Abstract
Proteasomal degradation of p53 is mediated by two alternative pathways that are either dependent or independent of both Mdm2 and ubiquitin. The ubiquitin-independent pathway is regulated by NAD(P)H: quinone oxidoreductase 1 (NQO1) that stabilizes p53. The NQO1 inhibitor dicoumarol induces ubiquitin-independent p53 degradation. We now show that, like dicoumarol, several other coumarin and flavone inhibitors of NQO1 activity, which compete with NAD(P)H for binding to NQO1, induced ubiquitin-independent p53 degradation and inhibited wild-type p53-mediated apoptosis. Although wild-type p53 and several p53 mutants were sensitive to dicoumarol-induced degradation, the most frequent “hot-spot” p53 mutants in human cancer, R175H, R248H, and R273H, were resistant to dicoumarol-induced degradation, but remained sensitive to Mdm2-ubiquitin-mediated degradation. The two alternative pathways for p53 degradation thus have different p53 structural requirements. Further mutational analysis showed that arginines at positions 175 and 248 were essential for dicoumarol-induced p53 degradation. NQO1 bound to wild-type p53 and dicoumarol, which induced a conformational change in NQO1, inhibited this binding. Compared with wild-type p53, the hot-spot p53 mutants showed increased binding to NQO1, which can explain their resistance to dicoumarol-induced degradation. NQO1 thus has an important role in stabilizing hot-spot p53 mutant proteins in human cancer.
Wild-type p53 is a labile protein and its cellular level is mainly regulated by the rate of its proteasomal degradation (reviewed in ref. 1). p53 degradation is mediated by two alternative pathways that either depend on Mdm2 and ubiquitin (2, 3) or are independent of both (4). The Mdm2- and ubiquitin-independent pathway is regulated by NAD(P)H: quinone oxidoreductase 1 (NQO1) (4-7). Our previous studies showed that NQO1 stabilizes p53 (5, 6) and that reducing the NQO1 level by small interfering RNA decreases the level of p53 (4). Dicoumarol is a competitive inhibitor of NQO1 activity that competes with NAD(P)H for binding to NQO1 (8) and induces ubiquitin-independent p53 degradation (4-7). We suggested that p53 stabilization requires the physical interaction of p53 with NQO1 (6), and recent studies have shown that NQO1 can bind to p53 (9). The tumor suppressor wild-type p53 is mutated in >50% of human cancers (1, 10, 11). We have now studied the control of ubiquitin-independent p53 degradation by using different NQO1 inhibitors, different p53 mutants, and binding of NQO1 to p53. Our results include the finding that the most frequent “hot-spot” p53 mutants in human cancer showed increased binding to NQO1 and resisted dicoumarol-induced degradation. These findings indicate that NQO1 plays a major role in stabilizing hot-spot p53 mutant proteins in human cancer cells.
Materials and Methods
Cells and Cell Culture. The cell lines used were: p53 null HCT116 human colon carcinoma cells (12), 293 human kidney cells, Huh 7 human hepatocellular carcinoma cells that carry the Y220P mutant 53 (13), mouse M1 myeloid leukemic cells stably transfected with the mouse p53 mutants C132F and A135V (14), and A31N-ts20 cells that have a temperature-sensitive E1 ubiquitin-activating enzyme, which is inactivated at 39°C (15). The p53 A135V is a temperature-sensitive protein that behaves like a tumor-suppressing wild-type p53 at 32°C and like a mutant p53 at 37°C (16). When cultured at 32°C, the M1 cells carrying A135V p53 (M1-t-p53 cells) undergo apoptosis (ref. 14; reviewed in ref. 17). HCT116, Huh 7, and 293 cells were grown in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin, and cultured at 37°C in a humidified incubator with 5.6% CO2. A31N-ts20 cells were cultured in DMEM and 10% FBS at 32°C. M1-t-p53 cells were grown in DMEM supplemented with 10% heat-inactivated (56°C, 30 min) horse serum and cultured at 37°C in an incubator with 10% CO2.
Compounds. Dicoumarol, phenindione, 7,8-dihydroxyflavone (DHF), warfarin, 4-hydroxycoumarin (Sigma), chrysin and esculetin (6,7-dihydroxycoumarin) (Aldrich) were dissolved in 0.13 N NaOH. Cibacron blue 3GA (Sigma) was dissolved in water.
Apoptosis and Cell Viability Assays. Apoptosis in M1-t-p53 cells was induced by culture for 23 h at 32°C, and the percent cell viability was determined as described (6).
Plasmids. The plasmids used were pRc/CMV human wild-type p53; pRc/CMV human p53 mutants R175H, R175K, R175D, R248H, R248L, R248P, and R273H; and pRc/CMV FLAG human p53 mutants R175H and R273H (obtained from M. Oren, Weizmann Institute of Science); pCOC-mouse mdm2 × 2 (18); pEFIRES human NQO1; pEFIRES human NQO1 FLAG; and pEFIRES FLAG p73b.
Transfection. Transfections were carried out by the calcium phosphate method. In the HCT 116 cells, transfection was followed by a 10% glycerol shock for 30 sec, 7 h after transfection. Whenever needed, an empty vector was used to maintain a constant amount of total DNA in each transfection mixture.
Immunoblot Analysis. Cell extracts and immunoblot analysis were carried out as described (5). The antibodies used were the following: monoclonal anti-p53 (Pab 248, Pab 421, and Pab 1801) (obtained from M. Oren); monoclonal anti-p53 (Pab 240), goat anti-NQO1, and rabbit anti-IκB (Santa Cruz Biotechnology); monoclonal anti-actin and monoclonal anti-FLAG (Sigma); and hamster anti-mouse Bcl-2 (Pharmingen).
Coimmunoprecipitation of p53 and NQO1. In vitro coimmunoprecipitation experiments were carried out with in vitro reticulocyte lysate-translated, [35S]methionine-labeled wild-type p53 and NQO1 incubated in Nonidet P-40 buffer (100 mM Tris·HCl, pH 7.5/150 mM NaCl/2 mM EDTA/1% Nonidet P-40) in the absence or presence of 300 μM dicoumarol. Mouse monoclonal anti-p53 antibody (Pab 1801) was added, and mixtures were incubated at 4°C for 12 h. Samples were then incubated with Trisacril beads (LKB) conjugated to anti-mouse IgG antibody (obtained from T. Rimon, Weizmann Institute of Science, Rehovot, Israel) for an additional 2 h. Coimmunoprecipitation was also carried out with extracts from cells transiently transfected with pEFIRES FLAG p73β, pRc/CMV FLAG human wild-type p53, FLAG human p53 R175H, or FLAG human p53 R273H with or without pEFIRES NQO1. The cells were lysed in radioimmunoprecipitation assay lysis buffer [150 mM NaCl/1% Nonidet P-40/0.5% AB-deoxycholate/0.1% SDS/50 mM Tris·HCl, pH 8/1 mM DTT and 1 μM each of leupeptin, aprotinin, and pepstatin (Sigma mixture)], and samples were incubated for 4 h with anti-FLAG agarose beads (Sigma). In both coimmunoprecipitation methods the beads were collected by centrifugation and washed four times with Nonidet P-40 buffer. The beads were mixed with Laemmli sample buffer and heated at 95°C for 5 min, and samples were loaded on a SDS-12.5% polyacrylamide gel. Coimmunoprecipitation was detected either by autoradiography in the in vitro experiments or by immunoblotting with anti-NQO1 antibody.
Results
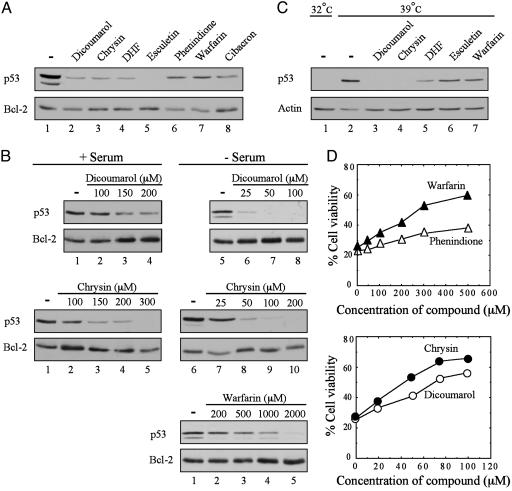
Degradation of p53 and Inhibition of p53-Mediated Apoptosis by Inhibitors of NQO1 Activity. Dicoumarol, an inhibitor of NQO1 activity that competes with NAD(P)H for binding to NQO1 (8), induces p53 degradation and inhibits p53-mediated apoptosis (4-7). Certain coumarins (19), flavones (20, 21), and the reactive dye cibacron blue (21, 22) are also competitive inhibitors of NQO1 activity that compete with NAD(P)H for binding to NQO1. Coumarins, flavones, and cibacron blue are less potent than dicoumarol and appear to bind differently to NQO1 (20). We have tested several flavones, coumarins, and cibacron blue for their ability to induce p53 degradation and inhibit p53-mediated apoptosis. Like dicoumarol, the flavones chrysin and DHF at 300 μM induced efficient p53 degradation in M1-t-p53 cells (Fig. 1A). Phenindione, a less potent NQO1-inhibiting flavone (20), the coumarins warfarin and esculetin (6,7-dihydroxycoumarin), and the dye cibacron blue also induced p53 degradation but required higher concentrations (1-2 mM) (Fig. 1 A). 4-Hydroxycoumarin did not cause p53 degradation even at 2 mM. Coumarins are known to bind to serum albumin, which reduces their effective concentration (19). We, therefore, compared the ability of dicoumarol, warfarin, and chrysin to induce p53 degradation in cells cultured in medium with or without serum. In a 5-h degradation assay carried out with M1-t-p53 cells, the concentrations of dicoumarol, chrysin, and warfarin that induced detectable p53 degradation in serum-free medium were ≈5-fold lower than in medium containing serum (Fig. 1B).
Fig. 1.
Degradation of p53 and inhibition of p53-dependent apoptosis with different NQO1 inhibitors. (A) M1-t-p53 cells were cultured in DMEM plus serum for 5 h without (-) or with 300 μM dicoumarol, chrysin, or DHF; 2 mM phenindione, esculetin, or warfarin; or 1 mM cibacron blue. (B) M1-t-p53 cells were cultured for 5 h in DMEM with (+) or without (-) serum and without (-) or with different concentrations of dicoumarol, warfarin, or chrysin. (C) A31N-ts20 cells were preincubated at 32°C (lane 1) or 39°C for 24 h (lanes 2-7). Cells (lanes 2-7) were then incubated at 39°C for additional 5 h without (-) or with 200 μM dicoumarol or chrysin; 500 μM DHF; and 2 mM esculetin or warfarin. Immunoblot analysis was carried out by using mouse monoclonal anti-p53 antibody (Pab 240 in A and B or Pab 248 plus Pab 421 in C), hamster anti-Bcl-2, or mouse monoclonal anti-actin. (D) M1-t-p53 cells were cultured at 32°C without or with different concentrations of chrysin, dicoumarol, phenindione, or warfarin, and cell viability was determined after 23 h.
Using A31N-ts20 cells that contain a temperature-sensitive E1 ubiquitin-activating enzyme that is inactivated at 39°C (15), we have shown that dicoumarol induces ubiquitin-independent degradation of p53 (4). We have now tested the effect of chrysin, DHF, esculetin, and warfarin on degradation of wild-type p53 that accumulates in A31N-ts20 cells cultured at 39°C for 24 h. Culture of A31N-ts20 cells at 39°C for an additional 5 h with dicoumarol or these other compounds induced p53 degradation (Fig. 1C). Because ubiquitination is defective in A31N-ts20 cells at 39°C (15), these results indicate that like dicoumarol, all these other tested compounds induce ubiquitin-independent p53 degradation.
Apoptosis assays using M1-t-p53 cells cultured for 23 h at 32°C, when p53 behaves like wild-type, showed that chrysin was as effective as dicoumarol in inhibiting p53-mediated apoptosis (Fig. 1D). Warfarin and phenindione also inhibited p53-mediated apoptosis but were less effective than dicoumarol or chrysin (Fig. 1D), as expected from their lower ability to induce p53 degradation. Although phenindione and warfarin showed a similar ability to induce p53 degradation (Fig. 1 A), the antiapoptotic effect of phenindione was weaker than warfarin (Fig. 1D), which can be explained by the higher toxicity of phenindione to M1-t-p53 cells. The data indicate that, like dicoumarol, several other competitive NQO1 inhibitors for binding of NAD(P)H to NQO1 induced p53 degradation and inhibited p53-mediated apoptosis.
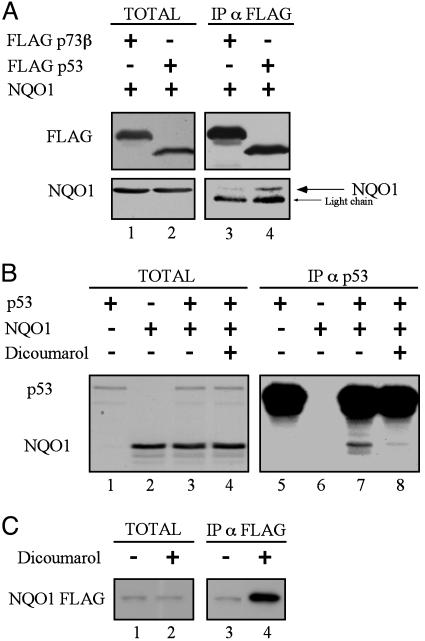
Binding of Wild-Type p53 to NQO1 Is Inhibited by Dicoumarol. NQO1 stabilizes p53 (6) and coimmunoprecipitation indicated that NQO1 binds to p53 in cells (Fig. 2A), as reported (9). The specificity of the interaction was shown by the inability of NQO1 to physically associate with p73β, which is structurally similar to p53 (Fig. 2 A). The NQO1 inhibitor dicoumarol induces p53 degradation (4-7). We therefore determined whether dicoumarol affects NQO1 binding to p53. Coimmunoprecipitation of in vitro translated NQO1 and p53 showed that NQO1 binds to wild-type p53 and that this binding was inhibited by dicoumarol (Fig. 2B). Incubation of in vitro translated NQO1 FLAG with dicoumarol followed by immunoprecipitation with anti-FLAG antibody showed an increased amount of immunoprecipitated NQO1 in the presence of dicoumarol (Fig. 2C). This result indicates that dicoumarol induced a conformational change in NQO1, increasing its recognition by the antibody. Dicoumarol thus induces p53 degradation by inducing a conformational change in NQO1, which inhibits its binding to p53.
Fig. 2.
Binding of wild-type p53 to NQO1 is inhibited by dicoumarol. (A) 293 human kidney cells were transiently transfected with pEFIRES FLAG p73β or pRc/CMV FLAG p53 with pEFIRES wild-type NQO1. Extracts were electrophoresed either before immunoprecipitation (TOTAL) or after immunoprecipitation of FLAG p73β or FLAG p53 with anti-FLAG agarose beads (IP α FLAG). Immunoblot analysis was carried out with mouse monoclonal anti-FLAG antibody, and the blots were then stripped and reprobed with goat anti-NQO1 antibody. (B) In vitro reticulocyte lysate translated [35S]methionine-labeled wild-type p53 and NQO1 were incubated alone or mixed together without (-) or with (+) 300 μM dicoumarol (TOTAL). p53 was immunoprecipitated (IP α p53) with mouse monoclonal Pab 1801 anti-human p53 and NQO1 and p53 were detected by autoradiography. (C) In vitro reticulocyte lysate-translated, [35S]methionine-labeled NQO1 FLAG was incubated for 1 h in Nonidet P-40 buffer without (-) or with (+) 300 μM dicoumarol (TOTAL). NQO1 FLAG was immunoprecipitated with anti-FLAG agarose beads (IP α FLAG), and NQO1 was detected by autoradiography.
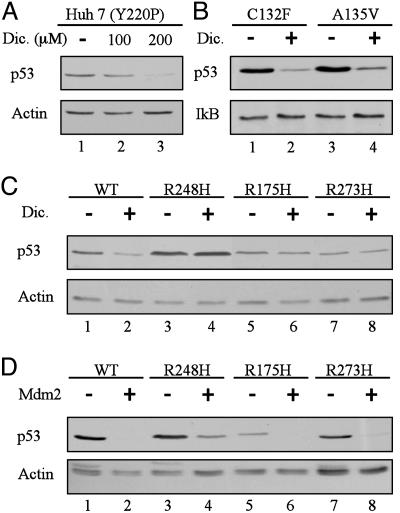
Hot-Spot p53 Mutants Are Resistant to Dicoumarol-Induced Degradation but Sensitive to Mdm2-Mediated Degradation. The NQO1 inhibitor dicoumarol induces proteasomal degradation of human and mouse wild-type p53 protein by a Mdm2- and ubiquitin-independent pathway (4-7). Dicoumarol also induces degradation of the mouse A135V mutant p53 (4-6) and the Mdm2-resistant human p53 mutant L22Q, W23S (p53[22,23]) (4). Two other p53 mutants, human Y220P (Fig. 3A) and mouse C132F (Fig. 3B), were also efficiently degraded by dicoumarol. The most frequent p53 mutations in human cancer are in codons 175, 248, and 273 (10, 11). Mutations in the p53 gene frequently result in accumulation of mutant p53 proteins in cancer cells (23). To determine whether these hot-spot p53 mutant proteins are sensitive to dicoumarol-induced degradation, HCT116 p53-null cells were transiently transfected with human p53 in which the arginines at codons 175, 248, or 273 were substituted by histidines. Unlike wild-type p53 (Fig. 3C, lanes 1 and 2), the R175H, R248H, and R273H p53 mutants were resistant to dicoumarol-induced degradation (Fig. 3C, lanes 3-8). However, all these p53 mutants were susceptible to Mdm2-ubiquitin-mediated degradation (Fig. 3D).
Fig. 3.
p53 mutants R248H, R175H, and R273H are resistant to dicoumarol-induced degradation but sensitive to Mdm2-mediated degradation. (A) Huh 7 cells that carry the Y220P mutant p53 were cultured for 5 h without (-) or with 100 or 200 μM dicoumarol (Dic.). (B) M1 myeloid leukemic cells that carry the C132F or A135V mouse mutant p53 cells were incubated without (-) or with (+) 300 μM dicoumarol for 5 h. (C) HCT116 p53 null cells were transiently transfected with pRc/CMV human wild-type p53 or the p53 mutants R248H, R175H, or R273H. Twenty-four hours after transfection, cells were cultured for 5 h without (-) or with (+) 300 μM dicoumarol. (D) HCT116 p53 null cells were transiently transfected with pRc/CMV human wild-type p53 or the p53 mutants R248H, R175H, or R273H without (-) or with (+) pCOC-mouse mdm2 × 2. Protein extraction and immunoblot analysis were carried out as described (5) by using mouse monoclonal anti-human p53 (Pab 1801) or anti-mouse and human p53 (Pab 240) antibody. The blots were then stripped and reprobed with mouse monoclonal anti-actin or rabbit anti-IκB antibody.
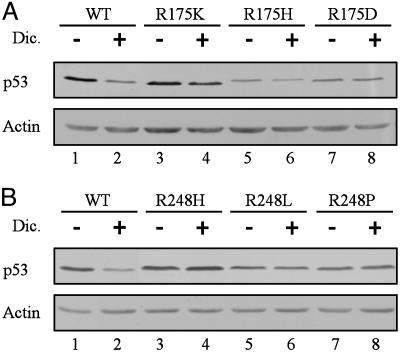
To determine whether the resistance of R175H, R248H, and R273H p53 mutants to dicoumarol-induced degradation is specific for the arginine-to-histidine substitution, we tested several other amino acid substitutions (10). The p53 mutants in which arginine 175 was substituted by lysine (R175K) or aspartic acid (R175D) and arginine 248 substituted by leucine (R248L) or proline (R248P) were all resistant to dicoumarol-induced degradation (Fig. 4). Thus, the arginines at positions 175 and 248 of human p53 are essential for susceptibility to dicoumarol-induced degradation.
Fig. 4.
Arginines at positions 175 or 248 are essential for dicoumarol-induced degradation of p53. HCT116 p53 null cells were transiently transfected with pRc/CMV human wild-type p53, or the p53 mutants R175K, R175H, or R175D (A) or with pRc/CMV human p53 mutants R248H, R248L, or R248P (B). Twenty-four hours after transfection, cells were cultured for 5 h without (-) or with (+) 300 μM dicoumarol. Immunoblot analysis was carried out by using mouse monoclonal anti-human p53 (Pab 1801) and mouse monoclonal anti-actin antibody.
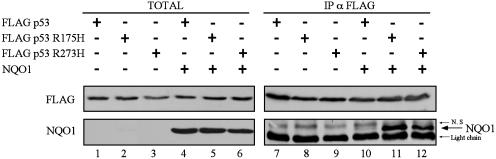
Increased Binding of NQO1 to Hot-Spot p53 Mutants. The resistance of the hot-spot p53 mutants to dicoumarol-induced degradation raised the question whether this property may be due to their altered binding to NQO1. Cells were transfected with wild-type or mutant p53 without or with NQO1. A similar level of expression occurred with the different transfected plasmids (Fig. 5, lanes 1-6). Coimmunoprecipitation from cell extracts of NQO1 with wild-type p53, p53 R175H, or p53 R273H showed increased binding of NQO1 to these hot-spot p53 mutants compared with wild-type p53 (Fig. 5, lanes 10-12). This increased binding of NQO1 to the hot-spot p53 mutants R175H and R273H can explain their resistance to degradation by dicoumarol.
Fig. 5.
p53 mutants R175H and R273H bind wild-type NQO1 in vivo with higher affinity than wild-type p53. 293 human kidney cells were transiently transfected with pRc/CMV FLAG wild-type p53 or with the p53 mutants R175H or R273H alone or cotransfected with pEFIRES NQO1. Extracts were electrophoresed either before immunoprecipitation (TOTAL) or after immunoprecipitation of FLAG p53 with anti-FLAG agarose beads (IP α FLAG). Immunoblot analysis was carried out with mouse monoclonal anti-FLAG antibody, and the blots were then stripped and reprobed with goat anti-NQO1 antibody. N.S, nonspecific band.
Discussion
Two alternative pathways regulate p53 degradation: one is ubiquitin-dependent and mediated by Mdm2 (2, 3) and the other is Mdm2 and ubiquitin-independent and regulated by NQO1 (4, 7). We have shown that the NQO1 inhibitor dicoumarol induces degradation of wild-type p53, the A135V p53 mutant, and the Mdm2-resistant p53[22,23] mutant (4-7). We have now found that two other p53 mutants, Y220P and C132F, are also sensitive to dicoumarol-induced degradation. Not all p53 mutants are dicoumarol-sensitive, and the three most frequent p53 mutants in human cancer, R175H, R248H, and R273H, that are susceptible to Mdm2-ubiquitin-mediated degradation, were resistant to dicoumarol-induced degradation. These findings indicate that, whereas wild-type p53 is sensitive to both the ubiquitin-dependent and -independent degradation pathways, different p53 mutants are differentially sensitive to these degradation pathways. These two degradation pathways thus have different p53 structural requirements that allow p53 degradation.
In addition to the dicoumarol resistance conferred to human p53 by an arginine-to-histidine substitution at codons 175, 248, or 273, substitution of arginines 175 or 248 by other charged or neutral amino acids, including lysine, aspartic acid, leucine, or proline, also resulted in dicoumarol resistance. These results suggest that each of the human p53 arginine residues 175, 248, and possibly also 273 is essential for dicoumarol-induced degradation. These arginines may regulate binding of p53 to NQO1. It is also possible that these arginines are involved in certain posttranslational modifications and/or in binding to other proteins that control p53 degradation. It will be interesting to determine whether other arginines in the p53 protein are also important for susceptibility to dicoumarol-induced degradation.
Many p53 mutant proteins have a longer half-life than wild-type p53 and thus accumulate in cancer cells (23). It was suggested that p53 mutants are more stable because, unlike wild-type p53, they do not induce expression of Mdm2 and, therefore, are not targeted by Mdm2 for degradation (24). However, studies with various hot-spot p53 mutant proteins have shown that they promote stabilization and accumulation of Mdm2 protein (25). In addition, we have now shown that the hot-spot p53 mutants R175H, R248H, and R273H retain susceptibility to Mdm2-mediated degradation. p53 deletion mutants lacking each of the p53 regions with these arginine residues are also susceptible to Mdm2-mediated degradation (26). Accumulation of mutant p53 in cancer cells thus cannot be explained only by lack of Mdm2 expression or resistance to Mdm2-mediated degradation. Mutant p53 may be stabilized in cancer cells by binding to other proteins, such as hsp90, which inhibits Mdm2-mediated degradation (27). Our findings that NQO1 stabilizes p53 and binds better to hot-spot p53 mutants than to wild-type p53 indicate that NQO1 plays an important role in stabilizing these p53 mutant proteins in cancer cells. The elevated levels of NQO1 in many cancer cells, compared with normal cells (28, 29), would further promote stabilization of these p53 mutants in cancer cells. It remains to be determined whether binding of NQO1 to p53 is sufficient for p53 stabilization. Studies with different NQO1 mutants, such as the structurally and enzymatically defective polymorphic C609T NQO1 (ref. 30; reviewed in ref. 31), which does not stabilize p53 (6), may reveal whether other NQO1 properties are also required for p53 stabilization.
ES936 is an inhibitor of NQO1 activity by alkylating tyrosine 127 or 129 in the active site of NQO1 (32). ES936 did not induce p53 degradation and did not inhibit p53-NQO1 binding (9). However, our experiments show that dicoumarol and several other competitive inhibitors of NQO1 enzymatic activity, including coumarins, flavones, and the dye cibacron blue, all induced p53 degradation. In contrast to the results with ES936 (9), our results show that dicoumarol inhibited NQO1 binding to p53. ES936 does not compete with NAD(P)H for binding to NQO1 (32), whereas dicoumarol and all the other tested NQO1 inhibitors are competitive inhibitors for NAD(P)H binding to NQO1 (8, 19-22). These results suggest that NAD(P)H plays a role in regulating NQO1-p53 binding and p53 stabilization. NAD(P)H may serve not only as an electron donor in the enzymatic activity of NQO1 but may also promote NQO1 binding to p53, and our in vitro coimmunoprecipitation experiments indicate that NADH enhanced p53-NQO1 binding (unpublished data). The conformational change induced in NQO1 by NADH (32) could be involved in promoting the binding of NQO1 to p53. Therefore, displacement of NAD(P)H from NQO1 by dicoumarol, which induces a conformational change in NQO1, would inhibit both NQO1's enzymatic activity and its binding to p53 and result in p53 degradation. In contrast, although ES936 inhibits NQO1's enzymatic activity (32), it does not displace NAD(P)H from NQO1 and thus would neither inhibit p53 binding nor induce p53 degradation.
Pyridine nucleotides such as NAD(P)H are also involved in the regulation of other protein-protein interactions. These interactions include the increased binding of the transcriptional repressor carboxyl-terminal binding protein to the transcription factor E1A in the presence of NADH (33, 34) and the increased binding of the metabolic enzyme GAPDH to the POU domain of the transcription factor OCT-1 by NAD+ (35). The NAD+-dependent binding of GAPDH to OCT-1 does not require the enzymatic activity of GAPDH (35). It has also been shown that stabilization of the lens quinone oxidoreductase ζ-crystallin and its binding to the chaperone α-crystallin require NADPH, which induces a conformational change in ζ-crystallin (36). Changes in metabolism that result in altered NAD+/NADH ratio may alter the interaction of NQO1 with p53 and affect p53 stability in cells. Sir2 is a NAD+-dependent histone deacetylase, which also deacetylates p53 and inhibits its activity (37, 38). Altered NAD+/NADH ratio may thus affect both the stability and the transcriptional activity of p53. In this way, transcription by different transcription factors, including p53, can be regulated by the metabolic state of the cell.
Acknowledgments
We thank S. Budilovsky for technical assistance, Dr. M. Oren for advice and the plasmids encoding the hot-spot human p53 mutants, and Dr. T. Rimon for the Trisacryl beads. This work was supported by the Benoziyo Institute of Molecular Medicine, the Dolfi and Lola Ebner Center for Biomedical Research, Mrs. Bernice Gershenson, the M. D. Moross Institute for Cancer Research at the Weizmann Institute of Science, and the Israel Academy of Sciences and Humanities.
Abbreviations: NQO1, NAD(P)H:quinone oxidoreductase 1; DHF, 7,8-dihydroxyflavone; Pab, monoclonal anti-p53 antibody.
References
- 1.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 2.Haupt, Y., Maya, R., Kazaz, A. & Oren, M. (1997) Nature 387, 296-299. [DOI] [PubMed] [Google Scholar]
- 3.Kubbutat, M. H., Jones, S. N. & Vousden, K. H. (1997) Nature 387, 299-303. [DOI] [PubMed] [Google Scholar]
- 4.Asher, G., Lotem, J., Sachs, L., Kahana, C. & Shaul, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 13125-13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asher, G., Lotem, J., Cohen, B., Sachs, L. & Shaul, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asher, G., Lotem, J., Kama, R., Sachs, L. & Shaul, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asher, G., Lotem, J., Sachs, L. & Shaul, Y. Methods Enzymol., in press. [DOI] [PubMed]
- 8.Hosoda, S., Nakamura, W. & Hayashi, K. (1974) J. Biol. Chem. 249, 6416-6423. [PubMed] [Google Scholar]
- 9.Anwar, A., Dehn, D., Siegel, D., Kepa, J. K., Tang, L. J., Pietenpol, J. A. & Ross, D. (2003) J. Biol. Chem. 278, 10368-10373. [DOI] [PubMed] [Google Scholar]
- 10.Hollstein, M., Rice, K., Greenblatt, M. S., Soussi, T., Fuchs, R., Sorlie, T., Hovig, E., Smith-Sorensen, B., Montesano, R. & Harris, C. C. (1994) Nucleic Acids Res. 22, 3551-3555. [PMC free article] [PubMed] [Google Scholar]
- 11.Prives, C. (1994) Cell 78, 543-546. [DOI] [PubMed] [Google Scholar]
- 12.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W. & Vogelstein, B. (1998) Science 282, 1497-1501. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, I. C., Tokiwa, T., Bennett, W., Metcalf, R. A., Welsh, J. A., Sun, T. & Harris, C. C. (1993) Carcinogenesis 14, 987-992. [DOI] [PubMed] [Google Scholar]
- 14.Yonish-Rouach, E., Resnitzky, D., Lotem, J., Sachs, L., Kimchi, A. & Oren, M. (1991) Nature 352, 345-347. [DOI] [PubMed] [Google Scholar]
- 15.Chowdary, D. R., Dermody, J. J., Jha, K. K. & Ozer, H. L. (1994) Mol. Cell. Biol. 14, 1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalovitz, D., Halevy, O. & Oren, M. (1990) Cell 62, 671-680. [DOI] [PubMed] [Google Scholar]
- 17.Lotem, J. & Sachs, L. (2002) Oncogene 21, 3284-3294. [DOI] [PubMed] [Google Scholar]
- 18.Barak, Y., Gottlieb, E., Juven-Gershon, T. & Oren, M. (1994) Genes Dev. 8, 1739-1749. [DOI] [PubMed] [Google Scholar]
- 19.Garten, S. & Wosilait, W. D. (1971) Biochem. Pharmacol. 20, 1661-1668. [DOI] [PubMed] [Google Scholar]
- 20.Chen, S., Hwang, J. & Deng, P. S. (1993) Arch. Biochem. Biophys. 302, 72-77. [DOI] [PubMed] [Google Scholar]
- 21.Chen, S., Wu, K., Zhang, D., Sherman, M., Knox, R. & Yang, C. S. (1999) Mol. Pharmacol. 56, 272-278. [DOI] [PubMed] [Google Scholar]
- 22.Prestera, T., Prochaska, H. J. & Talalay, P. (1992) Biochemistry 31, 824-833. [DOI] [PubMed] [Google Scholar]
- 23.Soussi, T. (2000) Ann. N.Y. Acad. Sci. 910, 121-139. [DOI] [PubMed] [Google Scholar]
- 24.Midgley, C. A. & Lane, D. P. (1997) Oncogene 15, 1179-1189. [DOI] [PubMed] [Google Scholar]
- 25.Peng, Y., Chen, L., Li, C., Lu, W., Agrawal, S. & Chen, J. (2001) J. Biol. Chem. 276, 6874-6878. [DOI] [PubMed] [Google Scholar]
- 26.Kubbutat, M. H., Ludwig, R. L., Ashcroft, M. & Vousden, K. H. (1998) Mol. Cell. Biol. 18, 5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng, Y., Chen, L., Li, C., Lu, W. & Chen, J. (2001) J. Biol. Chem. 276, 40583-40590. [DOI] [PubMed] [Google Scholar]
- 28.Malkinson, A. M., Siegel, D., Forrest, G. L., Gazdar, A. F., Oie, H. K., Chan, D. C., Bunn, P. A., Mabry, M., Dykes, D. J., Harrison, S. D., et al. (1992) Cancer Res. 52, 4752-4757. [PubMed] [Google Scholar]
- 29.Belinsky, M. & Jaiswal, A. K. (1993) Cancer Metastasis Rev. 12, 103-117. [DOI] [PubMed] [Google Scholar]
- 30.Traver, R. D., Horikoshi, T., Danenberg, K. D., Stadlbauer, T. H., Danenberg, P. V., Ross, D. & Gibson, N. W. (1992) Cancer Res. 52, 797-802. [PubMed] [Google Scholar]
- 31.Chen, S., Wu, K. & Knox, R. (2000) Free Radical Biol. Med. 29, 276-284. [DOI] [PubMed] [Google Scholar]
- 32.Winski, S. L., Faig, M., Bianchet, M. A., Siegel, D., Swann, E., Fung, K., Duncan, M. W., Moody, C. J., Amzel, L. M. & Ross, D. (2001) Biochemistry 40, 15135-15142. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Q., Piston, D. W. & Goodman, R. H. (2002) Science 295, 1895-1897. [DOI] [PubMed] [Google Scholar]
- 34.Fjeld, C. C., Birdsong, W. T. & Goodman, R. H. (2003) Proc. Natl. Acad. Sci. USA 100, 9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng, L., Roeder, R. G. & Luo, Y. (2003) Cell 114, 255-266. [DOI] [PubMed] [Google Scholar]
- 36.Rao, P. V., Horwitz, J. & Zigler, J. S., Jr. (1994) J. Biol. Chem. 269, 13266-13272. [PubMed] [Google Scholar]
- 37.Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L. & Gu, W. (2001) Cell 107, 137-148. [DOI] [PubMed] [Google Scholar]
- 38.Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. & Weinberg, R. A. (2001) Cell 107, 149-159. [DOI] [PubMed] [Google Scholar]