Abstract
Cytotoxicity of methylating agents is caused mostly by methylation of the O6 position of guanine in DNA to form O6–methylguanine (O6–meG). O6–meG can direct misincorporation of thymine during replication, generating O6–meG:T mismatches. Recognition of these mispairs by the mismatch repair (MMR) system leads to cell cycle arrest and apoptosis. MMR also modulates sensitivity to other antitumor drugs. The base excision repair (BER) enzyme MED1 (also known as MBD4) interacts with the MMR protein MLH1. MED1 was found to exhibit thymine glycosylase activity on O6–meG:T mismatches. To examine the biological significance of this activity, we generated mice with targeted inactivation of the Med1 gene and prepared mouse embryonic fibroblasts (MEF) with different Med1 genotype. Unlike wild-type and heterozygous cultures, Med1-/- MEF failed to undergo G2-M cell cycle arrest and apoptosis upon treatment with the methylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). Similar results were obtained with platinum compounds' 5-fluorouracil and irinotecan. As is the case with MMR-defective cells, resistance of Med1-/- MEF to MNNG was due to a tolerance mechanism because DNA damage accumulated but did not elicit checkpoint activation. Interestingly, steady state amounts of several MMR proteins are reduced in Med1-/- MEF, in comparison with Med1+/+ and Med1+/- MEF. We conclude that MED1 has an additional role in DNA damage response to antitumor agents and is associated with integrity of the MMR system. MED1 defects (much like MMR defects) may impair cell cycle arrest and apoptosis induced by DNA damage.
Methylating agents are a class of compounds that react with nucleophilic centers of organic macromolecules, including nucleic acids (1). The reaction of methylating agents with DNA is a unimolecular (SN1) nucleophilic substitution characterized by high affinity for oxygens (2). The biological effects of methylating agents, such as cell cycle arrest and apoptosis, are mostly linked to methylation of the O6 position of guanine to form O6–meG (3), a modification that tends to occur preferentially at GpG sites (4). The cytotoxic effects of methylating agents are exploited in chemotherapy of several malignancies; antitumor methylating agents are the oldest and most widely used class of anticancer drugs (5).
The protein O6–meG methyltransferase directly reverses O6–meG methylation damage by removing the methyl group in a suicidal reaction, but it is easily saturated (6, 7). O6–meG cytotoxicity is mediated mainly by the mismatch repair (MMR) system via the recognition of O6–meG:C and O6–meG:T mismatches, the latter originating by misincorporation of thymine opposite O6–meG during DNA synthesis (8, 9). Mammalian cell lines defective in the MMR proteins MSH2, MSH6, MLH1, and PMS2 are resistant to killing by methylating agents (10–13). In fact, in MMR-deficient lines, methylation damage accumulates but does not trigger cell death (14, 15). This effect has been named methylation ”tolerance,” a more appropriate term than ”resistance” (16).
It has been proposed that direct recognition of O6–meG:C and O6–meG:T mismatches by MMR proteins initiates signaling pathways leading to cell cycle arrest and apoptosis (17). Alternatively, MMR removes the paired C or T in the newly synthesized strand, but persistence of O6–meG in the template strand promotes subsequent futile cycles of excision/resynthesis, causing replication fork stalling and strand breaks, which ultimately result in cell cycle arrest and cell death (16).
In addition to methylation damage, MMR modulates sensitivity to other DNA-damaging molecules, such as cisplatin (a chemotherapeutic agent that forms intra- and interstrand adducts), topoisomerase inhibitors, the antimetabolite 5-fluorouracil (5-FU), and, to some extent, ionizing radiation (18). The degree of resistance/tolerance to these agents afforded by defective MMR is less pronounced than that toward methylating drugs (3, 18).
In vitro, O6–meG:T mismatches are a substrate of the base excision repair (BER) enzyme thymine DNA glycosylase (TDG), which removes the mismatched thymine via its DNA N-glycosylase activity. However, TDG does not seem to have any biological role in methylating agent cytotoxicity (8). The BER enzyme MED1 (also known as MBD4), identified in our laboratory as an interactor of the MMR protein MLH1 (19), displays biochemical activities similar to TDG (20, 21). Like TDG, MED1 acts as a thymine and uracil glycosylase specific for G:T and G:U mismatches originated by deamination of 5-methylcytosine and cytosine, respectively, at CpG sites (20, 21), and mice with targeted inactivation of Med1(Mbd4) show enhanced mutability at CpG sequences (22, 23).
Because of the biochemical similarity of MED1 to TDG, and the interaction between MED1 and MLH1, we investigated the role of MED1 in DNA damage induced by methylating agents and other antitumor drugs using biochemical and cell-based assays. Our results indicate that, unlike TDG, MED1 plays a role in cytotoxicity of methylating and other antitumor agents, and affects the integrity of some components of the MMR system in the response to DNA damage.
Materials and Methods
Glycosylase Assay. The following oligonucleotides were purified by 8.3 M urea/15–20% PAGE: CAATCCTAGCTGACAZ-O6meG-ATGTGGCCAATGGCATGACT (top strand), where Z is either C or G, and GAGTCATGCCATTGGCCACATYGTGTCAGCTAGGATT (bottom strand), where Y is either G or C. Double-strand oligonucleotide substrates were 32P-labeled at the 3′ end of the bottom strand (21). Substrate DNA (5 nM) was incubated with purified recombinant MED1 catalytic domain or full-length protein (5 nM) at 37°C for 30 min (21). Reactions were treated with NaOH at 90°C for 30 min and separated by 8.3 M urea/15% PAGE (21).
Generation of Med1 Mutant Embryonic Stem (ES) Cells and Mice. For construction of the Med1 Δ1–3 targeting vector, genomic clones from the mouse Med1 locus were obtained by screening a 129/SvJ lambda library (Stratagene) with human MED1 cDNA probes. A 4.5-kb HindIII fragment immediately upstream of exon 1 and a 4.5-kb BglII fragment immediately downstream of exon 3 were subcloned in pPNT (24), engineered to contain the diphtheria toxin gene driven by the pol2 promoter (P-DT) for negative selection. For construction of the Med1 Δ2–5 targeting vector, a murine exon 8 probe was used to isolate BAC 9N2 from the 129/SvJ library CITB (Research Genetics, Huntsville, AL). A 4.5-kb fragment containing exon 1 and the first half of intron 1 and a fully sequenced 2.5-kb PCR fragment containing intron 5 through part of intron 7 were inserted into pKO-NEO, along with the P-DT transcriptional unit. Targeting vectors were linearized with NotI and electroporated into W9.5 and R1 ES cells. ES colonies bearing gene replacement were selected with G418 (300 μg/ml) and further screened by Southern blot analysis by using 5′- and 3′-flanking probes. For the Δ1–3 vector, 1 positive ES clone was isolated out of 271; for the Δ2–5 vector, 3 positive ES clones were identified out of 119. Positive ES clones carrying the targeted Med1 locus were injected into C57BL/6 blastocysts to generate chimeric mice. Male chimeras obtained from all four ES clones were mated to C57BL/6 females, and the resulting Med1+/- F1 offspring were backcrossed to C57BL/6. F2 Med1+/- mice were interbred to obtain experimental embryos and animals. Thus, the mixed genetic background of the latter generation is ≈75% C57BL/6 and 25% 129SvJ. Mouse embryonic fibroblast (MEF) cultures from littermate embryos were used to minimize genetic variability.
Isolation of MEF. Embryos were isolated from the uteri of day 12.5 postcoitus pregnant mice. The embryo head was used for genotyping whereas the body was minced and incubated in 2 ml of 0.25% trypsin at 37°C for 10 min. DMEM (7 ml) was added, and the cells were gently pelleted, resuspended in medium, and plated for growth.
RT-PCR Analysis. RNA was prepared from mice and embryos of different genotype from both the Δ1–3 and Δ2–5 colonies by using guanidinium/phenol extraction (25). Total RNA was reverse-transcribed by using an oligo(dT) primer and SuperScript (GIBCO/BRL). Med1 RT-PCR analysis was conducted with primers located in exon 1 (GAGAGCCTAGTTCCAGACCCG) and 3 (GATGCTCCCTTTCGGCAGTAC). For analysis of MMR genes and Gapd, the following primers were used: Msh2: CTGTGATCAGAGTTTCGGG, CCGTGAAATGATCTCGTTTAC; Msh6: TTGAGTGAAACTGCTAGCATAC, TGTGTCCCTTCTGAATAACC; Mlh1: CAGTATATACTGGAGGAGTCGACCC, TGTATAGATCTGGCAGGTTGGC; Pms2: GTCACTGAAAGGGCTAAATTG, ACATCCAGATTGGCAACG; and Gapd: GTAGACAAAATGGTGAAGGTCG, GTTGTCATATTTCTCGTGGTTC. To obtain semiquantitative RT-PCR data, 25 cycles were used (35 cycles for Med1 and Msh2, due to low expression). Each cycle consisted of 94°C for 30 s, 52°C (for Gapdh, Med1, Mlh1 54°C, 56°C, and 58°C, respectively) for 1 min, and 72°C for 1 min.
Cell Culture and Drug Treatments. Primary MEF were cultured in DMEM supplemented with 15% FCS, 1 mM sodium pyruvate, 2 mM glutamine, 10 units/ml penicillin, and 10 μg/ml streptomycin. MEF cultures were kept on a 3T3 protocol, i.e., they were split one-third every 3 days. MNNG (N-methyl-N′-nitro-N-nitrosoguanidine, Aldrich) was dissolved in DMSO at a concentration of 10 mg/ml; all of the other drugs were used as supplied by the manufacturer: 5-FU (50 mg/ml, American Pharmaceutical Partners, Los Angeles), cisplatin (1 mg/ml, GensiaSicor Pharmaceuticals, Irvine, CA) irinotecan (20 mg/ml, Pharmacia & Upjohn), and oxaliplatin (reconstituted in DMSO at 5 mg/ml, Sanofi-Synthelabo Research, Malvern, PA).
Detection of Apoptosis. Mono- and oligonucleosomes generated during apoptosis were recognized by antibodies directed against DNA and histones in a colorimetric assay (Cell Death Detection Elisaplus Assay, Roche). Absorbance was measured with an ELISA reader (ThermoLabsystems, Cheshire, U.K.). Nucleosome enrichment was computed with respect to vehicle-treated cells.
Apoptosis-associated DNA strand breaks were measured by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay (Fluorescein-FragEL DNA Fragmentation Detection Kit, Oncogene Research, La Jolla, CA). Slides were observed under an Axioplan 2 microscope (Zeiss). Images were collected and processed by using the same settings, to allow semiquantitative comparisons.
Cell Cycle Analysis by Fluorescence-Activated Cell Sorter (FACS). Cells were seeded at 30,000 cells per well in six-well plates, and, after 24 h, they were treated with MNNG at the indicated concentrations. Apoptotic cells in the medium and attached cells were collected, washed with PBS, recentrifuged, and resuspended in PBS. Cells were fixed in 70% ethanol at 4°C for 15–20 min and collected by centrifugation. The pellet was resuspended in PBS plus propidium iodide (0.02 mg/ml) and Ribonuclease A (0.25 mg/ml). Samples were analyzed with a FACS (FACScan, Becton Dickinson) and FLOW-JO software (Tree Star, Ashland, OR).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-2H-Tetrazolium Bromide (MTT) Assay. Cells were seeded in 96-well plates (4,000 per well) and, after 24 h, were treated with the indicated drugs for 3 and 5 days. MTT (5 mg/ml solution in PBS) was added, and the plates were incubated for 2 h at 37°C. Cells were lysed in 20% SDS, 50% N,N-dimethylformamide, 2.5% glacial acetic acid, and 2.5% HCl (pH 4.7) (26). After overnight incubation at 37°C, plates were analyzed in an ELISA reader (570 nm). Surviving fraction was computed with respect to vehicle-treated cells.
Retroviral Infection. For retroviral infection, cells were plated onto six-well plates (105 per well). The following day, cells were infected with an amphotropic ASV vector carrying the GFP marker under control of the cytomegalovirus promoter (27). GM847 human fibroblasts expressing a dominant negative ATR mutant under the control of a doxycycline-inducible promoter (28) were stimulated with doxycycline (5 mg/ml) before infection. Multiplicity of infection ranged from 1 to 10. The virus was removed after 2 h, and the cells were fed with fresh medium. Five days after infection, cells were harvested and sorted by fluorescence-activated cell sorting to quantify GFP-positive cells. Apoptosis was evaluated by TUNEL assay.
Alkaline Elution. Alkaline elution was conducted essentially as described (29, 30). Cells were labeled for 24 h with [3H]thymidine [50 mCi/mol, 0.1 μCi/ml, NEN (1 Ci = 37 GBq)]. After MNNG treatment, cells were lysed with 0.2% Sarkosyl, 2 M NaCl, 0.02 M Na2EDTA (pH 10), and 0.5 mg/ml proteinase K. The lysate was allowed to flow by gravity through a 0.8-μm polyvinylchloride filter (Millipore) and was eluted at a flow rate of 0.06 ml/min by using 0.04 M Na2EDTA, 0.1% SDS (pH 12.1) with tetrapropylammonium hydroxide. The radioactivity in each eluted fraction and that remaining on the filter were used to calculate the percentage of DNA left on the filter.
Western Blot Analysis. Cells were lysed on ice in RIPA buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1% sodium deoxycholate/1% Triton X-100/0.1% SDS/10 mM NaF/1 mM each of sodium pyrophosphate, sodium orthovanadate, DTT, and EDTA), plus protease inhibitors. Lysates were separated by SDS/PAGE and transferred to polyvinylidene fluoride membranes (Millipore). Membranes were blocked in 4% nonfat dry milk in PBS (for phospho-p53 Ser-15, 5% nonfat dry milk in Tris-buffered saline, 0.1% Tween) and incubated with antibodies in 2% nonfat dry milk in PBS (for phospho-p53 Ser-15, in 5% BSA in Tris-buffered saline). Antibodies, used at 1/400 dilutions, were as follows: anti-Msh2, -Mlh1, and -β-actin (Santa Cruz Biotechnology), anti-Msh6 (Transduction Laboratories, Lexington, KY), anti-Pms2 (Oncogene Science), anti-Pcna (Sigma), anti-cyclin A (31), and anti-phospho-p53 Ser-15 (Cell Signaling Technology, Beverly, MA). Detection was carried out by using enhanced chemiluminescence (Amersham Pharmacia). Quantitation was performed with NIH IMAGE software.
Results
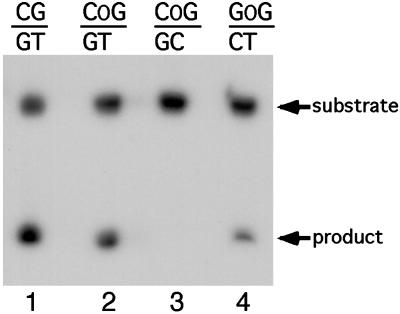
To investigate the activity of MED1 on O6–meG:T mismatches, purified recombinant human MED1 catalytic domain (32) was incubated with oligonucleotide substrates that carried an O6–meG:T mismatch in a GpG or CpG context and were 32P-labeled on the thymine-containing strand. The reaction products were treated with strong alkali to cleave at abasic sites and were separated by denaturing PAGE. As shown in Fig. 1, a cleavage product was detected with substrates that contained O6–meG:T in both sequence contexts, indicating that MED1 has thymine glycosylase activity for O6–meG:T mismatches (lanes 2 and 4). In contrast, O6–meG:C was not cleaved by the MED1-dependent reaction (lane 3). Virtually identical results were obtained with recombinant full-length protein (21) (data not shown).
Fig. 1.
MED1 thymine glycosylase activity for O6–meG:T mismatches. Double-stranded oligonucleotides, bearing O6–meG:T mismatches in a CpG or GpG context and 32P-labeled at the 3′ end on the bottom strand, were treated with purified recombinant MED1 protein at 37°C. The reactions were then treated with NaOH at 90°C, to cleave the sugar-phosphate backbone at the abasic site, and were separated by PAGE. OG: O6–meG. Lanes 1 is a positive control.
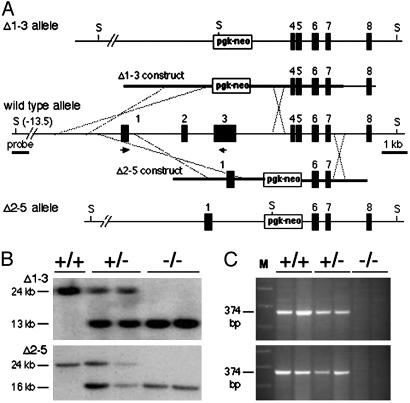
To investigate the biological significance of this finding, e.g., whether MED1 loss would interfere with cell cycle arrest and apoptosis induced by methylating agents, we used an isogenic cultured cell system: MEF cultures of different Med1 genotype derived from mice with targeted inactivation of the Med1 gene. Strains bearing two different mutant alleles were generated in our laboratory, one lacking exons 1 through 3 (designated Δ1–3) and the other lacking exons 2–5 (designated Δ2–5) (Fig. 2A). A detailed description of these mice will be published separately. Southern blot analysis confirmed production of the mutant alleles (Fig. 2B). The two alleles are null with respect to both the 5-methylcytosine binding domain, encoded in exons 1–3, and the catalytic domain, encoded in exons 3–8. In fact, the next initiator codon after exon 3 is located in exon 6 (Met-447) downstream of the region involved in the scissile thymine flipping required for activity (33); a truncated catalytic domain protein that starts at Met-447, is soluble when produced in E. coli, but is devoid of thymine glycosylase activity (data not shown). RT-PCR confirmed lack of Med1 expression in homozygous homologous recombinant cells (Fig. 2C).
Fig. 2.
Production of two novel murine Med1 alleles. (A) The Δ1–3 and Δ2–5 constructs were prepared by replacing exons (black boxes) 1–3 and 2–5, respectively, with a pgk-neo cassette; thicker lines indicate the recombination arms. The Δ1–3 and Δ2–5 alleles were produced in ES cells after homologous recombination. In the wild-type allele, the SpeI (S) restriction sites are located 13 kb upstream and 11 kb downstream of exon 1, respectively. (B) Southern blot analysis of genomic DNA from MEF of different Med1 genotype. After SpeI digestion and hybridization with a 1.5-kb SacI-HindIII probe located 6 kb upstream of exon 1 (panel A), the wild-type, Δ1–3, and Δ2–5 alleles show a 24-, 13-, and 16-kb fragment, respectively. (C) The expression of Med1 was monitored by RT-PCR analysis of MEF RNA by using primers located in exons 1 and 3 (A). No expression was detected in homologous recombinant cells.
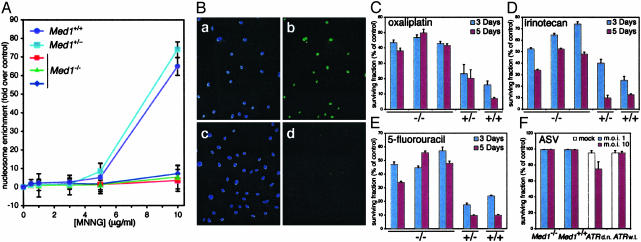
MEF were treated with increasing doses of the methylating agent MNNG, a potent inducer of O6–meG via the SN1 reaction of its putative decomposition product methyl-carbonium ion with guanine O6 (2). Med1-/- MEF displayed reduced apoptosis in comparison to wild type and heterozygous cultures, as measured by detection of mono- and oligonucleosomal DNA (Fig. 3A). This finding was confirmed with a TUNEL assay: Med1-/- MEF exhibited reduced apoptosis in comparison to wild type, and heterozygous, cultures (Fig. 3B and data not shown). We then assessed the generality of this observation by challenging MEF of different Med1 genotype with a variety of DNA damaging agents. Albeit with some differences in sensitivity, Med1-/- MEF showed increased survival on treatment with cisplatin (data not shown) and oxaliplatin, the topoisomerase inhibitor irinotecan, and 5-FU (Fig. 3 C–E).
Fig. 3.
Med1-/- MEF are resistant to cytotoxicity of antitumor agents. (A) Detection of apoptotic mono- and oligonucleosomes in MEF with different genotypes treated with increasing doses of MNNG for 48 h. Nucleosome enrichment is computed with respect to vehicle-treated cells. (B) TUNEL assay of Med1+/+ (a and b) and Med1-/- (c and d) MEF treated with 10 μg/ml MNNG for 6 h: a and c, DAPI staining; b and d, TUNEL staining. (C–E) Survival analysis (3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay) at 3 and 5 days of MEF with different genotypes treated with 20 μM oxaliplatin, 20 μM irinotecan, and 10 μM 5-FU, respectively. (F) Survival analysis (TUNEL assay) of MEF with different Med1 genotype infected with amphotropic ASV at the indicated multiplicity of infection. The negative and positive controls are uninduced (ATR wild type) GM847 human fibroblasts and GM847 cells induced with doxycycline to express a dominant negative ATR mutant (d.n.), respectively.
The wide spectrum of DNA damage to which Med1-null cells display reduced sensitivity is reminiscent of that of MMR-defective cells. Based on this similarity, we predicted that MEF of varying Med1 genotype would not show differential sensitivity to DNA double-strand breaks, a type of DNA damage that activates checkpoint pathways in a MMR-independent fashion (34, 35). To inflict DNA double-strand breaks in the absence of other DNA lesions, we used retroviral infection. It has been shown recently that retroviral DNA integration triggers a DNA damage response (36, 37). Cells deficient in some DNA repair components, such as nonhomologous end joining (NHEJ) proteins, or in the checkpoint protein ATR, show increased cell death as a result of retroviral infection. As a consequence, NHEJ- and ATR-deficient cells cannot be transduced efficiently by retroviral vectors. On retroviral infection of MEF cultures, we observed no induction of apoptosis, regardless of the Med1 genotype (Fig. 3F). To determine whether Med1-deficient cells are more resistant to retroviral transduction, we infected these and control cells (Med1+/+ and Med1+/-) with an amphotropic ASV-based vector carrying the GFP marker (27). Again, no significant differences were observed in transduction efficiencies of Med1-deficient and -proficient cells (see Table 1, which is published as supporting information on the PNAS web site). We conclude that the DNA damage induced by retroviral DNA integration does not trigger a Med1-dependent response.
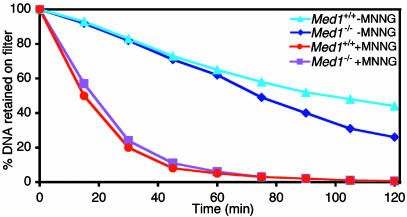
This and previous experiments point to a remarkable similarity in the phenotype of Med1-null cells and MMR-deficient cells. Resistance of MMR-deficient cells is due to a tolerance mechanism, in that DNA damage accumulates but does not elicit cell killing (14–16). To determine whether the resistance of Med1-/- MEF to MNNG was also due to tolerance to DNA damage, we used an alkaline elution assay, a classical method that measures the rate of DNA elution through a filter membrane under alkaline conditions. DNA damage, including alkylation, reduces the average size of DNA molecules, increasing the elution rate in this assay (29, 30). The results showed that DNA damage accumulates at nearly identical rates in Med1+/+ and Med1-/- MEF (Fig. 4), suggesting that the resistance of Med1-null cells to MNNG was due to a DNA damage tolerance mechanism, much as that which occurs in MMR-deficient cells.
Fig. 4.
Accumulation of DNA damage in MEF, irrespective of Med1 genotype. Alkaline elution analysis of [3H]thymidine-labeled genomic DNA from MEF of the indicated genotype treated or not with 10 μg/ml MNNG for 1 h. Percent of radioactivity remaining on filter is plotted as a function of elution time. Results are representative of three independent experiments.
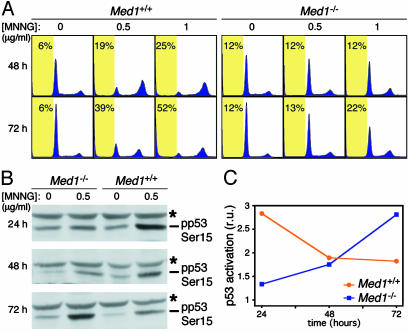
Tolerance to DNA damaging agents in MMR-defective cells is characterized by the loss of the G2–M checkpoint in response to methylating agents (38, 39). This prompted us to evaluate the cell cycle profile of Med1+/+ and Med1-/- MEF before and after exposure to MNNG. Unlike Med1+/+ MEF, Med1-/- cells did not readily exhibit G2–M arrest and failed to undergo apoptosis, as evidenced by cells with a subG1 DNA content (Fig. 5A). Only at high doses of MNNG did a fraction of the Med1-deficient cells show features of G2–M arrest and apoptosis (Fig. 5A). Consistent with these findings, a Western blot analysis revealed that the kinetics of p53 activation by MNNG, as measured by the appearance of the phospho-Ser-15 epitope, is delayed in Med1-deficient cells (Fig. 5 B and C).
Fig. 5.
Tolerance of Med1-deficient cells to alkylation damage. (A) Cell cycle analysis by flow cytometry of Med1+/+ and Med1-/- MEF treated with 0, 0.5 and 1 μg/ml MNNG for 48 and 72 h, and stained with propidium iodide. Each profile represents the analysis of 20,000 events. Yellow boxes mark the subG1 region with the indicated percent of apoptotic cells. (B) Western blot analysis with anti-phospho-p53 Ser-15 antibody of lysates from MEF treated or not with 0.5 μg/ml MNNG for 24–72 h. The asterisk marks a cross-reacting band. (C) Quantification of the anti-phospho-p53 Ser-15 Western blot (B). Relative units of p53 activation are computed with respect to untreated cells at the same time point.
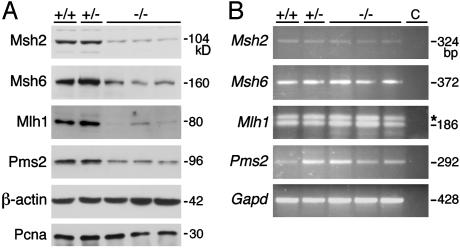
To define the potential mechanism(s) underlying a broad DNA damage resistance in Med1-deficient cells and clarify the functional relationship between Med1 and MMR, we chose to analyze the levels of MMR proteins. Although there are some differences among lines with the same genotype, Western blot analyses showed that the amounts of several MMR proteins are reduced in extracts of Med1-/- but not Med1+/+ or Med1+/- MEF (Fig. 6A). The down-regulation of Mlh1, Msh2, Pms2, and Msh6 is ≈5.8-, 5.6-, 2.6-, and 2.7-fold, respectively. Levels of other replication or S-phase associated proteins, such as Pcna (Fig. 6A) and cyclin A (data not shown) did not vary significantly among the different lines. This finding is consistent with observations in cell line models (40, 41), which indicate that reduced levels of the MMR proteins MLH1 and MSH2 impair the DNA damage response to methylating agents (see Discussion). The steady state mRNA levels of MMR genes assessed by RT-PCR do not significantly vary in lines with different Med1 genotype (Fig. 6B), indicating that down-regulation of the early MMR proteins in Med1-defective cells is apparently due to a posttranscriptional mechanism (e.g., at the level of protein translation or stability). Thus, absence of Med1 is associated with loss of integrity of the MMR system with respect to its role in the DNA damage response to methylating agents and antitumor drugs.
Fig. 6.
MMR protein levels are reduced in Med1-deficient cells by a posttranscriptional mechanism. (A) Western blot analysis of the indicated MMR proteins in MEF with different Med1 genotype. Probing with β-actin and PCNA antibodies revealed approximately equal loading of lysates. Size of the bands in kDa is shown. (B) Analysis of mRNA abundance of the indicated MMR genes in MEF with different Med1 genotype. Exonic primers for RT-PCR were separated by at least one intron. Size of the bands in base pairs is shown. Control lanes (c) are reactions in which cDNA was omitted. The asterisk marks a nonspecific band in Mlh1 reactions. Glyceraldehyde-3 phosphate dehydrogenase (Gapd) levels indicated the use of approximately equal amounts of RNA.
Discussion
In this article we present evidence that the BER glycosylase MED1 (MBD4) has a novel in vivo role in the response to methylating agents and other DNA damaging drugs. Results of our studies of murine cell lines with defined genotype indicate that homozygosity or heterozygosity for wild type Med1 is required for cytotoxicity of several classes of DNA damaging agents.
It is possible that MED1 may have a direct role in the DNA damage response to methylating agents via its glycosylase activity on O6–meG:T mismatches: repeated repair attempts by MED1 may trigger a long patch requiring a replicative polymerase and the associated MMR machinery; via its interaction with MLH1, MED1 may directly participate in the MMR-dependent checkpoint. Similar considerations may apply to 5-FU and platinum treatment, via the activity of MED1 on 5-FU:G mismatches (32) and, possibly, unspecified platinum lesions.
Alternatively, the requirement of MED1 for cytotoxicity may be indirect, and mediated by its effect on the integrity of the MMR system. In support of this interpretation, we observed that the amounts of several MMR proteins were reduced in extracts of Med1-/- but not Med1+/+ or Med1+/- MEF. This finding is consistent with previous observations in cell culture models, which indicate that reduction in the amounts of the MMR proteins MLH1 and MSH2 causes tolerance/resistance to methylating agents (40, 41). Interestingly, in these studies, reduced amounts of MLH1 and MSH2 did not compromise mismatch repair activity. It would seem that small amounts of endogenous DNA damage, e.g., base–base mismatches and short insertions/deletions originated as replication errors or recombination intermediates, can be dealt with effectively by reduced levels of MMR proteins/complexes, whereas the cytotoxic response to pharmacological doses of DNA damage requires normal amounts of MMR components. Thus, to cause cell cycle arrest and apoptosis, it seems that a threshold level of futile cycles of excision/resynthesis or sufficient signal strength must be crossed. Consistent with this interpretation, p53 activation kinetics was delayed in Med1-/- cells in comparison to Med1+/+ cells, despite significant accumulation of DNA damage.
Mice with targeted inactivation of Med1 generated in ours (data not shown) and other laboratories do not display spontaneous tumor formation or microsatellite instability (22, 23), both indications of defective MMR. This is consistent with the observations, noted above, that reduction in MMR protein levels in untreated cells does not seem to affect MMR proficiency per se. The Med1 knock-out mice therefore represent a valuable system to study the role of the MMR system in the DNA damage response in a setting of mismatch repair proficiency. Analysis of these mice may allow a dissection of the relevant signaling pathways in vivo, without the complications linked to deficient MMR (e.g., tumor incidence, reduced survival, and mutator phenotype with consequent mutations at unknown loci) (42).
The mechanism that results in reduced amounts of MMR proteins in Med1-defective cells remains to be determined. Our results are consistent with a posttranscriptional effect, because the steady state amounts of the MMR transcripts, assessed by RT-PCR, are not altered. Given the potential of MED1 to interact with MLH1 in mammalian cells (19), it is possible that Mlh1 and other MMR proteins are unstable in the absence of Med1. For instance, it is well known that PMS2 levels increase on re-expression of MLH1 in an MLH1-deficient cell line (43). Further study of the interaction between MED1 and MMR may increase our understanding of how MMR protein levels are coordinately regulated and how the MMR and BER systems cross talk in DNA repair and DNA damage response.
Our findings may have implications for human cancer. We and others showed that the MED1 gene is mutated in 25–40% of human colorectal, endometrial, and pancreatic carcinomas, exhibiting microsatellite instability (44, 45). Indeed, as a consequence of the generalized microsatellite instability resulting from MLH1 or MSH2 inactivation, two polyadenine microsatellites in the coding region of the MED1 gene contract or expand, causing frameshifts that predict the synthesis of truncated proteins that lack the C-terminal catalytic domain. In colorectal cancer specimens, we also detected loss of heterozygosity at the MED1 locus (45), suggesting that homozygous inactivation of this gene does occur in human cancer. It is tempting to speculate that this secondary inactivation of MED1 in tumors with a primary MMR defect leads to a more dramatic impairment of the DNA damage response. It is also possible that primary MED1 mutations (i.e., mutations other than at coding microsatellites) may occur in tumors with no primary MMR defect (no microsatellite instability). In both situations, it would be important to determine whether tumors with homozygous inactivation of MED1 are resistant/tolerant to treatment with DNA damaging agents. A recent clinical study indicated that colorectal tumors with MMR defects may be resistant to treatment with 5-FU (46). Our data suggest that these tumors, in particular cases with MED1 deficiency, may also be resistant to other agents commonly used in the treatment of colorectal cancer (i.e., irinotecan, oxaliplatin). Clinical validation of these hypotheses may have significant implications for treatment selection. Additional understanding of the mechanisms of MED1 and MMR inactivation may suggest methods to overcome drug resistance.
The observations reported here lend support to our proposal that MED1 acts as a tumor suppressor involved in genetic stability (35, 47). MED1 may not only effect genome surveillance at CpG sites (22, 23), but also participate in DNA damage checkpoints. Indeed, most of the DNA repair genes with a role in human cancer, i.e., MMR genes BRCA1, BRCA2, and ATM, are also involved in the regulation of cell cycle checkpoints and apoptosis (48). This dual role in DNA repair and DNA damage response may facilitate selection of inactivating mutations of these genes, provide a growth advantage during tumorigenesis and, later in the natural history of the tumor, shape response to chemotherapy.
Supplementary Material
Acknowledgments
We thank P. Adams, J. Chernoff, A. Knudson, Y. Matsumoto, E. Moss, M. Murphy, and J. Testa for critical reading of the manuscript and the anonymous reviewer for suggesting the model of the direct role of MED1 in triggering the DNA damage checkpoint. We acknowledge the following Fox Chase Cancer Center core facilities: Cell Culture, Transgenic and Knock-out, Laboratory Animal, and the Biotechnology Facility. This work was supported by National Institutes of Health Grants CA78412 and CA06927 and by an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. S.C. was supported in part by a Consiglio Nazionale delle Ricerche Fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: O6–meG, O6–methylguanine; MMR, DNA mismatch repair; BER, base excision repair; ES, embryonic stem; MEF, mouse embryonic fibroblast; MNNG, N-methyl-N′nitro-N-nitrosoguanidine; 5-FU, 5-fluorouracil; PAGE, polyacrylamide gel electrophoresis; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling.
See Commentary on page 14601.
References
- 1.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) in DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC), pp. 1-58.
- 2.Grover, P. L. (1979) Chemical Carcinogens and DNA (CRC, Boca Raton, FL).
- 3.Karran, P. & Bignami, M. (1999) in DNA Recombination and Repair, eds. Smith, P. J. & Jones, C. J. (Oxford Univ. Press, New York), pp. 66-159.
- 4.Mironov, N. M., Bleicher, F., Martel-Planche, G. & Montesano, R. (1993) Mutat. Res. 288, 197-205. [DOI] [PubMed] [Google Scholar]
- 5.DeVita, V. T., Hellman, S. & Rosenberg, S. A. (2001) CANCER, Principles and Practice of Oncology (Lippincott-Raven, Philadelphia).
- 6.Erickson, L. C. (1991) Semin. Cancer Biol. 2, 257-265. [PubMed] [Google Scholar]
- 7.Lindahl, T., Demple, B. & Robins, P. (1982) EMBO J. 1, 1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin, S., Branch, P., Xu, Y. Z. & Karran, P. (1994) Biochemistry 33, 4787-4793. [DOI] [PubMed] [Google Scholar]
- 9.Duckett, D. R., Drummond, J. T., Murchie, A. I., Reardon, J. T., Sancar, A., Lilley, D. M. & Modrich, P. (1996) Proc. Natl. Acad. Sci. USA 93, 6443-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aquilina, G., Hess, P., Branch, P., MacGeoch, C., Casciano, I., Karran, P. & Bignami, M. (1994) Proc. Natl. Acad. Sci. USA 91, 8905-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branch, P., Hampson, R. & Karran, P. (1995) Cancer Res. 55, 2304-2309. [PubMed] [Google Scholar]
- 12.Koi, M., Umar, A., Chauhan, D. P., Cherian, S. P., Carethers, J. M., Kunkel, T. A. & Boland, C. R. (1994) Cancer Res. 54, 4308-4312. [PubMed] [Google Scholar]
- 13.Reitmair, A. H., Risley, R., Bristow, R. G., Wilson, T., Ganesh, A., Jang, A., Peacock, J., Benchimol, S., Hill, R. P., Mak, T. W., et al. (1997) Cancer Res. 57, 3765-3771. [PubMed] [Google Scholar]
- 14.Branch, P., Aquilina, G., Bignami, M. & Karran, P. (1993) Nature 362, 652-654. [DOI] [PubMed] [Google Scholar]
- 15.Kat, A., Thilly, W. G., Fang, W.-H., Longley, M. J., Li, G.-M. & Modrich, P. (1993) Proc. Natl. Acad. Sci. USA 90, 6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karran, P. & Bignami, M. (1994) BioEssays 16, 833-839. [DOI] [PubMed] [Google Scholar]
- 17.Fishel, R. (1999) Nat. Med. 5, 1239-1241. [DOI] [PubMed] [Google Scholar]
- 18.Fink, D., Aebi, S. & Howell, S. B. (1998) Clin. Cancer Res. 4, 1-6. [PubMed] [Google Scholar]
- 19.Bellacosa, A., Cicchillitti, L., Schepis, F., Riccio, A., Yeung, A. T., Matsumoto, Y., Golemis, E. A., Genuardi, M. & Neri, G. (1999) Proc. Natl. Acad. Sci. USA 96, 3969-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrich, B., Hardeland, U., Ng, H. H., Jiricny, J. & Bird, A. (1999) Nature 401, 301-304. [DOI] [PubMed] [Google Scholar]
- 21.Petronzelli, F., Riccio, A., Markham, G. D., Seeholzer, S. H., Stoerker, J., Genuardi, M., Yeung, A. T., Matsumoto, Y. & Bellacosa, A. (2000) J. Biol. Chem. 275, 32422-32429. [DOI] [PubMed] [Google Scholar]
- 22.Wong, E., Yang, K., Kuraguchi, M., Werling, U., Avdievich, E., Fan, K., Fazzari, M., Jin, B., Brown, A. M., Lipkin, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millar, C. B., Guy, J., Sansom, O. J., Selfridge, J., MacDougall, E., Hendrich, B., Keightley, P. D., Bishop, S. M., Clarke, A. R. & Bird, A. (2002) Science 297, 403-405. [DOI] [PubMed] [Google Scholar]
- 24.Tybulewicz, V. L., Crawford, C. E., Jackson, P. K., Bronson, R. T. & Mulligan, R. C. (1991) Cell 65, 1153-1163. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, M. B., Nielsen, S. E. & Berg, K. (1989) J. Immunol. Methods 119, 203-210. [DOI] [PubMed] [Google Scholar]
- 27.Katz, R. A., Greger, J. G., Darby, K., Boimel, P., Rall, G. F. & Skalka, A. M. (2002) J. Virol. 76, 5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cliby, W. A., Roberts, C. J., Cimprich, K. A., Stringer, C. M., Lamb, J. R., Schreiber, S. L. & Friend, S. H. (1998) EMBO J. 17, 159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornace, A. J., Jr., & Little, J. B. (1979) Cancer Res. 39, 704-710. [PubMed] [Google Scholar]
- 30.Kohn, K. W. (1991) Pharmacol. Ther. 49, 55-77. [DOI] [PubMed] [Google Scholar]
- 31.Howard, C. M., Claudio, P. P., De Luca, A., Stiegler, P., Jori, F. P., Safdar, N. M., Caputi, M., Khalili, K. & Giordano, A. (2000) Cancer Res. 60, 2737-2744. [PubMed] [Google Scholar]
- 32.Petronzelli, F., Riccio, A., Markham, G. D., Seeholzer, S. H., Genuardi, M., Karbowski, M., Yeung, A. T., Matsumoto, Y. & Bellacosa, A. (2000) J. Cell. Physiol. 185, 473-480. [DOI] [PubMed] [Google Scholar]
- 33.Wu, P., Qiu, C., Sohail, A., Zhang, X., Bhagwat, A. S. & Cheng, X. (2003) J. Biol. Chem. 278, 5285-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, M., Narayanan, L., Xu, X. S., Prolla, T. A., Liskay, R. M. & Glazer, P. M. (2000) Cancer Res. 60, 4889-4893. [PubMed] [Google Scholar]
- 35.Bellacosa, A. (2001) Cell Death Differ. 8, 1076-1092. [DOI] [PubMed] [Google Scholar]
- 36.Daniel, R., Kao, G., Taganov, K., Greger, J. G., Favorova, O., Merkel, G., Yen, T. J., Katz, R. A. & Skalka, A. M. (2003) Proc. Natl. Acad. Sci. USA 100, 4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel, R., Katz, R. A. & Skalka, A. M. (1999) Science 284, 644-647. [DOI] [PubMed] [Google Scholar]
- 38.Buermeyer, A. B., Wilson-Van Patten, C., Baker, S. M. & Liskay, R. M. (1999) Cancer Res. 59, 538-541. [PubMed] [Google Scholar]
- 39.Hawn, M. T., Umar, A., Carethers, J. M., Marra, G., Kunkel, T. A., Boland, C. R. & Koi, M. (1995) Cancer Res. 55, 3721-3725. [PubMed] [Google Scholar]
- 40.Cejka, P., Stojic, L., Mojas, N., Russell, A. M., Heinimann, K., Cannavo, E., di Pietro, M., Marra, G. & Jiricny, J. (2003) EMBO J. 22, 2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claij, N. & Te Riele, H. (2002) Oncogene 21, 2873-2879. [DOI] [PubMed] [Google Scholar]
- 42.Prolla, T. A., Abuin, A. & Bradley, A. (1996) Semin. Cancer Biol. 7, 241-247. [DOI] [PubMed] [Google Scholar]
- 43.Raschle, M., Marra, G., Nystrom-Lahti, M., Schar, P. & Jiricny, J. (1999) J. Biol. Chem. 274, 32368-32375. [DOI] [PubMed] [Google Scholar]
- 44.Bader, S., Walker, M., Hendrich, B., Bird, A., Bird, C., Hooper, M. & Wyllie, A. (1999) Oncogene 18, 8044-8047. [DOI] [PubMed] [Google Scholar]
- 45.Riccio, A., Aaltonen, L. A., Godwin, A. K., Loukola, A., Percesepe, A., Salovaara, R., Masciullo, V., Genuardi, M., Paravatou-Petsotas, M., Bassi, D. E., et al. (1999) Nat. Genet. 23, 266-268. [DOI] [PubMed] [Google Scholar]
- 46.Ribic, C. M., Sargent, D. J., Moore, M. J., Thibodeau, S. N., French, A. J., Goldberg, R. M., Hamilton, S. R., Laurent-Puig, P., Gryfe, R., Shepherd, L. E., et al. (2003) N. Engl. J. Med. 349, 247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellacosa, A. (2001) J. Cell. Physiol. 187, 137-144. [DOI] [PubMed] [Google Scholar]
- 48.Heinen, C. D., Schmutte, C. & Fishel, R. (2002) Cancer Biol. Ther. 1, 477-485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.