Abstract
The 8q24 locus, which contains the thyroglobulin (Tg) gene, was previously shown to be strongly linked with autoimmune thyroid disease (AITD). We sequenced all 48 exons of the Tg gene and identified 14 single-nucleotide polymorphisms (SNPs). Case control association studies demonstrated that an exon 10-12 SNP cluster and an exon 33 SNP were significantly associated with AITD (P < 0.01). Haplotype analysis demonstrated that the combination of these two SNP groups was more significantly associated with AITD (P < 0.001). Gene-gene interaction studies provided evidence for an interaction between HLA-DR3 and the exon 33 SNP, giving an odds ratio of 6.1 for Graves' disease. We then sequenced exons 10,12, and 33 of the mouse Tg gene in 19 strains of mice. Fifty percent of the strains susceptible to thyroiditis had a unique SNP haplotype at exons 10 and 12, whereas none of the mouse strains that were resistant to thyroiditis had this SNP haplotype (P = 0.01). We concluded that Tg is a susceptibility gene for AITD, both in humans in and in mice. A combination of at least two Tg SNPs conferred susceptibility to human AITD. Moreover, the exon 33 SNP showed evidence for interaction with HLA-DR3 in conferring susceptibility to Graves' disease.
Keywords: Graves' disease, Hashimoto's disease, thyroiditis
The autoimmune thyroid diseases (AITDs), including Graves' disease (GD) and Hashimoto's thyroiditis (HT), are among the most common human autoimmune diseases with recent data showing a prevalence of clinical AITD of up to 1% in the U.S. population (1, 2). The hallmark of GD is the production of thyroid-stimulating hormone receptor (TSHR)-stimulating antibodies causing hyperthyroidism, whereas HT is characterized by the apoptosis of thyrocytes, leading to hypothyroidism (reviewed in refs. 3 and 4). However, despite their contrasting clinical presentations, GD and HT share many features in common, mainly, infiltration of the thyroid by T cells and production of antithyroid autoantibodies [antithyroglobulin and anti-thyroid peroxidase (TPO) antibodies] (3-5). AITDs are complex diseases, which are caused by an interaction between susceptibility genes (6-8) and nongenetic factors, such as infection (9-12). This paradigm is based on solid epidemiologic evidence demonstrating a genetic predisposition to AITDs, including: (i) familial clustering (13); (ii) a sibling risk ratio (λs) of >10 (7, 14); (iii) a high concordance rate in monozygotic twins when compared with dizygotic twins (15-18); and (iv) the presence of thyroid autoantibodies, which are markers of sub-clinical AITD, in up to 50% of siblings of patients with AITD (19, 20).
We have previously performed a whole-genome scan in a dataset of 102 multiplex, multigenerational AITD families (540 individuals; refs. 21-23). Three loci on chromosomes 6p, 8q, and 10q showed evidence for linkage with the entire AITD dataset giving maximum multipoint heterogeneity logarithm of odds (lod) scores of 2.0, 3.5, and 4.1, respectively. The 8q24 locus was also reported to be linked with AITD in a dataset of 123 Japanese sibling pairs (24). The 8q24 locus contains the thyroglobulin (Tg) gene, one of the major autoantigens in AITDs.
The AITDs are characterized by cellular and humoral immune responses to Tg, and, thus, the Tg gene is a strong positional candidate gene for AITD (5). We therefore proceeded to analyze the Tg gene directly. Sequence alignment searches revealed three new microsatellites inside introns 10, 27, and 23 of the Tg gene (designated Tgms1, 2, and 3, respectively). Tgms2 was the most informative microsatellite and it showed strong evidence for linkage with AITD (lod score = 2.9; ref. 23). Moreover, association studies showed a significant association of Tgms2 with AITD (23; P = 0.004). These results have been replicated recently in a U.K. dataset (25). As in our study, the U.K. study also showed a significant association between Tgms2 and AITD (P < 0.001). Moreover, the same Tgms2 allele that we found to be associated with AITD was found to be associated by Collins et al. (25). The fact that a microsatellite inside the Tg gene was linked and associated with AITD suggested that the Tg gene was the AITD susceptibility gene on 8q24. We therefore sequenced the thyroglobulin gene in AITD patients and controls, as well as in mice susceptible and resistant to autoimmune thyroiditis induced by Tg immunization. Here, we report the identification of amino acid substitutions in the thyroglobulin gene predisposing to human AITD, as well as to murine experimental autoimmune thyroiditis (EAT).
Materials and Methods
Patients and Controls. AITD patients. The project was approved by the Mount Sinai School of Medicine Institutional Review Board. Two hundred eighty-five Caucasian AITD patients were studied, 99 of them were probands from our AITD families, which were used in our whole-genome scan (22). The probands came from Caucasian families that were multiplex for AITD (more than one affected) and/or multigenerational. There were a total of 193 GD patients and 92 HT patients.
Clinical assessment. Graves' disease was diagnosed by (i) documented clinical and biochemical hyperthyroidism requiring treatment, (ii) a diffuse goiter, or (iii) presence of TSHR antibodies and/or a diffuse thyroid scan. TSHR antibodies were measured by using a radioreceptor assay (Kronus, Boise, ID). HT was diagnosed by documented clinical and biochemical hypothyroidism requiring thyroid hormone replacement and the presence of anti-TPO, with or without anti-Tg antibodies, with or without goiter. Anti-Tg and anti-TPO antibodies were measured by specific RIA (Kronus). For all subjects, phenotype was determined with the clinician blinded to the individual's genotype.
Controls. One hundred fifty age- and sex-matched healthy Caucasian volunteers served as controls in our association studies. All controls had no personal or family history of thyroid disease, and no goiter on examination; they had normal thyroid functions and were negative for thyroid autoantibodies.
Sequencing the Human Tg Gene. The Tg gene has 48 exons (26), and the primers used for amplifying the Tg exons are given in Table 7, which is published as supporting information on the PNAS web site. Each exon was sequenced by using flanking primers that were ≈100 base pairs upstream from 5′ intron-exon junction or downstream from the 3′ intron-exon junction. This approach enabled us to sequence all regions that could affect the amino acid sequence of Tg, and potential alternative splicing sites of Tg (the binding sites for the spliceosome) (27-29). Genomic DNA was amplified by using these primers and the PCR product was sequenced as described (30). We sequenced each exon of the Tg gene in 10 unrelated normal controls to identify common single-nucleotide polymorphisms (SNPs) (i.e., SNPs that show in healthy individuals with a frequency of >10%). In addition, we sequenced the Tg gene in five affected sib pairs from five families that showed positive lod scores at the 8q24 region.
Analysis of the Human Tg SNPs. Fourteen SNPs were identified within the Tg gene (see Results). These SNPs were analyzed either by a fluorescent-based restriction fragment length polymorphism method (30) or by the SNAPshot method (PE Applied Biosystems, Foster City, CA). The details of the primers, restriction enzymes, and fragment size for SNP genotyping are available in Table 8, which is published as supporting information on the PNAS web site.
Analysis of a Large Insertion/Deletion (INDEL) Polymorphism of 1,464 Base Pairs in the Tg Gene. Recently (31), a large INDEL polymorphism was identified in intron 18 of the Tg gene. The INDEL polymorphism was also analyzed for association with AITD. The INDEL polymorphism was analyzed by amplification with sequence specific primers. The primer sequences are available in Table 8.
Sequencing Exons 10, 12, and 33 of the Mouse Tg Gene. Genomic DNA from 19 strains of mice, CBA/J (H-2k), C3H/HeJ (H-2k), C3HeB/FeJ (H-2k), CE/J (H-2k), AKR/J (H-2k), B10.BR/SgSnJ (H-2k), MA/MyJ (H-2k), RF/J (H-2k), ST/bJ (H-2k), B6C3F1/J (H-2b, k), C57BL/6J (H-2b), 129/J (H-2b), C57L/J (H-2b), LP/J (H-2b), BALB/cJ (H-2d), DBA/2J (H-2d), C57BLKS/J (H-2d), PL/J (H-2a), and DBA/1J (H-2q), was purchased from The Jackson Laboratory. Exons 10, 12, and 33 of the mouse Tg (mTg) gene, which show 75-80% homology with the corresponding exons in the human Tg gene (32), were amplified and sequenced. The primers used for amplification and sequencing of these mTg exons are given in Table 9, which is published as supporting information on the PNAS web site. All primers were intronic at an average distance of 100 bases from the 3′ or 5′ end of the exons. Mouse genomic DNA was amplified by using these primers and the PCR product was sequenced as described (30).
Association Analyses. Case control association analyses were performed by using the χ2 and Fisher's exact tests with Yates correction. The odds ratio (OR) was calculated by the method of Woolf (33). A P value of <0.05 was considered significant.
Results
Sequence Analysis of the Tg Gene and SNP Identification. The National Center for Biotechnology Information Tg gene transcript (accession no. NM 003235) and all sequence fragments from the Celera database (255 fragments) that matched the Tg transcript were aligned together by using the CAP3 program (34) to identify mismatches that could potentially reflect true SNPs. In the case of two exons (9 and 36), no matching fragments were found and the relevant portion of the Celera gene sequence was used instead. Two or more mismatches at a specific nucleotide (indicating high probability for the existence of a true SNP and less likely sequencing errors) were identified in 11 exons, demonstrating the existence of a large number of sequence polymorphisms in the Tg gene. We therefore sequenced the entire Tg gene (48 exons) in 10 affected individuals (five sib pairs) from five families that showed positive lod scores at the 8q24 region, and 10 randomly selected healthy controls.
Fourteen Tg gene SNPs were identified; 10 exonic and 4 intronic (Table 1). Eight of the 10 exonic SNPs were predicted by our sequence alignment, demonstrating that sequence alignment alone is a powerful tool for SNP discovery. Of the 10 exonic SNPs, 2 were silent (i.e., did not cause an amino acid substitution), and 8 caused an amino acid substitution. Three of the SNPs were nonconservative (Table 1). Linkage disequilibrium (LD) testing by using the snphap software (www-gene.cimr.cam.ac.uk/clayton/software/snphap.txt), demonstrated the existence of three LD groups of SNPs (exons 10-12 cluster, exons 18-21 cluster, and exon 34 cluster).
Table 1. Fourteen SNPs of the human Tg gene.
| Marker name | Allele | SNP location | Nucleotide position (mRNA) | Amino acid position | Amino acid substitution |
|---|---|---|---|---|---|
| E10SNP24 | T/G | Exon 10 | 2200 | 734 | Ser/Ala |
| E10SNP158 | T/C | Exon 10 | 2334 | 778 | Pro/Pro |
| E12SNP | A/G | Exon 12 | 3082 | 1027 | Met/Val |
| E18SNP-20 | T/C | Intron 17 | — | — | — |
| E18SNP88 | A/G | Exon 18 | 3935 | 1293 | Asp/Gly |
| E21SNP | T/C | Exon 21 | 4506 | 1501 | Ala/Ala |
| E27SNP-30 | C/T | Intron 26 | — | — | — |
| E29SNP | G/A | Exon 29 | 5512 | 1837 | Asn/Asp |
| E33SNP | C/T | Exon 33 | 5995 | 1980 | Arg/Trp |
| E34SNP+33 | A/G | Intron 34 | — | — | — |
| E34SNP+73 | G/A | Intron 34 | — | — | — |
| E43SNP4 | C/T | Exon 43 | 7408 | 2469 | Leu/Leu |
| E43SNP97 | T/C | Exon 43 | 7501 | 2500 | Arg/Trp |
| E46SNP | C/T | Exon 46 | 7920 | 2621 | Tyr/Tyr |
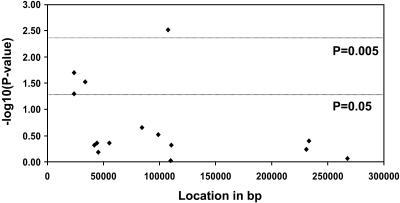
Identifying the Tg SNPs Associated with AITD. Case control association studies were performed for the 14 discovered Tg SNPs in 285 AITD patients and 150 controls. One SNP cluster (the exons 10-12 cluster) and the exon 33 SNP showed significant associations with AITD (Fig. 1 and Tables 10-12, which are published as supporting information on the PNAS web site). Subset analysis showed that these four SNPs were associated, with both GD and HT giving similar ORs (Tables 10-12). We then analyzed interactions between these SNPs and examined whether certain combinations conferred stronger susceptibility for AITD. The analysis demonstrated that (i) any of the three SNPs of the exons 10-12 cluster in combination with the exon 33 SNP were strongly associated with AITD; and (ii) the association was strongest when an individual was homozygous for the exon 33 SNP susceptibility allele, and were either homozygous or heterozygous for the exons 10-12 SNP susceptibility alleles (Tables 2, 3, 4). Thus, susceptibility to AITD was conferred by inheritance of both copies of the susceptibility allele of exon 33 SNP (recessive inheritance) and one or two copies of the susceptibility allele of either one of the SNPs of the exons 10-12 cluster (dominant inheritance). While we cannot determine which SNP of the exons 10-12 SNP cluster is the AITD-susceptibility SNP because of the strong LD between them, it is unlikely to be E10SNP158, which is a silent SNP, and is more likely to be either E10SNP24 or E12 SNP. Separate analyses for GD and HT showed that both GD and HT were associated with the exons 10-12 and exon 33 SNPs with similar ORs (Tables 2, 3, 4).
Fig. 1.
Association analysis comparing the frequencies of 14 Tg SNPs and a large INDEL polymorphism in 285 AITD patients and 150 controls. Shown are case control association plots [-log10 (P value) versus location in base pairs] for our population. The x axis shows the relative position (in base pairs) of each SNP in the genomic sequence of the Tg gene.
Table 2. Association of genotype combinations in the Tg gene with AITD.
| E10SNP24 (T/G) | E10SNP158 (T/C) | E12SNP (A/G) | E33SNP (C/T) | AITD, % (n = 277) | Control, % (n = 140) | P value | OR |
|---|---|---|---|---|---|---|---|
| T allele | T allele | A allele | CC | 91 (32.9) | 23 (16.2) | 0.0004 | 2.49 |
| T allele | — | — | CC | 92 (33.0) | 24 (16.7) | 0.0005 | 2.42 |
| — | T allele | — | CC | 93 (33.1) | 23 (16.4) | 0.0003 | 2.52 |
| — | — | A allele | CC | 93 (32.9) | 24 (16.6) | 0.0004 | 2.43 |
Table 3. Association of genotype combinations in the Tg gene with GD.
| E10SNP24 (T/G) | E10SNP158 (T/C) | E12SNP (A/G) | E33SNP (C/T) | GD, % (n = 186) | Control, % (n = 140) | P value | OR |
|---|---|---|---|---|---|---|---|
| T allele | T allele | A allele | CC | 62 (33.3) | 23 (16.2) | 0.0006 | 2.54 |
| T allele | — | — | CC | 62 (33.2) | 24 (16.9) | 0.0009 | 2.44 |
| — | T allele | — | CC | 64 (33.7) | 23 (16.4) | 0.0004 | 2.58 |
| — | — | A allele | CC | 63 (33.0) | 24 (16.8) | 0.0008 | 2.44 |
Table 4. Association of genotype combinations in the Tg gene with HT.
| E10SNP24 (T/G) | E10SNP158 (T/C) | E12SNP (A/G) | E33SNP (C/T) | HT, % (n = 91) | Control, % (n = 140) | P value | OR |
|---|---|---|---|---|---|---|---|
| T allele | T allele | A allele | CC | 29 (31.9) | 23 (16.2) | 0.006 | 2.38 |
| T allele | — | — | CC | 30 (32.6) | 24 (16.9) | 0.005 | 2.38 |
| — | T allele | — | CC | 29 (31.9) | 23 (16.4) | 0.006 | 2.38 |
| — | — | A allele | CC | 30 (32.6) | 24 (16.8) | 0.005 | 2.40 |
Recently, a novel large INDEL polymorphism of 1,464 base pairs was identified in intron 18 of the Tg gene (31). We found no significant association between AITD and the INDEL polymorphism (Fig. 1 and Tables 10-12).
Sequence Analysis for Rare Sequence Variants. It was possible that additional rare sequence variants (not detected in our sequencing of 10 patients and 10 controls) in tight LD with E10SNP24, E12SNP, and E33SNP might be the causative polymorphisms for AITD in the Tg gene. Therefore, we sequenced exons 10, 12, and 33 in 190 AITD patients to look for these possible rare variants. Only one rare SNP was identified in exon 12, but it was a silent SNP (did not cause an amino acid change) and was not associated with AITD (data not shown).
Interaction Between HLA-DR3 and the Tg SNPs. Because HLA-DR3 is associated with GD (35, 36), we analyzed for gene-gene interaction between the associated Tg SNPs and HLA-DR3. Case control association analysis of the Tg SNPs in combination with HLA-DR3 showed evidence for an interaction between HLA-DR3 and the exon 33 SNP, giving an OR of 6.1 for GD (Table 5).
Table 5. Interraction between the Tg exon 33 SNP and HLA-DR3.
| Polymorphism | Allele | GD patients (n = 191) | Controls (n = 150) | OR |
|---|---|---|---|---|
| Tg-E33SNP | CC | 81 | 38 | 2.0 |
| Others | 110 | 105 | ||
| HLA-DR3 | DR3 | 69 | 22 | 3.3 |
| Others | 120 | 128 | ||
| Tg-E33SNP + HLA-DR3 | CC + DR3 | 34 | 5 | 6.1 |
| Others | 154 | 138 |
Tg SNPs Are Associated with EAT in Mice. Because mTg has 71.8% homology (77.3% for exon 10, 76.1% for exon 12, and 81.2% for exon 33) with human Tg (32), we hypothesized that sequence variants in mTg predisposing to EAT may be localized close to the positions where we found the causative SNPs for human AITD; i.e., in exons 10, 12, and 33 of the mTg gene. We therefore sequenced exons 10, 12, and 33 of mTg in 19 strains of mice, 10 susceptible to EAT (CBA/J, C3H/HeJ, C3HeB/FeJ, AKR/J, B10.BR/SgSnJ, CE/J, MA/MyJ, RF/J, ST/bJ, and B6C3F1/J), seven resistant (C57BL/6J, 129/J, C57L/J, LP/J, BALB/cJ, DBA/2J, and C57BLKS/J), and two partially resistant (PL/J and DBA/1J; refs. 37-39; Table 6). Seven SNPs were discovered in and around exon 10 of mTg (two intronic, five exonic, and two of them silent); four SNPs were discovered in and around exon 12 (two intronic, two exonic, and both of them silent); and no SNPs were discovered in exon 33 (as for human Tg, all of the exon 10 and 12 SNPs were in tight LD). Thus, amino acid substitutions were identified only in exon 10 of the mTg gene. There was a significant association between the exon 10 SNP haplotypes and susceptibility to thyroiditis in mice. Five of 10 (50%) mouse strains susceptible to thyroiditis had the haplotype Ser-Met-Thr for exon 10, whereas all of the mouse strains that were resistant to thyroiditis had the haplotype Asn-Val-Ile for exon 10 (χ2 = 6.1, P = 0.01; Table 6). One of the mouse strains susceptible to thyroiditis (RF/J) had the haplotype Asn-Met-Ile for exon 10 (Table 6). Therefore, Met at position 808 of the mTg gene was found in 6/10 (60%) of susceptible mouse strains and in none of the resistant strains (χ2 = 7.9, P = 0.005). In summary, SNPs in exons 10 and 12 of the Tg gene correlated with the susceptibility to autoimmune thyroid disease in both humans and in mice.
Table 6. Exon 10 haplotypes of the mouse Tg gene.
| Mouse strain | MHC | EAT | Exon 10 haplotype |
|---|---|---|---|
| CBA/J | H-2k | Susceptible | Ser-Met-Thr* |
| C3H/HeJ | H-2k | Susceptible | Ser-Met-Thr |
| C3HeB/FeJ | H-2k | Susceptible | Ser-Met-Thr |
| CE/J | H-2k | Susceptible | Ser-Met-Thr |
| B6C3F1/J | H-2b,k | Susceptible | Ser-Met-Thr/Asn-Val-Ile |
| RF/J | H-2k | Susceptible | Asn-Met-Ile |
| AKR/J | H-2k | Susceptible | Asn-Val-Ile† |
| B10.BR/SgSnJ | H-2k | Susceptible | Asn-Val-Ile |
| MA/MyJ | H-2k | Susceptible | Asn-Val-Ile |
| ST/bJ | H-2k | Susceptible | Asn-Val-Ile |
| C57BL/6J | H-2b | Resistant | Asn-Val-Ile |
| 129/J | H-2b | Resistant | Asn-Val-Ile |
| C57L/J | H-2b | Resistant | Asn-Val-Ile |
| LP/J | H-2b | Resistant | Asn-Val-Ile |
| BALB/cJ | H-2d | Resistant | Asn-Val-Ile |
| DBA/2J | H-2d | Resistant | Asn-Val-Ile |
| C57BLKS/J | H-2d | Resistant | Asn-Val-Ile |
| PL/J | H-2a | Partial resistance | Asn-Val-Ile |
| DBA/1J | H-2a | Partial resistance | Asn-Val-Ile |
The amino acid positions are as follows: Ser-757, Met-808, Thr-919
The amino acid positions are as follows: Asn-757, Val-808, Ile-919
Discussion
This study provided further evidence that the Tg gene is an important susceptibility gene for AITD. Tg is one of the main autoantigens in AITD and anti-Tg antibodies are common in AITD (reviewed in ref. 5). There is abundant evidence that Tg plays an important role in the etiology of AITD, including: (i) anti-Tg antibodies are detected in almost all patients with AITD (5), and these Tg antibodies are restricted in their epitope specificity in contrast to the polyclonal nature of Tg antibodies found in healthy individuals (40); (ii) immunization with Tg induces autoimmune thyroiditis in experimental animals (37-39); and (iii) spontaneous models of autoimmune thyroiditis are also characterized by the development of anti-Tg antibodies (41). This article extended these observations and demonstrated that Tg was directly involved in the genetic etiology of AITD. The Tg gene region has been previously shown to be linked with AITD (23, 24), and our data identified the sequence changes in the Tg gene, which conferred this susceptibility to AITD. Case control association studies for 14 Tg SNPs in AITD patients and controls showed that one SNP cluster (the exons 10-12 cluster) and an exon 33 SNP were significantly associated with AITD (Fig. 1). Moreover, SNPs in exons 10 and 12 of mTg were also associated with EAT. Taken together, these data strongly supported the Tg gene as an important susceptibility gene for AITD.
Tg is one of the three genes encoding major disease-specific thyroid autoantigens, including the TPO and TSHR genes. Previous studies examining the TPO gene (42, 43) and the TSHR gene (44, 45) did not find linkage and failed to support them as major susceptibility genes for AITD in Caucasians. Thus, Tg is the first thyroid-autoantigen gene to be shown to confer susceptibility for AITD.
To date, three immunomodulatory genes have been shown to confer susceptibility to AITD: the HLA genes (reviewed in ref. 35), the CTLA-4 gene (30, 46, 47), and the CD40 gene (48, 49). Thus, it is likely that susceptibility to AITD involves an interaction between immune regulatory genes and at least one thyroid autoantigen-specific gene, namely Tg, as well as environmental factors. Indeed, our analysis for interaction between the associated Tg SNPs and HLA-DR3 showed evidence for an interaction between HLA-DR3 and the exon 33 SNP (Table 5). This proposed mechanism of immuneregulatory genes interacting with autoantigen-specific genes, may be a more general mechanism for the development of organ-specific autoimmune diseases. Indeed, this mechanism has been shown to play a central role in the etiology of type 1 (autoimmune) diabetes (T1D). In T1D, an immune-related gene, HLA-DQ, and an autoantigen gene, the insulin gene, VNTR, were shown to be linked and associated with disease (50). The VNTR polymorphism influences insulin gene expression (51), and was postulated to predispose to T1D by influencing the induction of tolerance to insulin in the thymus (52, 53), whereas HLA-DQ is believed to predispose to T1D by influencing the presentation of insulin peptides to T cells (54, 55). Thus, these two genes may interact and induce tolerance to insulin. In view of our findings, one may postulate a similar mechanism in AITD, where the susceptibility SNPs in Tg predispose to AITD by influencing the formation of immunogenic peptides and their presentation to T cells. However, further structure-function studies are required to substantiate this model.
Susceptibility to EAT in mice was traditionally thought to be influenced only by the mouse class II MHC genes (39, 56). However, further studies have shown that other genes outside the MHC gene complex must be involved in the susceptibility to EAT in mice (57, 58). Moreover, one study (59) has shown that induction of EAT by immunization with Tg from different strains of mice had a profound effect on the degree of thyroiditis developed, suggesting that alterations in the sequence/structure of mTg were important for the development of EAT. We therefore tested whether SNPs in mTg could affect the susceptibility to EAT and our results showed an association between SNPs in exon 10 of mTg and susceptibility to thyroiditis. Five of the ten (50%) mouse strains susceptible to thyroiditis had the haplotype Ser-Met-Thr for exon 10 SNPs, whereas all nine mouse strains that were resistant to thyroiditis had the haplotype Asn-Val-Ile for exon 10 SNPs (P = 0.01; the two exon 12 SNPs were also associated with EAT, but they were both silent SNPs; Table 6). This finding suggested that SNPs in exons 10 and/or 12 of mTg influence susceptibility and resistance to murine EAT. Interestingly, one of the mouse strains susceptible to thyroiditis (RF/J) was a recombinant and had the haplotype Asn-Met-Ile for exon 10 (Table 6), suggesting that V808M may be the susceptibility polymorphism in the mTg gene. It should be mentioned that EAT is not a perfect model for human AITD because it is induced by immunization with Tg and does not develop spontaneously. Moreover, one mouse strain, which is resistant to EAT (BALB/c), was recently shown to be susceptible to induction of a Graves'-like disease by DNA immunization with the TSH R gene (60).
The Tg gene may predispose to AITD by a number of mechanisms; for example: (i) sequence changes in Tg may change its antigenicity making it more immunogenic; (ii) sequence changes in Tg may change its interaction with HLA class II molecules; and (iii) sequence changes in Tg may influence its degradation by cathepsin S in endosomes, a process which has been recently shown to play an important role in development of autoimmunity (61).
While our data strongly suggest that Tg is an AITD susceptibility gene on chromosome 8q24, we cannot rule out the possibility that it may be in tight LD with another nearby gene. There is no absolute way of proving that a polymorphism in a gene actually causes disease, but structure-function studies provide strong evidence. For example, structure-function studies substantiated the role of HLA-DQ and the insulin gene, VNTR, in the etiology of T1D. Hence, it is important to note that the exon 33 SNP we identified caused a nonconservative amino acid substitution (Trp to Arg). The change from a hydrophobic amino acid (Trp) to a positively charged hydrophilic amino acid (Arg) would be expected to change the structure of Tg at this region.
In conclusion, Tg was confirmed as a susceptibility gene for AITD. A combination of amino acid substitutions at exons 10 and/or 12 and at exon 33 conferred susceptibility to AITD. The exon 33 amino acid substitution showed evidence for interaction with HLA-DR3 in conferring susceptibility to GD.
Supplementary Material
Acknowledgments
We thank Dr. Terry F. Davies for expert comments, advice, and critical review of the manuscript, and the Institute for Computational Biomedicine at Mount Sinai School of Medicine for bioinformatics support and infrastructure. This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK61659 and DK58072 (to Y.T.) and DK31775, NS27941, and MH48858 (to D.A.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AITD, autoimmune thyroid disease; Tg, thyroglobulin; SNP, single-nucleotide polymorphism; GD, Graves' disease; HT, Hashimoto's thyroiditis; TSH, thyroid-stimulating hormone; TSHR, TSH receptor; lod, logarithm of odds; EAT, experimental autoimmune thyroiditis; TPO, thyroid peroxidase; INDEL, insertion/deletion; LD, linkage disequilibrium; mTg, mouse Tg; T1D, type 1 diabetes; OR, odds ratio.
References
- 1.Jacobson, D. L., Gange, S. J., Rose, N. R. & Graham, N. M. (1997) Clin. Immunol. Immunopathol. 84, 223-243. [DOI] [PubMed] [Google Scholar]
- 2.Hollowell, J. G., Staehling, N. W., Flanders, W. D., Hannon, W. H., Gunter, E. W., Spencer, C. A. & Braverman, L. E. (2002) J. Clin. Endocrinol. Metab. 87, 489-499. [DOI] [PubMed] [Google Scholar]
- 3.Davies, T. F. (2000) in Werner and Ingbar's The Thyroid, eds. Braverman, L. E. & Utiger, R. D. (Lippincott, Williams & Wilkins, Philadelphia), pp. 518-530.
- 4.Weetman, A. P. (2000) in Werner and Ingbar's The Thyroid, eds. Braverman, L. E. & Utiger, R. D. (Lippincott-Raven, Philadelphia), pp. 721-732.
- 5.Tomer, Y. (1997) Clin. Immunol. Immunopathol. 82, 3-11. [DOI] [PubMed] [Google Scholar]
- 6.Weetman, A. P. & McGregor, A. M. (1994) Endocr. Rev. 15, 788-830. [DOI] [PubMed] [Google Scholar]
- 7.Brix, T. H., Kyvik, K. O., and Hegedus, L. (1998) Thyroid 8, 727-734. [DOI] [PubMed] [Google Scholar]
- 8.Tomer, Y. & Davies, T. F. (2003) Endocr. Rev. 24, 694-717. [DOI] [PubMed] [Google Scholar]
- 9.Tomer, Y. & Davies, T. F. (1993) Endocr. Rev. 14, 107-120. [DOI] [PubMed] [Google Scholar]
- 10.Huang, M. J., Tsai, S. L., Huang, B. Y., Sheen, I. S., Yeh, C. T. & Liaw, Y. F. (1999) Clin. Endocrinol. (Oxford) 50, 503-509. [DOI] [PubMed] [Google Scholar]
- 11.Onodera, T. & Awaya, A. (1990) Immunology 71, 581-585. [PMC free article] [PubMed] [Google Scholar]
- 12.Weetman, A. P. (1992) Clin. Endocrinol. (Oxford) 36, 307-323. [DOI] [PubMed] [Google Scholar]
- 13.Mather, B. A., Roberts, D. F., Scanlon, M. F., Mukhtar, E. D., Davies, T. F., Smith, B. R. & Hall, R. (1980) Clin. Endocrinol. (Oxford) 12, 155-163. [DOI] [PubMed] [Google Scholar]
- 14.Villanueva, R. B., Greenberg, D. A., Davies, T. F. & Tomer, Y. (2003) Thyroid 13, 761-764. [DOI] [PubMed] [Google Scholar]
- 15.Brix, T. H., Kyvik, K. O., Christensen, K. & Hegedus, L. (2001) J. Clin. Endocrinol. Metab 86, 930-934. [DOI] [PubMed] [Google Scholar]
- 16.Brix, T. H., Kyvik, K. O. & Hegedus, L. (2000) J. Clin. Endocrinol. Metab. 85, 536-539. [DOI] [PubMed] [Google Scholar]
- 17.Ringold, D. A., Nicoloff, J. T., Kesler, M., Davis, H., Hamilton, A. & Mack, T. (2002) Thyroid 12, 647-653. [DOI] [PubMed] [Google Scholar]
- 18.Phillips, D. I., Osmond, C., Baird, J., Huckle, A. & Rees-Smith, B. (2002) Thyroid 12, 377-380. [DOI] [PubMed] [Google Scholar]
- 19.Hall, R., Dingle, P. R. & Roberts, D. F. (1972) Clin. Genet. 3, 319-324. [DOI] [PubMed] [Google Scholar]
- 20.Burek, C. L., Hoffman, W. H. & Rose, N. R. (1982) Clin. Immunol. Immunopathol. 25, 395-404. [DOI] [PubMed] [Google Scholar]
- 21.Tomer, Y., Barbesino, G., Greenberg, D. A., Concepcion, E. S. & Davies, T. F. (1999) J. Clin. Endocrinol. Metab. 84, 4656-4664. [DOI] [PubMed] [Google Scholar]
- 22.Tomer, Y., Ban, Y., Concepcion, E. S., Barbesino, G., Villanueva, R. B., Greenberg, D. A. & Davies, T. F. (2003) Am. J. Hum. Genet. 73, 736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomer, Y., Greenberg, D. A., Concepcion, E., Ban, Y. & Davies, T. F. (2002) J. Clin. Endocrinol. Metab. 87, 404-407. [DOI] [PubMed] [Google Scholar]
- 24.Sakai, K., Shirasawa, S., Ishikawa, N., Ito, K., Tamai, H., Kuma, K., Akamizu, T., Tanimura, M., Furugaki, K., Yamamoto, K., et al. (2001) Hum. Mol. Genet. 10, 1379-1386. [DOI] [PubMed] [Google Scholar]
- 25.Collins, J. E., Heward, J. M., Carr-Smith, J., Daykin, J., Franklyn, J. A. & Gough, S. C. (2003) J. Clin. Endocrinol. Metab. 88, 5039-5042. [DOI] [PubMed] [Google Scholar]
- 26.Mendive, F. M., Rivolta, C. M., Vassart, G. & Targovnik, H. M. (1999) Thyroid 9, 903-912. [DOI] [PubMed] [Google Scholar]
- 27.Le Hir, H., Izaurralde, E., Maquat, L. E. & Moore, M. J. (2000) EMBO J. 19, 6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichert, V. L., Le Hir, H., Jurica, M. S. & Moore, M. J. (2002) Genes Dev. 16, 2778-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graves, P. N. & Davies, T. F. (1990) Mol. Endocrinol. 4, 155-161. [DOI] [PubMed] [Google Scholar]
- 30.Tomer, Y., Greenberg, D. A., Barbesino, G., Concepcion, E. S. & Davies, T. F. (2001) J. Clin. Endocrinol. Metab. 86, 1687-1693. [DOI] [PubMed] [Google Scholar]
- 31.Moya, C. M., Varela, V., Rivolta, C. M., Mendive, F. M. & Targovnik, H. M. (2003) Thyroid 13, 319-323. [DOI] [PubMed] [Google Scholar]
- 32.Caturegli, P., Vidalain, P. O., Vali, M., Aguilera-Galaviz, L. A. & Rose, N. R. (1997) Clin. Immunol. Immunopathol. 85, 221-226. [DOI] [PubMed] [Google Scholar]
- 33.Woolf, B. (1955) Ann. Hum. Genet. 19, 251-253. [DOI] [PubMed] [Google Scholar]
- 34.Huang, X. & Madan, A. (1999) Genome Res. 9, 868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenszky, V., Kozma, L., Balazs, C., Rochlitz, S., Bear, J. C. & Farid, N. R. (1985) J. Clin. Endocrinol. Metab. 61, 735-740. [DOI] [PubMed] [Google Scholar]
- 36.Ban, Y., Davies, T. F., Greenberg, D. A., Concepcion, E. S. & Tomer, Y. (2002) Clin. Endocrinol. (Oxford) 57, 81-88. [DOI] [PubMed] [Google Scholar]
- 37.Braley-Mullen, H., Johnson, M., Sharp, G. C. & Kyriakos, M. (1985) Cell. Immunol. 93, 132-143. [DOI] [PubMed] [Google Scholar]
- 38.Charreire, J. (1989) Adv. Immunol. 46, 263-334. [DOI] [PubMed] [Google Scholar]
- 39.Vladutiu, A. O. & Rose, N. R. (1971) Science 174, 1137-1139. [DOI] [PubMed] [Google Scholar]
- 40.Kuppers, R. C., Bresler, H. S., Lynne Burek, C., Gleason, S. L. & Rose, N. R. (1992) in Molecular Immunobiology of Self-Reactivity, eds. Bona, C. A. & Kaushik, A. K. (Dekker, New York), pp. 247-284.
- 41.Braley-Mullen, H., Sharp, G. C., Medling, B. & Tang, H. (1999) J. Autoimmun. 12, 157-165. [DOI] [PubMed] [Google Scholar]
- 42.Pirro, M. T., De Filippis, V., Di Cerbo, A., Scillitani, A., Liuzzi, A. & Tassi, V. (1995) Thyroid 5, 461-464. [DOI] [PubMed] [Google Scholar]
- 43.Tomer, Y., Barbesino, G., Keddache, M., Greenberg, D. A. & Davies, T. F. (1997) J. Clin. Endocrinol. Metab. 82, 1645-1648. [DOI] [PubMed] [Google Scholar]
- 44.De Roux, N., Shields, D. C., Misrahi, M., Ratanachaiyavong, S., McGregor, A. M. & Milgrom, E. (1996) J. Clin. Endocrinol. Metab. 81, 3483-3486. [DOI] [PubMed] [Google Scholar]
- 45.Ban, Y., Greenberg, D. A., Concepcion, E. S. & Tomer, Y. (2002) Thyroid 12, 1079-1083. [DOI] [PubMed] [Google Scholar]
- 46.Yanagawa, T., Hidaka, Y., Guimaras, V., Soliman, M. & DeGroot, L. J. (1995) J. Clin. Endocrinol. Metab 80, 41-45. [DOI] [PubMed] [Google Scholar]
- 47.Ueda, H., Howson, J. M., Esposito, L., Heward, J., Snook, H., Chamberlain, G., Rainbow, D. B., Hunter, K. M., Smith, A. N., Di Genova, G., et al. (2003) Nature 423, 506-511. [DOI] [PubMed] [Google Scholar]
- 48.Tomer, Y., Concepcion, E. & Greenberg, D. A. (2002) Thyroid 12, 1129-1135. [DOI] [PubMed] [Google Scholar]
- 49.Kim, T. Y., Park, Y. J., Hwang, J. K., Song, J. Y., Park, K. S., Cho, B. Y. & Park, D. J. (2003) Thyroid 13, 919-926. [DOI] [PubMed] [Google Scholar]
- 50.Pociot, F. & McDermott, M. F. (2002) Genes Immun. 3, 235-249. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed, S., Bennett, S. T., Huxtable, S. J., Todd, J. A., Matthews, D. R. & Gough, S. C. (1999) Diabet. Med 16, 910-917. [DOI] [PubMed] [Google Scholar]
- 52.Puglise, A., Zeller, M., Fernandez, A., Jr., Zalcberg, L. J., Bartlett, R. J., Ricordi, C., Pietropaolo, M., Eisenbarth, G. S., Bennett, S. T. & Patel, D. D. (1997) Nat. Genet. 15, 293-297. [DOI] [PubMed] [Google Scholar]
- 53.Vafiadis, P., Bennett, S. T., Todd, J. A., Nadeau, J., Grabs, R., Goodyer, C. G., Wickramasinghe, S., Colle, E. & Polychronakos, C. (1997) Nat. Genet. 15, 289-292. [DOI] [PubMed] [Google Scholar]
- 54.McFarland, B. J. & Beeson, C. (2002) Med. Res. Rev. 22, 168-203. [DOI] [PubMed] [Google Scholar]
- 55.Lee, K. H., Wucherpfennig, K. W. & Wiley, D. C. (2001) Nat. Immunol. 2, 501-507. [DOI] [PubMed] [Google Scholar]
- 56.Beisel, K., David, C. S., Giraldo, A. A., Kong, Y.-C. M. & Rose, N. R. (1982) Immunogenetics 15, 427-430. [DOI] [PubMed] [Google Scholar]
- 57.Beisel, K. W., Kong, Y.-C. M., Babu, K. S., David, C. S. & Rose, N. R. (1982) J. Immunogenet. 9, 257-265. [DOI] [PubMed] [Google Scholar]
- 58.Kuppers, R. C., Epstein, L. D., Outschoorn, I. M. & Rose, N. R. (1994) Immunogenetics 39, 404-411. [DOI] [PubMed] [Google Scholar]
- 59.Tomazic, V. & Rose, N. R. (1976) Immunology 30, 63-68. [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, C. R., Pichurin, P., Nagayama, Y., Latrofa, F., Rapoport, B. & McLachlan, S. M. (2003) J. Clin. Invest 111, 1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saegusa, K., Ishimaru, N., Yanagi, K., Arakaki, R., Ogawa, K., Saito, I., Katunuma, N. & Hayashi, Y. (2002) J. Clin. Invest 110, 361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.