Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) exhibits specific tumoricidal activity and is under development for cancer therapy. Mismatch-repair-deficient colonic tumors evade TRAIL-induced apoptosis through mutational inactivation of Bax, but chemotherapeutics including Camptosar (CPT-11) restore TRAIL sensitivity. However, the signaling pathways in restoring TRAIL sensitivity remain to be elucidated. Here, we imaged p53 transcriptional activity in Bax-/- carcinomas by using bioluminescence, in vivo, and find that p53 is required for sensitization to TRAIL by CPT-11. Small interfering RNAs directed at proapoptotic p53 targets reveal TRAIL receptor KILLER/DR5 contributes significantly to TRAIL sensitization, whereas Bak plays a minor role. Caspase 8 inhibition protects both CPT-11 pretreated wild-type and Bax-/- HCT116 cells from TRAIL-induced apoptosis, whereas caspase 9 inhibition only rescued the wild-type HCT116 cells from death induced by TRAIL. The results suggest a conversion in the apoptotic mechanism in HCT116 colon carcinoma from a type II pathway involving Bax and the mitochondria to a type I pathway involving efficient extrinsic pathway caspase activation. In contrast to Bax-/- cells, Bak-deficient human cancers undergo apoptosis in response to TRAIL or CPT-11, implying that these proteins have nonoverlapping functions. Our studies elucidate a mechanism for restoration of TRAIL sensitivity in MMR-deficient Bax-/- human cancers through p53-dependent activation of KILLER/DR5 and reconstitution of a type I death pathway. Efforts to identify agents that up-regulate DR5 may be useful in cancer therapies restoring TRAIL sensitivity.
Keywords: in vivo bioluminescence imaging, TRAIL, chemosensitivity, DR5, drug resistance
The tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the tumor necrosis factor superfamily and has unique cytotoxic effects against a wide range of tumor cells but not most normal cells (1, 2). TRAIL induces apoptosis through activating its proapoptotic receptors, death receptor 4 (DR4) and KILLER/DR5 (3, 4). Preclinical studies in mice and nonhuman primates have shown that administration of soluble TRAIL suppresses growth of TRAIL-sensitive tumor xenografts without apparent systemic toxicity (1, 5). Therefore, TRAIL remains a promising therapeutic agent under development. We have previously demonstrated that a proapoptotic member of the Bcl-2 family, Bax, is required for TRAIL-mediated apoptosis in HCT116 human colon cancer cells (6). Mismatch repair-deficient tumors can acquire resistance to TRAIL through mutational inactivation of Bax, but preexposure to chemotherapy rescues tumor sensitivity (7). However, the mechanisms of sensitizing Bax-/- HCT116 cancer cells to TRAIL remain unclear.
p53 is the most commonly altered gene in human cancer (8). In response to DNA damage, p53 transactivates genes leading to cell cycle arrest or apoptosis (9, 10). It is well established that p53-dependent cell cycle arrest requires activation of the cyclin-CDK inhibitor p21WAF1 for G1 arrest (11). The pathway of apoptosis by p53 is more complex involving transcriptional regulation of multiple p53-target genes, as well as a transcription-independent mechanisms (9, 12). A number of targets have been proposed to mediate p53-dependent apoptosis through transcriptional activation, including KILLER/DR5, Bax, Bak, Bid, and Caspase 6 (4, 7, 13-17). In addition to transcriptional activation, down-regulated expression of some genes by p53 may also contribute to apoptosis (18, 19). In CD95-mediated apoptosis, two cell type-specific apoptosis signaling pathways have been identified, so-called type I and type II pathways (20). In type I (extrinsic) cell death, caspase 8 activation sufficient to kill cells occurs as a direct consequence of death receptor ligation, with activation of downstream (effector) procasapses such as caspase 3. This death is independent of mitochondria and is not blocked by overexpression of Bcl-2. In contrast, type II (intrinsic) cell death depends on amplification of death receptor signals via the mitochondria and can be blocked by Bcl-2 or caspase 9 inhibitors. We have previously shown that HCT116 human colon cancer cells behave like type II cells (6, 21). Most chemotherapeutic agents and irradiation trigger tumor apoptosis through the intrinsic pathway. The extrinsic pathway triggers apoptosis in response to death receptors by their ligands (e.g., TRAIL) (22, 23). Given that death-receptor targeting and conventional agents induce tumor-cell apoptosis through distinct signaling pathways, combination of the two approaches might lead to synergistic apoptosis activation and reduce the probability that tumor cells will develop resistance to therapy. A recent report showed that conventional chemotherapeutic agents restored TRAIL sensitivity in Bax-/- human colon cancer cells (7).
To explore the mechanisms underlying the combined antitumor activity of TRAIL and chemotherapeutic agents in the resensitization of Bax-/- human colon cancer cells to TRAIL, we investigated the importance of p53 and its induced target genes. It has been shown that human papillomavirus (HPV)-E6 protein targets p53 protein for degradation by ubiquitin-mediated proteolysis, and infection of human cancer cells by E6-expressing adenovirus (Ad-E6) leads to efficient degradation of either wild-type or mutant p53 protein (24). By using Ad-E6 and directly visualizing functional p53 transcriptional activity by in vivo imaging (25), we found that degradation of p53 abolished apoptosis induced by TRAIL plus chemotherapy in Bax-null HCT116 cells. These results indicate that p53 may be required for sensitizing Bax-/- cells to TRAIL after pretreatment with chemotherapy. Furthermore, we knocked down the expression of the two p53 targets, KILLER/DR5 and Bak, which are most likely involved in sensitizing Bax-/- human colon cancer cells to TRAIL, by employing small interfering RNAs (siRNAs) (26, 27). We found that silencing KILLER/DR5 in Bax-/- cells significantly inhibited TRAIL sensitivity, whereas blockade of Bak in Bax-/- cells did not significantly confer resistance to TRAIL in the presence of chemotherapy. Notably, caspase 8 inhibition protected both wild-type and Bax-/- HCT116 cells from TRAIL-induced apoptosis, whereas caspase 9 inhibition only prevented the wild-type HCT116 cells from death induced by TRAIL plus Camptosar (CPT-11). We also found that Bax is a determinant for apoptosis induced by multiple stimuli, whereas Bak deficiency did not confer prominent resistance to apoptotic stimuli in human cells. Taken together, our results support the hypothesis that resensitization of Bax-/- human HCT116 cells to TRAIL involves p53 activation of KILLER/DR5, and that chemotherapy reconstitutes the extrinsic apoptotic-signaling pathway thereby converting HCT116 into type I cells.
Materials and Methods
Cell Line and Cell Culture. Bax-/- HCT116 and p53-/- HCT116 cells were a kind gift of B. Vogelstein (Johns Hopkins University, Baltimore). Parental HCT116 cells were obtained from American Type Culture Collection. Cells were maintained in McCoy's 5A medium supplemented with 10% FBS and antibiotics.
Adenoviral Infections. Ad-LacZ and Ad-E6 have been described (17, 24). Cells were grown to 70% confluence and infected with recombinant adenovirus at a multiplicity of infection of 50.
siRNAs. The KILLER/DR5 and BAK siRNA duplexes (option C) were 21 bp including a 2-base deoxynucleotide overhang (synthesized by Dharmacon Research, Lafayette, CO). The sequence of the KILLER/DR5 siRNA oligos was 5′-AAGACCCUUGUGCUCGUUGUC-3′, and the sequence of BAK siRNA oligos was 5′-AACCGACGCUAUGACUCAGAG-3′. The control siRNA used was 5′-A ACGUACGCGGA AUACUUCGA-3′ against LacZ. Cells were transfected with siRNAs by using Lipofectamine 2000 (Invitrogen) following the manufacturer's instruction.
Plasmids, Transfection, and Luciferase Assay. The PG13-luc, a p53 reporter with a firefly luciferase gene under the control of 13 p53 response elements, has been described (11). pRL-SV40 (simian virus 40) vector was purchased from Promega, and this vector contains a cDNA encoding Renilla luciferase, which was originally cloned from the marine organism Renilla reniformis. Transfections were carried out as described (6, 13, 16). In brief, before transfection, 5 × 105 cells per six wells were seeded. The cells were transfected by Lipofectamine 2000-DNA conjugates suspended in Opti-MEM serum-free media (GIBCO/BRL). Serum containing medium was added at 4 h after transfection. Cells were harvested and assayed for luciferase following the manufacturer's instruction.
In Vivo Imaging Studies. Stable transfection of Bax-/- HCT116 cells was performed with PG13-Luc, pRL-SV40, and pcDNA 3.1 myc-His(-) vector carrying a neomycin selection marker, using Lipofectamine 2000 transfection reagent and culture in 0.5 mg/ml G418 (Invitrogen). The cells were infected with Ad-E6 or Ad-LacZ for 12 h, and then treated with 50 μg/ml etoposide (Sigma) or 10 μg/ml CPT-11 (Amersham Pharmacia/Upjohn) for 16 h followed by 10 ng/ml his-tagged TRAIL and 1 μg/ml anti-his6 antibody for 2 h. Cells were replated in 96-well black microplates (Fisher Scientific) after trypsinization. 150 μg/ml d-luciferin (Biotium, Hayward, CA) or 10 μg/ml coelenterazine (Native Coelenterazine, Biotium, Hayward, CA) was added to each well. Bioluminesence images were acquired by using the cooled charge-coupled device camera of an in vivo imaging system (IVIS, Xenogen, Alameda, CA) (25). For the mouse xenograft experiments, 1 × 106 stably transfected Bax-/- HCT116 cells were infected with Ad-LacZ or Ad-E6 for 16 h. The infected cells were implanted s.c. into the forearm region of female athymic nu/nu mice (Charles River Breeding Laboratories). CPT-11 (80 mg/kg) was administered by i.p. injection. At 0 or 16 h after treatment, coelenterazine (0.7 mg/kg of body weight) was injected into the tail-vein 3 min before imaging. At 2 h later, d-luciferin (150 mg/kg of body weight) was injected into the tail-vein 5 min before imaging. We have previously shown that these imaging times allow detection of firefly and Renilla luciferase fluorescence intensities at their respective peaks (28). The supine mice were placed in a light-tight chamber, with a field of view set at 15 cm above the sample shelf. A grayscale reference image was obtained under low-level illumination. Photons emitted from cells implanted in the mice were collected and integrated for a period of 1 min. Images were obtained with a binning of medium (8 × 8) by using the LIVING IMAGE software (Xenogen). Each image was acquired at the same time, relative to the injected substrate. All of the images are shown at the same scale.
Flow Cytometric Cell-Death Analysis. Cells were harvested after the indicated treatments and time periods, and prepared for detection of active caspase 3 by flow cytometry or stained with propidium iodide and analyzed by flow cytometry for sub-G1 content as described (6, 13, 16). Cell sorting was performed on a Coulter Epics Elite (Beckman Coulter).
Western Blot. Western blotting was carried out as described (6, 13, 16) with mouse anti-human p53 monoclonal antibody (pAb1801; Oncogene, San Diego) and mouse anti-human actin antibody (C-2; Santa Cruz Biotechnology). Polyclonal antibody against caspase 9 was purchased from Imgenex (San Diego).
Northern Blot. Total RNA was isolated by using RNeasy total miniprep kit (Qiagen, Valencia, CA) following the manufacturer's instruction. Northern blot was carried out as described (16). Full-length KILLER/DR5 and a partial BAK cDNA were used as probes.
Results
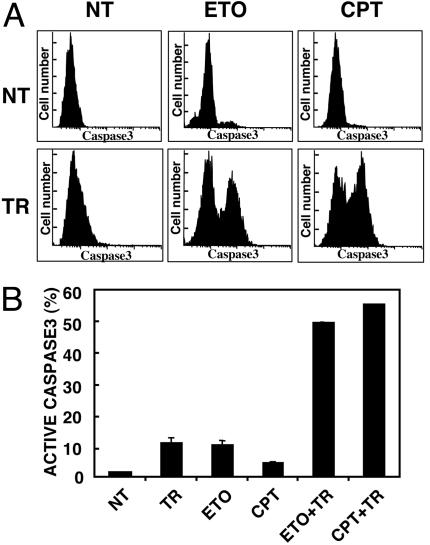
Restoring TRAIL Sensitivity by Chemotherapy. Bax is a target for mutation in a subset of human tumors harboring DNA mismatch repair deficiency (29). Inactivation of Bax can confer TRAIL resistance, but preexposure to chemotherapy rescues killing (6, 7). In the present study, we tested whether exposing Bax-/- HCT116 human colon cancer cells to etoposide or CPT-11 could resensitize cells to TRAIL. The results indicate that TRAIL effectively killed Bax-/- HCT116 cells by 4 h after pretreatment with either etoposide or CPT-11 for 16 h whereas treatment with etoposide, CPT-11 or TRAIL alone did not induce apparent apoptosis (Fig. 1). These results confirm earlier observations (7) and suggest that pretreatment with DNA-damaging agents restores TRAIL-induced cell death in Bax-null cells.
Fig. 1.
Etoposide and CPT-11 sensitize Bax-/- cells to TRAIL. Bax-/- HCT116 human colon cancer cells were not treated (NT) or treated for 4 h with 10 ng/ml his-tagged TRAIL (TR), for 16 h with etoposide (ETO) or CPT-11 (CPT), or for 16 h with etoposide or CPT-11 followed by TRAIL for 4 h (ETO+TR or CPT+TR). Cells were harvested after treatment for flow cytometric analysis. (A) Example of flow cytometric analysis for active caspase 3-expressing cells. (B) The percentage of cells expressing activated caspase 3 is shown (10,000 cells were examined for each sample).
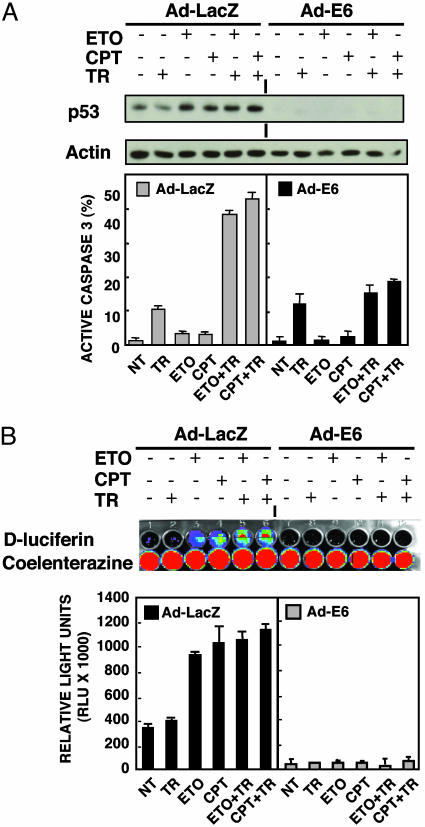
p53 Is Required for Chemotherapy-Induced Sensitization of Bax-/- HCT116 Cells to TRAIL. Given that etoposide and CPT-11 are topoisomerase inhibitors that activate p53 (30), we examined whether p53 is involved in the resensitization of Bax-/- HCT116 cells to TRAIL. We infected the Bax-/- cells with Ad-E6, which effectively degrades p53, or Ad-LacZ as a control. Western blot and luciferase assay by luminometer as well as in vivo imaging showed that preexposure of Ad-LacZ-infected Bax-null cells to either etoposide or CPT-11 stabilized p53, whereas Ad-E6 infection completely degraded p53 even in the presence of etoposide or CPT-11 (Fig. 2). Ad-LacZ-infected Bax-/- cells overcame TRAIL resistance in the presence of chemotherapy to the same extent as uninfected cells. In contrast, Ad-E6 infection abolished the apoptosis induced by TRAIL plus chemotherapy (Fig. 2 A). This provides strong evidence that p53 is required for etoposide- or CPT-11-dependent sensitization of Bax-null HCT116 cells to TRAIL.
Fig. 2.
p53 is required for sensitizing Bax-/- cells to TRAIL. Bax-/- HCT116 cells were infected with Ad-E6 or Ad-LacZ for 16 h. The infected cells were treated as described in the Fig. 1 legend. (A) p53 protein was determined by Western blot and active caspase 3 was analyzed by flow cytometry. (B) Infected cells were transfected with PG13-Luc and pRL-SV40 plasmids. Luciferase activity was determined by Fisher luminometer (Lower). PG13-Luc and pRL-SV40 stably transfected Bax-/- HCT116 cells were treated as described above, images were obtained by using the cooled charge-coupled camera of the in vivo imaging system (Upper).
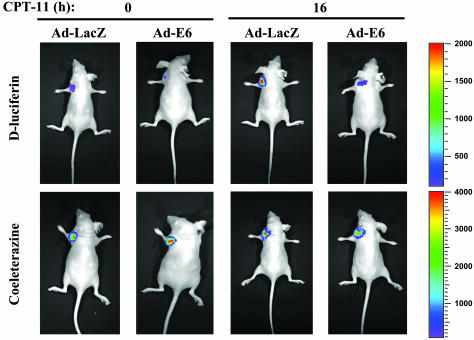
In Vivo Imaging of p53 Transcriptional Activity. Several imaging technologies and new reporter genes are being studied for noninvasive imaging and quantification of gene expression in living subjects (25, 31). To image p53 transcriptional activity in vivo, Bax-/- HCT116 cells were stably transfected with the p53 reporter plasmid PG13-Luc (firefly luciferase) and pRL-SV40 (Renilla luciferase) as an internal control. Two positive clones were isolated and both clones exhibited increased firefly luciferase activity without a change in Renilla luciferase activity upon CPT-11 treatment (data not shown). There was no crossreactivity of d-luciferin with Renilla luciferase or of coelenterazine with firefly luciferase as demonstrated previously (25). The stably transfected cells were infected with Ad-LacZ or Ad-E6 for 16 h. The infected cells were implanted at the forearm region of the mice, and CPT-11 was administered by i.p. injection. At 0 or 16 h after CPT-11 injection, the substrates for firefly and Renilla luciferase were injected into the same mice, 2 h apart, giving sufficient time for the first signal from coelenterazine to dissipate completely, followed by d-luciferin injection. We observed a significantly increased level of bioluminescence from the implanted site of Ad-LacZ infected cells after treatment with CPT-11, and a decreased level from the Ad-E6 infected cells when d-luciferin was injected. As expected, there were no acute changes after CPT-11 in the bioluminescence level among the different implanted sites when coelenterazine was injected (Fig. 3). We have therefore visualized p53 transcriptional activity in living mice in response to systemic administration of chemotherapy. Taken together with the cell culture data (Fig. 2B), the results implicate p53 transcriptional activity in the CPT-11 sensitization effect.
Fig. 3.
Optical imaging of p53 activity in living mice. PG13-Luc and pRL-SV40 stably transfected Bax-/- HCT116 cells were infected with Ad-LacZ or Ad-E6 for 16 h. Cells (1 × 106) were s.c. implanted into left or right forearm. CPT11 (80 mg/kg) was administered by i.p. injection. After 0 or 16 h of treatment, coelenterazine was injected 3 min before imaging, 2 h apart, followed by d-luciferin injection (5 min before imaging). A whole body image was acquired by using the cooled charge-coupled device camera. Each image was acquired at the same time, relative to the injected substrate, and all of the images are shown at the same scale.
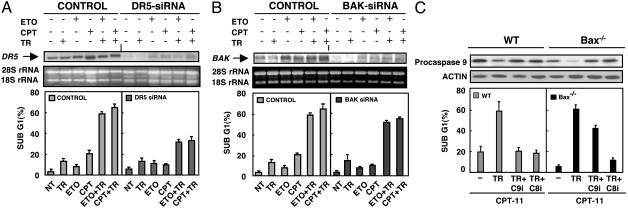
The KILLER/DR5-Mediated Type I Death Receptor Pathway Contributes to DNA Damage-Induced Resensitization of Bax-/- HCT116 Cells to TRAIL. Etoposide or CPT-11 activate p53, which consequently induces transcription of several proapoptotic genes (4, 7, 13-17, 30). We tested whether expression of two p53 targets, TRAIL receptor KILLER/DR5 and Bak, which are most likely involved in sensitizing Bax-/- cells to TRAIL, could be induced by either etoposide or CPT-11 treatment. The results indicate that KILLER/DR5 and Bak mRNAs were up-regulated in response to etoposide or CPT-11 pretreatment (Fig. 4 A and B). This finding is consistent with previous observations (7).
Fig. 4.
The effects of KILLER/DR5, Bak inhibition, and caspase 8 or 9 inhibition on sensitization of Bax-/- cells to TRAIL after DNA damage. Bax-/- HCT116 cells were transfected with KILLER/DR5 siRNA (A) or Bak siRNA (B). LacZ siRNA oligos were used as a control. Twenty-four hours after transfection, cells were treated as described in the legend to Fig. 1. KILLER/DR5 and Bak expression were determined by Northern blots. Cells were harvested, stained with propidium iodide, and analyzed by flow cytometry. The sub-G1 content of 20,000 cells was examined for each sample. (C) Differential effects of caspase 8 (Z-IETD-FMK) and caspase 9 (Z-LEHD-FMK) inhibitors on TRAIL-induced apoptosis. Wild-type or Bax-/- HCT116 cells were pretreated with 10 μg/ml CPT-11 for 16 h, after which 20 μM caspase 8 or 9 inhibitor was added; after 2 h, TRAIL (10 ng/ml) plus 1 μg/ml anti-His-6 was added for 4 h before sub-G1 analysis. Extracts were collected and analyzed by Western blotting for the expression of caspase 9.
Death receptor targeting and conventional chemotherapeutic agents induce tumor-cell apoptosis through different (intrinsic and extrinsic) signaling pathways (32, 33). To directly dissect whether elimination of KILLER/DR5 or Bak, respectively, would confer resistance to etoposide or CPT-11-induced resensitization of Bax-/- HCT116 cells to TRAIL, we used siRNA to silence the expression of KILLER/DR5 or Bak. Both KILLER/DR5 and Bak siRNAs efficiently and specifically knocked down the expression of these two genes (data not shown). We first examined the effects of silencing of KILLER/DR5 on etoposide or CPT-11-induced resensitization of Bax-/- HCT116 cells to TRAIL. The data indicate that suppression of KILLER/DR5 by siRNA prevents apoptosis induced by TRAIL plus either etoposide or CPT-11 (Fig. 4A). Surprisingly, knock-down of Bak did not significantly confer TRAIL resistance to Bax-/- HCT116 cells in response to DNA-damaging stimuli (Fig. 4B). Neither DR5 nor Bak siRNA alone was toxic toward Bax-/- HCT116 cells (Fig. 4 A and B). These data provide evidence that KILLER/DR5 plays a major role in restoration of TRAIL sensitivity and indicate that a type I death receptor pathway might contribute significantly to the rescue of TRAIL sensitivity by chemotherapy in (originally type II) Bax-/- HCT116 cells. To further substantiate this claim, we examined the effect of caspase 8 or caspase 9 inhibitors on the sensitization of Bax-/- cells to TRAIL by chemotherapy. We found that caspase 8 inhibition protects both CPT-11 pretreated wild-type and Bax-/- HCT116 cells from TRAIL-induced apoptosis, whereas caspase 9 inhibition only rescued the wild-type HCT116 cells from death induced by TRAIL (Fig. 4C). The caspase 9 depletion was fully inhibited by Z-LEHD-FMK inhibitor, and the extent of caspase 9 rescue was equivalent in the wild-type versus Bax-/- HCT116 cells (Fig. 4C). We conclude that chemotherapy restores an extrinsic type I apoptotic-signaling pathway activated by TRAIL in TRAIL-resistant Bax-/- HCT116 cells.
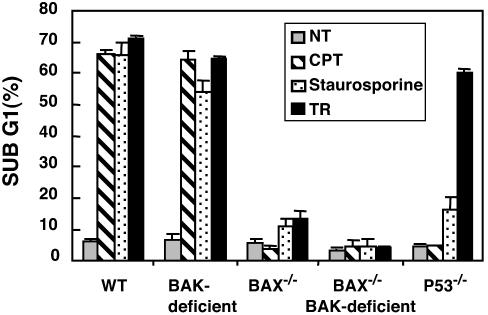
A Critical Requirement of Bax but Not Bak for Apoptosis Induced by Multiple Stimuli in Human Colorectal Cancers. Earlier studies with Bax, Bak knockout murine embryonic fibroblasts have shown that activation of Bax or Bak appears to be an essential gateway to mitochondria dysfunction required for cell death in response to diverse stimuli and that Bax and Bak are necessary, albeit mutually redundant, for apoptosis (34). However, whether these findings can be extrapolated to other cell types, including human cells, and additional death stimuli was not known. Here, we transfected wild-type and Bax-/- HCT116 human colon cancer cells with Bak siRNA or LacZ siRNA as a control, and determined the responses of wild-type, Bak-deficient (Bak siRNA transfectant), Bax-/-, Bax-/- Bak-deficient, and p53-/- HCT116 human colon cancer cells to intrinsic or extrinsic apoptotic stimuli. The results revealed that loss of both Bax and Bak or Bax alone can confer resistance to multiple death signals, whereas Bak deficiency does not abrogate apoptosis, suggesting that Bax but not Bak is a determinant of chemosensitivity in response to diverse apoptotic stimuli in human HCT116 cells (Fig. 5).
Fig. 5.
Susceptibility of human colon cancer cells to multiple apoptotic stimuli. Wild-type (WT) and Bax-/- HCT116 cells were transfected with Bak siRNA or LacZ control siRNA oligos. Twenty-four hours after transfection, cells (WT, Bak-deficient, Bax-/-, Bax-/- Bak-deficient, and p53-/-) were not treated or treated with 50 μg/ml CPT-11, 0.5 μM staurosporine, or 100 ng/ml TRAIL and 1 μg/ml anti-His-6 antibody for 24 h. Cell death was quantified by flow cytometric detection of propidium iodide staining.
In conclusion, these results suggest that p53-dependent up-regulation of KILLER/DR5 contributes dramatically to restoration of TRAIL sensitivity to Bax-/- HCT116 cells upon DNA damage. Cells lacking proapoptotic Bax and Bak molecules leading to inhibition of mitochondria-dependent events promoting cell death are resistant to multiple DNA-damaging agents, but surprisingly remain susceptible to resensitization to killing by TRAIL plus chemotherapy. In addition, Bax plays a critical role in the apoptosis induced by a number of apoptotic agents, and Bax and Bak may not possess fully overlapping functions in human colorectal cancer cells.
Discussion
In the present study, we determined the importance of p53 in sensitizing Bax-/- human colon cancer cells to TRAIL in the presence of chemotherapeutic agent (Fig. 2). A second advance was to employ in vivo bioluminescence imaging of p53 transcriptional activity to support the conclusion that p53 activity is required for sensitizing Bax-/- HCT116 tumors to TRAIL plus chemotherapy. Optical imaging of gene expression in living subjects is a rapidly evolving area of molecular imaging research and is crucial for many applications, including gene therapy, tumor pathogenesis and metastasis, and transgenic models (25, 31, 35). Here, we directly observed the involvement of functional p53 transcriptional activity in reversal of TRAIL sensitivity in Bax-/- HCT116 cells in culture and in living nude mice (Figs. 2B and 3). Our results indicated that p53 activation is a prerequisite for restoration of TRAIL sensitivity in mismatch repair-deficient Bax-/- HCT116 cells.
Most chemotherapeutic agents activate the intrinsic apoptotic pathway, which usually requires p53. Mutations in p53 or in the p53 pathway can produce multiple drug resistance phenotype both in vitro and in vivo, and reintroduction of wild-type p53 into p53-/- tumor cells can reestablish chemosensitivity (36). The contribution of the death receptor pathway in chemotherapeutic drug-induced cell death is not well established. Treatment of tumor cells with drugs can induce CD95 and KILLER/DR5. It has been argued that certain cell types (type I cells) require both the death receptor and mitochondrial pathways for drug-induced apoptosis, whereas others (type II cells) require only mitochondrial pathway (22). Overexpression of Bcl-2 is known to block death receptor-mediated apoptosis in type II but not type I cells. The same applies for type I versus type II tissues in vivo. Moreover, either increased expression of antiapoptotic Bcl-2 family or loss of proapoptotic Bax and Bak suffices to block apoptosis induced by death receptors in type II cells, as demonstrated by a recent study showing that hepatocytes of Bax/Bak double knockout mice resist death receptor-induced apoptosis in vivo (34). We previously demonstrated that HCT116 human colon cancer cells behave in a type II manner, such that inhibition of caspase 9 led to the prevention of TRAIL-induced apoptosis (21). In this study, we investigated the abilities of Bax-/- Bak-deficient human HCT116 to undergo apoptosis induced by multiple stimuli. The results revealed that Bax-/- Bak-deficient HCT116 cells evaded apoptosis induced by intrinsic as well as extrinsic apoptotic stimuli, whereas wild-type cells were extremely sensitive to these stimuli (Fig. 5). These results further confirm that HCT116 cells are type II cells. Our current studies with siRNAs indicated that the death receptor pathway (type I) contributes significantly to the rescue of TRAIL sensitivity by chemotherapy in Bax-/- HCT116 cells, because blockade of KILLER/DR5 expression by siRNA dramatically inhibited TRAIL-induced apoptosis after DNA damage in Bax-/-HCT116 cells (Fig. 4A). Surprisingly, silencing the expression of the Bak gene, which cooperates with Bax to promote cytochrome c release from the mitochondria, only slightly decreased apoptosis (Fig. 4B). In addition, caspase 8 inhibition protects both CPT-11 pretreated wild-type and Bax-/- HCT116 cells from TRAIL-induced apoptosis, whereas caspase 9 inhibition only rescued the wild-type HCT116 cells from death induced by TRAIL (Fig. 4C). The failure of the caspase 9 inhibitor to suppress apoptosis as potently in Bax-/- cells as it does in wild-type cells was not caused by differences in the extent of caspase 9 suppression in the two cell types (Fig. 4C). Thus, although the caspase 9 inhibitor rescued procaspase 9 from depletion after TRAIL plus CPT-11 exposure of either wild-type or Bax-/- HCT116 cells, the inhibitor did not significantly rescue the Bax-/- cells from death. These findings suggest that chemotherapy restores the extrinsic apoptotic-signaling pathway activated by TRAIL, implying a conversion of HCT116 cells from a type II into a type I cell line. This conversion in the apoptotic mechanism from a type II pathway requiring Bax and mitochondrial events to a type I pathway involving cytoplasmic caspase activation suggests a novel approach for reversing TRAIL resistance.
Earlier studies with murine embryonic fibroblasts (MEFs) showed that MEFs lacking both Bax and Bak, but not cells lacking only one of these components, exhibited complete resistance to multiple apoptotic stimuli, suggesting that both Bax and Bak are functionally overlapping, and that these proapoptotic “multidomains” constitute an essential “gateway” to apoptosis (34). Surprisingly, our observation that silencing the expression of Bak in Bax-/- human colon cancer cells appears not to significantly confer resistance to apoptosis when cells are exposed to TRAIL plus genotoxic agents (Fig. 4B) implies that the requirement of multidomain proteins may be cell type or species dependent (37). Therefore, we also investigated the relative importance of Bax and Bak for apoptosis induced by multiple stimuli in human colon cancer cells with different genetic backgrounds. It has been previously shown that wild-type HCT116 cells are very sensitive to TRAIL treatment alone and that chemotherapy greatly enhanced TRAIL-induced apoptosis in the cells. The results showed that wild-type and Bak-deficient HCT116 cells were extremely sensitive to apoptosis with similar efficiency, whereas the p53-/-, Bax-/-, and Bax-/- Bak-deficient HCT116 cells remained largely viable upon exposure to different apoptotic stimuli (Fig. 5). Hence, we conclude that Bax plays a major role in promoting cell death under circumstances where apoptosis proceeds through the mitochondrial pathway in human cells. Bax appears to be a major determinant of chemosensitivity, whereas Bak does not substitute for Bax during apoptosis of HCT116 human colon carcinoma cells. A recent report showed that Bak deficiency is associated with delayed kinetics of Bax translocation, but does not affect either the oligomerization of translocated Bax or the leakage of cytochrome c (38).
In summary, our loss-of-function studies reveal that p53 is required to sensitize Bax-/- human colon cancer cells to TRAIL. p53-dependent up-regulation of KILLER/DR5 contributes significantly to restoration of TRAIL sensitivity in Bax-/- cells upon DNA damage. siRNA directed at proapoptotic targets downstream of p53 reveals a critical role for TRAIL receptor KILLER/DR5 and a less important role for proapoptotic gene Bak which cooperates with Bax in promoting mitochondrial cytochrome c release. Our results support the hypothesis that resensitization of mismatch repair-deficient Bax-/- cells to TRAIL involves p53 activation of KILLER/DR5 and reconstitution of an extrinsic type I death pathway. Therefore, the present studies provide further insights into the mechanisms of restoration of TRAIL resistance, and may aid the development and application of TRAIL-based combinatorial regimens for the treatment of colorectal cancers. In particular, it is suggested that mitochondrial apoptotic defects do not necessarily represent an obstacle for the future design of anticancer therapy that redirects death signaling toward the death receptor cascade. Our results also indicate that it would be useful to identify small molecules that can reverse TRAIL resistance in cancer cells containing mitochondrial apoptotic defects as well as p53 mutations. Because the majority of human tumors including colorectal cancers contain p53 mutations, it is necessary to perform such screening by using high throughput bioluminescence imaging strategies. Reversal of TRAIL resistance in cancers with mitochondrial apoptotic defects and p53 mutations appears to be a viable strategy and is expected to impact in the future on anticancer therapeutic design.
Acknowledgments
We thank Wenge Wang for assistance with tail-vein injections and helpful comments on the manuscript. S.W. received the American Association for Cancer Research Scholar-in-Training Award. This work has been supported in part through funds provided by the Howard Hughes Medical Institute and National Institutes of Health Grants CA 75138 (to W.S.E.-D.) and CA 75454 (to W.S.E.-D.). W.S.E.-D. is an Assistant Investigator of the Howard Hughes Medical Institute.
This work was presented at the 94th Annual American Association for Cancer Research meeting, Washington, DC, July 11-14, 2003.
Abbreviations: TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; DR5, death receptor 5; siRNA, small interfering RNA; Ad-E6, E6-expressing adenovirus.
References
- 1.Walczak, H., Miller, R. E., Ariail, K., Gliniak, B., Griffith, T. S., Kubin, M., Chin, W., Jones, J., Woodward, A., Le, T., et al. (1999) Nat. Med. 5, 157-163. [DOI] [PubMed] [Google Scholar]
- 2.Takeda, K., Hayakawa, Y., Smyth, M. J., Kayagaki, N., Yamaguchi, N., Kakuta, S., Iwakura, Y., Yagita, H. & Okumura, K. (2001) Nat. Med. 7, 94-100. [DOI] [PubMed] [Google Scholar]
- 3.Ashkenazi, A. & Dixit, V. M. (1998) Science 281, 1305-1308. [DOI] [PubMed] [Google Scholar]
- 4.Wu, G. S., Burns, T. F., McDonald, E. R., III, Jiang, W., Meng, R., Krantz, I. D., Kao, G., Gan, D. D., Zhou, J. Y., Muschel, R., et al. (1997) Nat. Genet. 17, 141-143. [DOI] [PubMed] [Google Scholar]
- 5.Ichikawa, K., Liu, W., Zhao, L., Wang, Z., Liu, D., Ohtsuka, T., Zhang, H., Mountz, J. D., Koopman, W. J., Kimberly, R. P. & Zhou, T. (2001) Nat. Med. 7, 954-960. [DOI] [PubMed] [Google Scholar]
- 6.Burns, T. F. & El-Deiry, W. S. (2001) J. Biol. Chem. 276, 37879-37886. [DOI] [PubMed] [Google Scholar]
- 7.LeBlanc, H., Lawrence, D., Varfolomeev, E., Totpal, K., Morlan, J., Schow, P., Fong, S., Schwall, R., Sinicropi, D. & Ashkenazi, A. (2002) Nat. Med. 8, 274-281. [DOI] [PubMed] [Google Scholar]
- 8.Ryan, K. M., Phillips, A. C. & Vousden, K. H. (2001) Curr. Opin. Cell Biol. 13, 332-337. [DOI] [PubMed] [Google Scholar]
- 9.El-Deiry, W. S. (2001) Cell Death Differ. 8, 1066-1075. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt, C. A., Fridman, J. S., Yang, M., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cancer Cell 1, 289-298. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W. & Vogelstein, B. (1993) Cell 75, 817-825. [DOI] [PubMed] [Google Scholar]
- 12.Haupt, Y., Rowan, S., Shaulian, E., Vousden, K. H. & Oren, M. (1995) Genes Dev. 9, 2170-2183. [DOI] [PubMed] [Google Scholar]
- 13.Burns, T. F., Bernhard, E. J. & El-Deiry, W. S. (2001) Oncogene 20, 4601-4612. [DOI] [PubMed] [Google Scholar]
- 14.Wu, G. S., Burns, T. F., McDonald, E. R., III, Meng, R. D., Kao, G., Muschel, R., Yen, T. & El-Deiry, W. S. (1999) Oncogene 18, 6411-6418. [DOI] [PubMed] [Google Scholar]
- 15.Miyashita, T. & Reed, J. C. (1995) Cell 80, 293-299. [DOI] [PubMed] [Google Scholar]
- 16.Sax, J. K., Fei, P., Murphy, M. E., Bernhard, E., Korsmeyer, S. J. & El-Deiry, W. S. (2002) Nat. Cell Biol. 4, 842-849. [DOI] [PubMed] [Google Scholar]
- 17.MacLachlan, T. K. & El-Deiry, W. S. (2002) Proc. Natl. Acad. Sci. USA 99, 9492-9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman, W. H., Biade, S., Zilfou, J. T., Chen, J. & Murphy, M. (2002) J. Biol. Chem. 277, 3247-3257. [DOI] [PubMed] [Google Scholar]
- 19.Wu, Y., Mehew, J. W., Heckman, C. A., Arcinas, M. & Boxer, L. M. (2001) Oncogene 20, 240-251. [DOI] [PubMed] [Google Scholar]
- 20.Scaffidi, C., Fulda, S., Srinivasan, A., Friesen, C., Li, F., Tomaselli, K. J., Debatin, K. M., Krammer, P. H. & Peter, M. E. (1998) EMBO J. 17, 1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozoren, N., Kim, K., Burns, T. F., Dicker, D. T., Moscioni, A. D. & El-Deiry, W. S. (2000) Cancer Res. 60, 6259-6265. [PubMed] [Google Scholar]
- 22.Ashkenazi, A. (2002) Nat. Rev. Cancer 2, 420-430. [DOI] [PubMed] [Google Scholar]
- 23.Green, D. R. (2000) Cell 102, 1-4. [DOI] [PubMed] [Google Scholar]
- 24.Prabhu, N. S., Somasundaram, K., Satyamoorthy, K., Herlyn, M. & El-Deiry, W. S. (1998) Int. J. Oncol. 13, 5-9. [DOI] [PubMed] [Google Scholar]
- 25.Bhaumik, S. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tijsterman, M., Ketting, R. F. & Plasterk, R. H. (2002) Annu. Rev. Genet. 36, 489-519. [DOI] [PubMed] [Google Scholar]
- 27.Chiu, Y. L. & Rana, T. M. (2002) Mol. Cell 10, 549-561. [DOI] [PubMed] [Google Scholar]
- 28.Wang, W. & El-Deiry, W. S. (2003) Cancer Biol. Ther. 2, 196-202. [DOI] [PubMed] [Google Scholar]
- 29.Rampino, N., Yamamoto, H., Ionov, Y., Li, Y., Sawai, H., Reed, J. C. & Perucho, M. (1997) Science 275, 967-969. [DOI] [PubMed] [Google Scholar]
- 30.Evan, G. & Littlewood, T. (1998) Science 281, 1317-1322. [DOI] [PubMed] [Google Scholar]
- 31.Paulmurugan, R., Umezawa, Y. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 15608-15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashkenazi, A. & Dixit, V. M. (1999) Curr. Opin. Cell Biol. 11, 255-260. [DOI] [PubMed] [Google Scholar]
- 33.Wajant, H. (2002) Science 296, 1635-1636. [DOI] [PubMed] [Google Scholar]
- 34.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B. & Korsmeyer, S. J. (2001) Science 292, 727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spotts, J. M., Dolmetsch, R. E. & Greenberg, M. E. (2002) Proc. Natl. Acad. Sci. USA 99, 15142-15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace-Brodeur, R. R. & Lowe, S. W. (1999) Cell Mol. Life Sci. 55, 64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theodorakis, P., Lomonosova, E. & Chinnadurai, G. (2002) Cancer Res. 62, 3373-3376. [PubMed] [Google Scholar]
- 38.Mikhailov, V., Mikhailova, M., Degenhardt, K., Venkatachalam, M. A., White, E. & Saikumar, P. (2003) J. Biol. Chem. 278, 5367-5376. [DOI] [PubMed] [Google Scholar]