodies have been shown to neutralize the suppression of CD3-mediated proliferation of CD4+CD25- T cells by CD4+CD25+ regulatory T cells (4, 5). We asked whether the purified mGITRL could do the same. Inhibition of proliferation by CD4+CD25+ cells was completely neutralized with recombinant mGITRL but not with control, recombinant hCD40L (Fig. 2A, anti-CD3 Ab). We found that mGITRL could also neutralize the suppression of H-Y antigen (male-specific antigen) monospecific CD4+ T cells from naïve female TCR transgenic mice (Fig. 2 A, H-Y peptide).
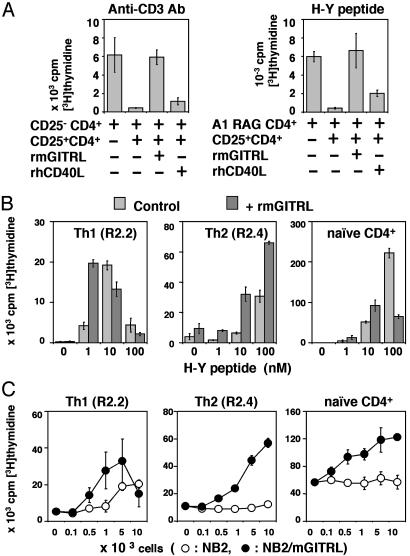
Fig. 2.
Enhancement and inhibition of proliferation of TCR-stimulated T cells with mGITRL. (A) Proliferation assays were performed by using CD4+CD25- cells stimulated with mitomycin C-treated T cell-depleted female spleen cells and anti-CD3 antibody (Anti-CD3 Ab) or H-Y peptide as antigen (H-Y peptide). CD4+CD25+ cells, recombinant mGITRL (rmGITRL), and/or recombinant hCD40L (rhCD40L) were added (marked +). For peptide assays, CD4+CD25+ were preactivated. Control cultures where CD4+CD25- from CBA/Ca mice were added failed to induce suppression (data not shown). (B) Proliferation assays were performed by using Th1 (R2.2), Th2 (R2.4) clones, and naïve CD4+ cells from A1(M)RAG-1-/- mice, with or without recombinant mGITRL. These T cells were stimulated with mitomycin C-treated female spleen cells and different amounts of H-Y peptide as antigen (0-100 nM). (C) Proliferation assays were performed by using Th1 (R2.2), Th2 (R2.4) clones, and naïve CD4+ cells from A1(M)RAG-1-/- mice with or without mitomycin C-treated mGITRL transfectants (NB2/mGITRL) or its parent cells (NB2) (0-104 cells). These T cells were stimulated with mitomycin C-treated female spleen cells and 10 nM of H-Y peptide as antigen.
As CD4+CD25- T cells (5) and Th cell clones (Fig. 8, which is published as supporting information on the PNAS web site) also express mGITR to varying extents, we investigated whether these clones and naïve CD4+ cells from H-Y antigen-specific TCR transgenic mice were able to respond to signals through mGITR. The H-Y antigen-specific populations were cultured with antigen-presenting cells, recombinant mGITRL, and varying amounts of antigen (H-Y peptide, 0-100 nM). In the case of the Th1 clone without mGITRL, maximal proliferation was observed with 10 nM H-Y peptide, with reduced responses at higher peptide doses (Fig. 2B). This finding is consistent with activation-induced apoptosis that we have previously found to be IFN-γ and NO dependent (unpublished work). Interestingly, mGITRL increased proliferation of Th1 cells with 1 nM peptide but tended to have the opposite effect at the higher 10- and 100-nM concentrations, suggesting GITR signaling had lowered the threshold for activation. The Th2 clone showed enhancement of proliferation with mGITRL and H-Y peptide across the range of peptide concentrations tested (Fig. 2B). mGITRL also enhanced proliferation of naïve CD4+ cells from the TCR transgenic mice (Fig. 2B) when challenged with 10 nM H-Y peptide, yet just as for the Th1 clone, reduced proliferation with the 100-nM peptide dose.
These results suggested that mGITR might function physiologically as a costimulatory molecule. To test this idea, we repeated the experiments with a fixed antigen concentration (10 nM) but using mGITRL-transfected cells, to ensure a more “natural” ligation of GITR (presumably as a trimer on the cell surface). Compared with nontransfected cells, the proliferation of both Th1 and Th2 clones was enhanced in a dose-dependent manner, although the Th1 clone showed evidence of inhibition at the highest dose of transfectants (Fig. 2C). Proliferation of naïve H-Y antigen-specific T cells from the TCR-transgenic mice was also enhanced in a dose-dependent manner (Fig. 2C). These results confirm our findings obtained with recombinant mGITRL (Fig. 2B). Taken together these results clearly suggest that the mGITRL engagement of mGITR acts as a costimulatory signal for T cells.
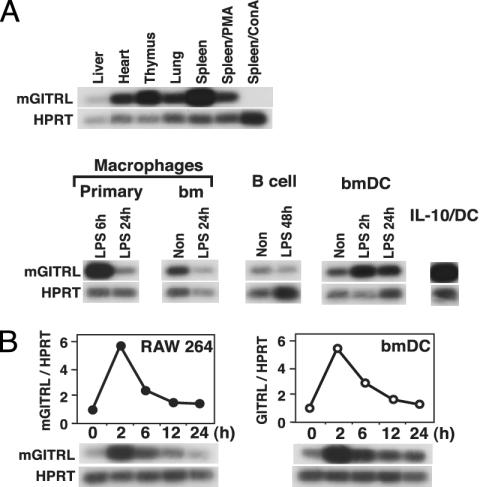
Distribution of mGITRL. Because mGITRL can neutralize the suppressive activity of CD4+CD25+ regulatory T cells and is costimulatory for T cells, it is important to know which cells likely to interact with T cells expressing the ligand. We therefore investigated mGITRL expression by RT-PCR (Fig. 3A). High levels of mGITRL mRNA were detected in spleen, and these levels were reduced after phorbol 12-myristate 13-acetate or Con A activation. Macrophages, B cells, and DC expressed mGITRL mRNA at high levels (Fig. 3A). We also analyzed mGITRL mRNA expression in resting and anti-CD3 antibody-activated T cells [CD4+, CD8+, Th1 and Th2 clones (6), regulatory T cell CD4+CD25+ cells, other regulatory T cell Tr1-like cells (13), and Tr1-like clones (14)], but mGITRL mRNA was not detected (data not shown). LPS stimulation resulted in reduced expression levels of mGITRL mRNA in macrophages, B cells, and DC. To investigate the nature of this regulation, we analyzed levels of mGITRL mRNA in LPS-stimulated (0-24 h) RAW 264 (macrophage cell line) and bmDC (Fig. 3B). mGITRL mRNA expression was transiently up-regulated, peaking at 2 h after stimulation and then declining.
Fig. 3.
Expression levels of mGITRL mRNA. Expression levels of mGITRL mRNA were analyzed by RT-PCR. cDNAs were amplified with mGITRL-specific or HPRT-specific primers. To compare expression levels and minimize PCR artifacts, the number of PCR cycles was kept low, and PCR products were detected by Southern blot hybridization using specific probes. (A) cDNAs were prepared by using RNA from indicated organs and cells with an oligo(dT) primer. Preparation of primary and bm macrophages, splenic B cell-enriched fraction, bmDC, and IL-10/DC was described in Materials and Methods. If required, cells were stimulated with LPS (10 μg/ml). RT-PCR using RNA from nonstimulated primary macrophage was not performed because it is extremely difficult to purify nonstimulated macrophages by the method used for purification of LPS-stimulated macrophages. (B) RT-PCR was performed by using RNA from nonstimulated (0 h) and LPS-stimulated (2-24 h) RAW 264 cells or bmDC. LPS stimulation times are indicated above the blot. RT-PCR results were also analyzed by using PhosphorImaging, allowing mRNA levels of mGITRL to be compared with those of HPRT (shown above the blot).
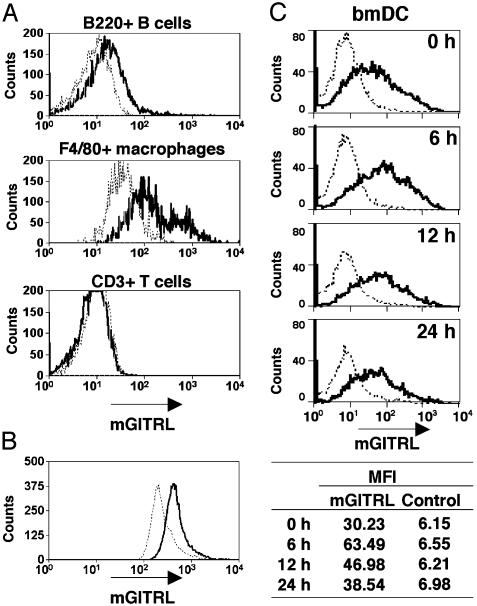
Cell surface expression of mGITRL was also analyzed by flow cytometry. In splenic populations, mGITRL expression was observed on CD3-B220+ B cells and F4/80+ macrophages (Fig. 4A). mGITRL was also detected on F4/80+ peritoneal macrophages (Fig. 4B). However, no mGITRL was detected on splenic T cells (Fig. 4A) or Th1 and Th2 clones (Fig. 8). Cell surface expression of mGITRL seems to correspond to the levels of mRNA, as demonstrated below for DC. mGITRL was detected on nonstimulated bmDC and increased on 6-h LPS-stimulated bmDC, and the proportion of highly expressing cells declined after 12 and 24 h of LPS stimulation (Fig. 4C). Because mGITRL protein is more stable than its mRNA, changes of protein expression levels of mGITRL were less pronounced than that of mGITRL mRNA, but similar outcomes were observed on both mGITRL mRNA and protein expression (Figs. 3B and 4C).
Fig. 4.
Cell surface expression of mGITRL. (A) Spleen cells were stained with anti-mGITRL antibody YGL386 (solid line) or an isotype control antibody (dotted line). These cells were costained with anti-CD3, anti-B220, or anti-F4/80 antibody and then positive cells were gated. Median fluorescence intensity (MFI) is as follows: B220+ B cell (control, 7.7; mGITRL, 14.9), F4/80+ macrophages (control, 25.4; mGITRL low, 56.2; mGITRL high, 673.2), and CD3+ T cell (control, 11.2; mGITRL, 9.65). (B) Peritoneal cells were also stained with anti-mGITRL antibody YGL386 (solid line) or an isotype control antibody (dotted line). Cells were costained with anti-F4/80 antibody and then positive cells were gated. MFI are: control, 254.8 and mGITRL, 421.7. (C) Nonstimulated (0 h) and LPS-stimulated (6, 12, and 24 h) bmDCs were stained with anti-mGITRL antibody YGL 386 (solid line) or an isotype control antibody (dotted line). bmDC were costained with an anti-CD11c antibody (DC maker), and positive cells were gated. MFI values are indicated under the histograms.
Transcription of mGITRL Is Regulated by the Transcription Factor NF-1. mGITRL expression seems to depend heavily on its transcriptional regulation. We therefore investigated mGITRL promoter activity. To determine the location of the promoter and gene structure, 5′ and 3′ RACE were performed. A total of 215 5′ RACE clones were analyzed (Fig. 9, which is published as supporting information on the PNAS web site), and the major transcription start site was defined as position +1 (Fig. 5A). Seventy-five percent of 5′ ends were mapped between -3 and +3. Ten of 13 3′ RACE clones contained 1.46-kb sequences found immediately downstream of the stop codon. In two clones, however, 0.65 kb of the 3′ UTR (0.68 kb) is located 1.9 kb further downstream, indicating that this mRNA is generated by alternative splicing. The gene structure is shown in Fig. 5A and Fig. 10, which is published as supporting information on the PNAS web site.
A typical TATA box sequence was found in a region ≈30 bp upstream of a major cluster of transcription start sites (Fig. 5A), suggesting that mGITRL gene expression is regulated by a TATA type promoter. A luciferase reporter assay was performed by using deletion mutants of this promoter (Fig. 5 A and B) in RAW 264 cells. Significant reduction of promoter activity was observed by deletion of a 27-bp sequence from -120 (D3) to -94 (D2) in both nonstimulated and LPS-stimulated cells. Transcription factor binding to this region was investigated by an EMSA (Fig. 6) using three probes (P1-P3) (Fig. 5A). A very strong complex formation was detected with 32P-labeled probe P2 (Fig. 6A) that was inhibited with a 100-fold excess of unlabeled P2, and not with P1 and P3 competitors, or mutant oligos M1 and M2 (Fig. 6B), suggesting that P2 contains a critical sequence similar to NF-1 consensus (TTGGCNNNNNGCCAA). A super shift assay confirmed that transcription factor NF-1 binds to this promoter (Fig. 6C), and mutation of the NF-1 consensus (TGCCA to GAATT) resulted in a large reduction of promoter activity (Fig. 5C).
Fig. 6.
Binding of transcription factor NF-1 to the mGITRL promoter. (A) The presence of cis-acting elements between -120 and -94 is suggested by luciferase assays (Fig. 5B). Oligo probes P1 to P3 for EMSA were designed in this identified region and its flanking regions. The locations of these probes are indicated in Fig. 5A. EMSA was performed by using the probes (P1-P3) and nuclear extract from RAW 264 cells. (B) A competition assay was performed by using a 100-fold excess of unlabeled competitor with 32P-labeled P2 probe. The competitors used are indicated above the gel. Probe P2 sequence and mutated sequences in M1 and M2 are shown under the gel. Sequence similar to NF-1 consensus in P2 is indicated with underlines. (C) Super shift assay was performed by using probe P2 and an anti-NF-1 antibody (marked +). (D) EMSA was performed by using probe P2 and nuclear extracts from nonstimulated (0 h) and LPS-stimulated (2-24 h) RAW 264 cells. NF-1 and probe complexes are indicated. (E) EMSA was performed by using probe P2 and nuclear extracts from nonstimulated (0 h) and LPS-stimulated (2-24 h) bmDC (EMSA). NF-1 in nuclear extracts used for EMSA was detected by immunoblotting using anti-NF-1 antibody (IB).
EMSA was also performed by using nuclear extracts from RAW 264 cells and bmDC that were stimulated with LPS for different amounts of time (Fig. 6 D and E, EMSA). The NF-1 complex formation with probe P2 increased in both cell types stimulated with LPS for 2 h, and then decreased in cells with longer stimulation times. Immunoblotting revealed a similar result to the EMSA (Fig. 6E, IB), indicating that the amount of NF-1 in nuclei is regulated by LPS stimulation. Taken together, these results suggest that NF-1 is a key transcription factor controlling mGITRL gene expression.
Discussion
We have identified mGITRL and its gene. mGITRL seems to be costimulatory for both naïve and primed T cells. By analogy with other members of the TNFR family, costimulatory activity of GITR on CD4+CD25- cells might be maintained through an NF-κB activation pathway. However, it is not clear how signaling through GITR on CD4+CD25+ can neutralize the suppressive activity of these cells. We ruled out ligand-induced cell death, by analyzing mixed cultures of CD3-activated CD4+CD25- (Thy1.1) and CD4+CD25+ [hCD52 (15), Thy1.2], with and without recombinant mGITRL and monitoring death by 7-aminoactinomycin D uptake. We found no evidence for any increased cell death of CD4+CD25+ (hCD52+, Thy1.2) cells with mGITRL (data not shown). Blockade of the suppressive activity of CD4+CD25+ seems, therefore, to be mediated by signaling either via an NF-κB activation pathway or an unidentified signaling mechanism through the GITR endodomain.
Although NF-1 is known to be a constitutively expressed transcription factor, the amount in nuclei could be regulated, both positively and negatively, by LPS stimulation. Nuclear translocation of this NF-1 may be controlled by posttranslational regulation such as phosphorylation and/or dephosphorylation as observed with NF-AT. At least four NF-1 isoforms (NF-1a, NF-1b, NF-1c, and NF-1x) with different activities are known (16). However, we are unable to determine with currently available reagents which NF-1 isoforms are the key transcription factors for mGITRL expression.
mGITRL expression on the cell surface seems to broadly reflect levels of mGITRL mRNA. However, mGITRL expression may also be controlled by posttranslational regulation. The expression level of mGITRL mRNA in bmDC was similar at 0 and 24 h post-LPS stimulation (Fig. 3), but by flow cytometry, the proportion of highly expressing cells was reduced (Fig. 4). Surface mGITRL may be regulated by a transport protein, as observed with Fas ligand (17), or shed by a metalloproteinase like TNF-α-converting enzyme (18, 19).
The major mGITRL mRNA contains potential RNA destabilization signal AUUUA + AU-rich sequences in the 3′ noncoding region (near 3′ end) (Fig. 10) (20, 21). An isoform mRNA, lacking the putative destabilization signal by alternative splicing, was detected in IL-10-treated bmDC that express particularly high levels of mGITRL mRNA (Fig. 3A). Levels of mGITRL mRNA might be controlled by posttranscriptional regulation.
Based on our findings, we suggest the following sequence of events for T cell stimulation: (i) In resting antigen-presenting cells (APCs), constitutive expression of mGITRL mRNA is determined by NF-1, with mGITRL expressed on the cell surface. (ii) Activated APCs initially up-regulate mGITRL to act as a costimulator in T cell interactions. In addition, mGITRL would tend to reverse any suppression by CD4+CD25+ T cells in the local microenvironment. (iii) At later stages of APC activation, mGITRL mRNA and protein would be down-modulated. This would limit any further costimulatory activity, but would release CD4+CD25+ regulatory T cells so that they might curtail the ongoing immune response.
In conclusion, our findings clearly suggest that GITR and mGITRL are key players in regulating T cell-mediated immunity. As such they may offer potential therapeutic targets for modulation of immune responses.
Supplementary Material
Acknowledgments
We thank S. Cartland, P. Taylor, L. Poulton, L. Martinez-Pomares, and M. Hieda for technical advice. This work was supported by grants from the Medical Research Council, the E. P. Abraham Research Foundation, and Herman Waldmann.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GITR, glucocorticoid-induced tumor necrosis factor receptor; mGITR, mouse GITR; mGITRL, mouse GITR ligand; hGITR, human GITR; hCD40L, human CD40 ligand; EMSA, electrophoretic mobility-shift assay; HPRT, hypoxanthine phosphoribosyltransferase; LPS, lipopolysaccharide; TCR, T cell receptor; bm, bone marrow-derived; DC, dendritic cells; Th1, T helper 1; Th2, T helper 2.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ577579, AJ577580, and AJ577581).
References
- 1.Nocentini, G., Giunchi, L., Ronchetti, S., Krausz, L. T., Bartoli, A., Moraca, R., Migliorati, G. & Riccardi, C. (1997) Proc. Natl. Acad. Sci. USA 94, 6216-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurney, A. L., Marsters, S. A., Huang, R. M., Pitti, R. M., Mark, D. T., Baldwin, D. T., Gray, A. M., Dowd, A. D., Brush, A. D., Heldens, A. D., et al. (1999) Curr. Biol. 9, 215-218. [DOI] [PubMed] [Google Scholar]
- 3.Kwon, B., Yu, K.-Y., Ni, J., Yu, G.-L., Jang, I.-K., Kim, Y.-J., Xing, L., Liu, D., Wang, S.-X. & Kwon, B. S. (1999) J. Biol. Chem. 274, 6056-6061. [DOI] [PubMed] [Google Scholar]
- 4.McHugh, R. S., Whitters, M. J., Piccirillo, C. A., Young, D. A., Shevach, E. M., Collins, M. & Byrne, M. C. (2002) Immunity 16, 311-323. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu, J., Yamazaki, S., Takahashi, T., Ishida, Y. & Sakaguchi, S. (2002) Nat. Immunol. 3, 135-142. [DOI] [PubMed] [Google Scholar]
- 6.Zelenika, D., Adams, E., Mellor, A., Simpson, E., Chandler, P., Stockinger, B., Waldmann, H. & Cobbold, S. P. (1998) J. Immunol. 161, 1868-1874. [PubMed] [Google Scholar]
- 7.Frohman, M. A., Dush, M. K. & Martin, G. R. (1988) Proc. Natl. Acad. Sci. USA 85, 8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tone, M., Tone, Y., Babik, J. M., Lin, C.-Y. & Waldmann, H. (2002) J. Biol. Chem. 277, 8890-8897. [DOI] [PubMed] [Google Scholar]
- 9.Tone, M., Tone, Y., Fairchild, P. J., Wykes, M. & Waldmann, H. (2001) Proc. Natl. Acad. Sci. USA 98, 1751-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelissen, I. C., Brown, A. J., Mander, E. L., Kritharides, L., Dean, R. T. & Jessup, W. (1996) J. Biol. Chem. 271, 17852-17860. [DOI] [PubMed] [Google Scholar]
- 11.Tone, M., Powell, M. J., Tone, Y., Thompson, S. A. J. & Waldmann, H. (2000) J. Immunol. 165, 286-291. [DOI] [PubMed] [Google Scholar]
- 12.Graca, L., Thompson, S., Lin, C.-Y., Adams, E., Cobbold, S. P. & Waldmann, H. (2002) J. Immunol. 168, 5558-5565. [DOI] [PubMed] [Google Scholar]
- 13.Barrat, F. J., Cua, D. J., Boonstra, A., Richards, D. F., Crain, C., Savelkoul, H. F., de Waal-Maleyfyt, R., Coffman, R. L., Hawrylowicz, C. M. & O'Garra, A. (2002) J. Exp. Med. 195, 603-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelenika, D., Adams, E., Humm, S., Graca, L., Thompson, S., Cobbold, S. P. & Waldmann, H. (2002) J. Immunol. 168, 1069-1079. [DOI] [PubMed] [Google Scholar]
- 15.Gilliland, L. K., Walsh, L. A., Frewin, M. R., Wise, M. P., Tone, M., Hale, G., Kioussis, D. & Waldmann, H. (1999) J. Immunol. 162, 3663-3671. [PubMed] [Google Scholar]
- 16.Chaudhry, A. Z., Vitullo, A. D. & Gronostajski, R. M. (1998) J. Biol. Chem. 273, 18538-18546. [DOI] [PubMed] [Google Scholar]
- 17.Bossi, G. & Griffiths, G. M. (1999) Nat. Med. 5, 90-96. [DOI] [PubMed] [Google Scholar]
- 18.Black, R. A., Rauch, C. T., Kozlosky, C. J., Peschon, J. J., Slack, J. L., Wolfson, M. F., Castner, B. J., Stocking, K. L., Reddy, P., Srinivasan, S., et al. (1997) Nature 385, 729-733. [DOI] [PubMed] [Google Scholar]
- 19.Moss, M. L., Jin, S.-L. C., Milla, M. E., Burkhart, W., Carter, H. L., Chen, W.-J., Clay, W. C., Didsbury, J. R., Hassler, D., Hoffman, C. R., et al. (1997) Nature 385, 733-736. [DOI] [PubMed] [Google Scholar]
- 20.Chen, C.-Y. A. & Shyu, A.-B. (1995) Trends Biochem. Sci. 20, 465-470. [DOI] [PubMed] [Google Scholar]
- 21.Powell, M. J., Thompson, S. A. J., Tone, Y., Waldmann, H. & Tone, M. (2000) J. Immunol. 165, 292-296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.