Abstract
We have proposed a cancer treatment modality based on poliovirus chimeras replicating under the translational control of an internal ribosomal entry site (IRES) derived from human rhinovirus type 2. Insertion of the heterologous IRES causes a neuron-specific propagation deficit and eliminates neurovirulence inherent in poliovirus without affecting viral growth in cells derived from malignant gliomas. We now report the elucidation of a molecular mechanism responsible for the cell type-specific defect mediated by the rhinovirus IRES. Rhinovirus IRES function in neuronal cell types depends on specific structural elements within the 3′ non-translated region of the viral genome. Our observations suggest long-range interactions between the IRES and the 3′ terminus that control IRES-mediated gene expression and virus propagation.
Initiation of eukaryotic mRNA translation occurs on assembly of the 43S preinitiation complex at the 5′ cap structure and 5′-3′ ribosomal scanning until the initiation codon is encountered. Efficient translation depends on the interaction between the poly(A)-binding protein (PABP) and the eukaryotic initiation factor [(eIF)4G] (1, 2). The interaction of eIF4G with eIF4E (binding to the cap structure) and PABP results in circularization of mRNAs (3). Bridging of poly(A) and the initiation complex stimulates translation, possibly by favoring 3′–5′ shunting of ribosomes or promoting initiation factor activity (reviewed in refs. 4, 5).
Picornavirus and Hepatitis C virus plus-stranded RNA genomes lack a 5′ cap structure and use alternative means, namely an internal ribosomal entry site (IRES; refs. 6–8), to initiate viral protein synthesis. IRES-mediated translation occurs despite proteolytic cleavage of eIF4G (ref. 9; reviewed in ref. 10) and PABP (11, 12) by entero- and rhinoviral 2A proteinases. Cleavage of eIF4G and PABP and the absence of a cap prevent circularization of picornaviral RNA genomes in the manner observed for eukaryotic mRNAs. However, a stimulating role for poly(A) alone (13, 14) or synergistic activation by PABP via eIF4G interactions (15, 16) has been proposed for picornavirus RNAs as well. It is currently unclear how the cleavage of eIF4G and PABP during infection may affect this proposed stimulatory function.
Previously, we described a striking cell type-specific deficit of IRES function in neuronal cell types, demonstrated by exchange of the cognate IRES of poliovirus (PV) with its counterpart from human rhinovirus type 2 (HRV2) to generate PV-RIPO (17). Although replicating as well as PV in HeLa cells, PV-RIPO experiences severe translation defects and depressed propagation in Sk-N-Mc neuroblastoma cells. Attenuation of PV neurovirulence correlates with reduced gene expression and propagation in neuroblastoma cells, making these cell lines an appropriate in vitro model for studies of PV neurovirulence (18, 19). Accordingly, depressed propagation rates in Sk-N-Mc cells render PV-RIPO nonpathogenic in mice transgenic for the human PV receptor CD155 (17) and in non-human primates (20).
Cell type-specific function of picornavirus IRES elements may depend on structural features of the IRES itself (21, 22) or may reflect the availability of sets of eukaryotic IRES transacting factors in infected cells. Several such factors have been identified, including the polypyrimidine tract-binding protein (PTB; ref. 23); La autoantigen (24); poly(C)-binding proteins 1 and 2 (25, 26); upstream of n-ras (27); and ITAF-45 (28). The requirement of PTB for picornavirus IRES function has recently been confirmed in vivo (P. F., O. Sessions, E. Wagner, M. G., and M. Garcia-Blanco, unpublished results), and a CNS-specific function of the Theiler's murine encephalomyelitis virus IRES has been proposed to codepend on the presence of a neural PTB isoform in that organ (29). Recently, cell type-specific regulation of IRES activity has also been proposed to control the expression of eukaryotic mRNAs (30, 31).
Although the reason for tissue type-specific IRES function remains under investigation, this phenomenon has been exploited for a novel treatment modality for CNS malignancies (32). PV chimeras replicating under control of the HRV2 IRES destroy cells derived of malignant glioma while unable to cause cytopathogenicity in the normal human brain (ref. 32; M. Merrill, G. Bernhardt, J. H. Sampson, C. Wikstrand, and M.G., unpublished results). Studies in cultured cells indicate that conditions prevail in malignant cell types that permit efficient HRV2 IRES function, while restricting it in normal neuronal cells (ref. 32; M. Merrill, G. Bernhardt, J. H. Sampson, C. Wikstrand, and M.G., unpublished results).
Here we report that neuronal propagation of enteroviruses containing a heterologous HRV2 IRES is codetermined by specific stem–loop structures in the 3′ nontranslated region (NTR) of the viral genome.
Materials and Methods
Construction of Virus Recombinants and One-Step Growth Curves. A coxsackievirus B3 (CBV3) chimera (CBV-RICO), containing the HRV2 IRES in a CBV3 background, was constructed as follows: plasmid pCBV3–0 (kindly provided by N. Chapman, University of Nebraska, Omaha) was digested with NotI and BglII and ligated with three PCR-amplified fragments. These fragments, encompassing the T7 promoter and 5′ 100 nt of the CBV3 genome, the HRV2 IRES element, and a capsid segment of CBV3, were amplified with primers (i) 5′-CCGCGGCCGCTAATACGACTCACTATAGGTTAAAACAGCCTGTGGGTTGATCC-3′ and (ii) 5′-GGACGCGTTGGGGGAGGGGGTATAAAACAGG-3′; (iii) 5′-CCACGCGTAACTTAGAAGTTTTTCACAAAGACC-3′ and (iv) 5′-GGGAGCTCCCATGGTGCCAATATATATATTG-3′; and (v) 5′-GGGAGCTCAAGTATCAACGCAAAAGACTGG-3′ and (vi) 5′-CAAGATCTCTCCTAGGAGCGTCCG-3′, respectively. To construct CBV-RICO chimeras with heterologous 3′ NTRs, an XbaI restriction site spanning the termination codon of the CBV3 polyprotein was created through silent mutagenesis. Fragments corresponding to the CBV3 P3 region and the PV type 1 Sabin [PV1(S)] 3′ NTR were amplified by using primers (vii) 5′-GGTCTAGACATGCTAGTCACCG-3′ and (viii) 5′-GGTCTAGAAGGAGTCCAACCACTTCC-3′; and (ix)5′-GGTCTAGAAACCCTACCTCAGTCG-3′ and (x) 5′-GGATCGATGC. (T)12CTCCGAATTA-3′, respectively, and ligated into CBV-RICO digested with XbaI and ClaI. The CBV3 3′ NTR without domain Z (CBV3 ΔZ) was PCR-amplified with primers (xi) 5′-CCTCTAGAAACCCTACTGTGCTAACCG-3′ and (xii) 5′-GGATCGATGGG(T)18CCGCACCGAATGC-3′. The HRV2 3′ NTR was generated by using complementary synthetic oligonucleotides (xiii) 5′-CTAGAGATATAGAAATAGTAAACTGATAGTTTATTAGTTTTAT(A)11T and (xiv) 5′-CGA(T)11ATAAAACTAATAAACTATCAGTTTACTATTTCTATATCT-3′. Annealing of primers xiii and xiv formed a double-stranded DNA fragment with XbaI and ClaI overhangs used for ligation. The CBV3 Z-loop was replaced with the HRV2 3′ NTR through digestion of CBV-RICO containing the HRV2 3′ NTR with PsiI and ClaI and ligation with the CBV3 ΔZ fragment PCR-generated with primers (xii) and (xv), 5′-CCTTATAACCCTACTGTGCTAACCGAACC-3′. The PV1(S)+Z3′ NTR was PCR-generated with primers (x) and (xvi), 5′-GGTCTAGATTAGAGACAATTTGAAATAATTTAGATTGGCTTAACCCTACCTCAGTCGAATTGG-3′. In vitro transcription of viral cDNA, RNA transfection of HeLa cells for virus recovery, and one-step growth kinetics were performed as described (17).
Luciferase Expression Assays. Luciferase reporter vectors were constructed as follows. CBV-RICO was digested with SacI and ClaI and ligated with two PCR-amplified fragments encompassing the coding region for Renilla luciferase (rLuc) [primers (xvii) 5′-CCGAGCTCAGACTTCGAAAGTTTATGATCC-3′ and (xviii) 5′-GGTCTAGAATTGTTCATTTTTGAGAACTCG-3′], and the CBV3 3′ NTR with or without SLD Z [(primers (xii) and (xviv), 5′-CCTTCTAGATTAGAGACAATTTG-3′, or (xi)], respectively.
rLuc expression vectors linearized with ClaI were used as templates for in vitro transcription with T7 RNA polymerase (17). In vitro transcription reactions were treated with RQ1 DNase (Promega), and transcript RNAs were purified (RNeasy; Qiagen, Chatsworth, CA), quantified by UV spectrophotometry, and 2 μg of RNA was used to transfect 1 × 106 HeLa or 2.5 × 106 Sk-N-Mc cells with DMRIE-C reagent (Invitrogen). The transfection efficiencies for HeLa and Sk-N-Mc cells, determined with radiolabeled in vitro transcript RNA (see below), were found to be similar for both at ≈10% and 8% of input RNA, respectively. At different intervals after transfection, the cells were washed with PBS and lysed with 0.2 ml of luciferase assay lysis buffer (Promega). rLuc activity assays were performed according to Promega's instructions in a Berthold luminometer (no. LB9507).
RNA Stability Assays. For RNA stability assays, cell monolayers were transfected with 0.5 μgof[α32P]UTP-labeled viral genomic or reporter in vitro RNA transcript. At the specified time points, total RNA was isolated from transfected cells by using Trizol reagent (Invitrogen) and subjected to denaturing gel electrophoresis followed by autoradiography.
Western Blotting. Synchronized infections of Sk-N-Mc cells were performed as described (17). At various time points, infected cells were washed with PBS and lysed with 0.1 ml of SDS/PAGE sample buffer (Invitrogen). Cell lysates were resolved by electrophoresis in 4–12% Bis-Tris NuPAGE Gels (Invitrogen) and transferred to PROTRAN nitrocellulose membrane (Schleicher & Schuell). After blocking in 3% nonfat milk in TBST (100 mM Tris/150 mM NaCl/0.05% Tween 20, pH 8.0) for 1 h, membranes were incubated with primary anti-PV 2BC/2C monoclonal antibodies (kindly provided by E. Wimmer) or anti-CBV3 3AB/3A polyclonal antibodies (kindly provided by K. Klingel). After three washes with TBST, membranes were treated with secondary biotinylated antispecies antibody (Vector Laboratories), rinsed again, finally developed with streptavidin–horseradish peroxidase conjugate (Roche Applied Science, Indianapolis), and visualized with enhanced chemiluminescent Western blotting detection reagents (Amersham Pharmacia).
Results
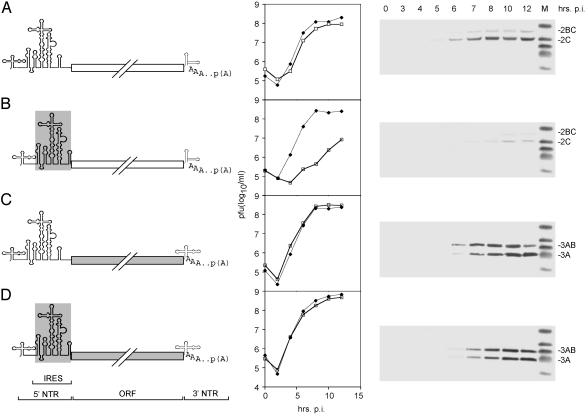
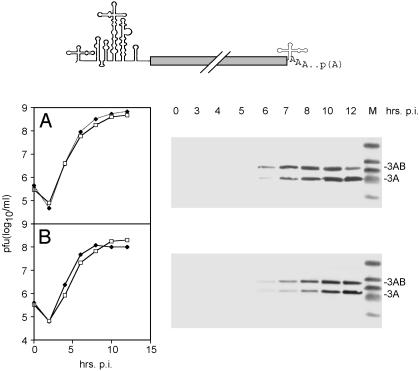
Replacement of the PV IRES with its counterpart from HRV2, yielding PV-RIPO, was shown to result in a neuron-specific replication deficit (17) and the elimination of viral neuropathogenicity in mice transgenic for the PV receptor CD155 (17) and non-human primates (20). The growth kinetics of the chimeric PV-RIPO in HeLa cells were equal to its parent PV (17), but proliferation in a neuroblastoma cell line (Sk-N-Mc) was significantly diminished (compare Fig. 1 A to B), consistent with poor accumulation of viral proteins in this cell line of neuronal origin (Fig. 1B).
Fig. 1.
Genetic structure (Left), growth properties (Center), and viral gene expression in Sk-N-Mc neuroblastoma cells (Right) of PV (A), CBV3 (C), and their derivatives PV-RIPO (B) and CBV-RICO (D), containing the HRV2 IRES (indicated by a gray box; B and D). One-step growth curves were established in HeLa (solid diamonds) and Sk-N-Mc neuroblastoma (open squares) cells. The kinetics of viral gene expression in Sk-N-Mc cells were assayed through Western blot detection of polioviral gene products 2BC and 2C (A and B) and coxsackieviral proteins 3AB and 3A (C and D) at the indicated intervals postinfection (p.i.).
A PV relative in the enterovirus genus, CBV3, exhibited parallel replication in HeLa and Sk-N-Mc cells and viral protein synthesis rates similar to PV (Fig. 1C). Surprisingly, when we inserted the HRV2 IRES into CBV3 (yielding CBV-RICO; Fig. 1D), viral replication and protein synthesis were at WT levels in both HeLa and Sk-N-Mc neuroblastoma cells (Fig. 1D). Thus, insertion of the HRV2 IRES, which reduced PV propagation yields and gene expression in neuronal cell types to <2% of WT levels (Fig. 1B), failed to have any effect on CBV3 gene expression and replication (Fig. 1D). Considering that the IRES element performs identical tasks in all enteroviruses, this discrepancy was highly unexpected. Because cell type-specific function of the HRV2 IRES depended on the origin of the viral genome, we speculated that cis-acting elements outside the 5′ NTR might influence translation activity.
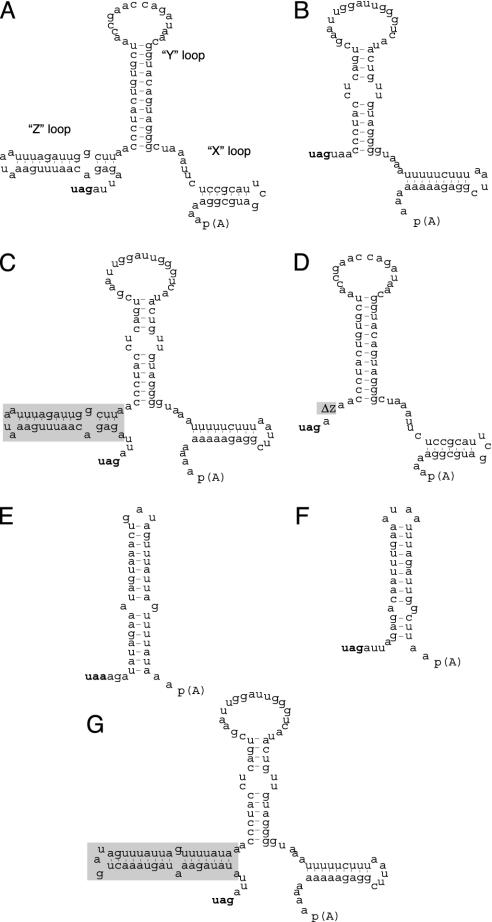
The presence of translation regulatory elements within the 3′ NTRs of eukaryotic mRNAs (33) and the known structural divergences among picornavirus 3′ NTRs (34) prompted us to investigate an influence of the viral 3′ NTR on IRES activity. The 3′ NTR of CBVs is ≈100 nt long and forms three distinct stem loops (X, Y, and Z; Fig. 2A), whereas the PV 3′ NTR (68 nt in length) lacks stem–loop domain (SLD) Z (Fig. 2B). Composition and secondary structure of the HRV2 3′ NTR (39 nt long) are reminiscent of the CBV Z-loop (Fig. 2 E and F).
Fig. 2.
Sequence and predicted secondary structure of the 3′ NTRs of CBV3 (A), PV1(S) (B), HRV2 (E), CBV3 SLD Z (F), and engineered variants (C, D, and G). The structure and position of the three distinct SLDs in the CBV3 3′ NTR are indicated (34). The PV1(S) 3′ NTR was altered by insertion of the CBV3 Z domain (boxed in gray) [PV1(S) + Z; C]. The CBV3 3′ NTR was modified by deletion of the Z domain (CBV3 ΔZ; D) or exchange thereof with the entire 3′ NTR of HRV2 (CBV3 ΔZ + HRV2; G).
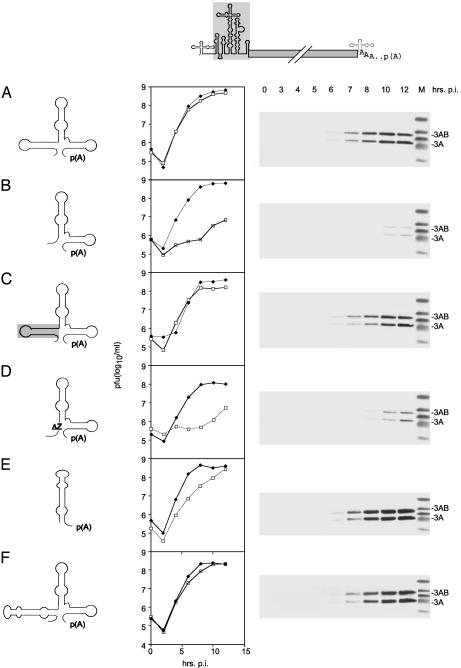
To evaluate the functional significance of structural differences in the 3′ NTR for IRES function, we engineered chimeric viruses based on CBV-RICO (Fig. 1D) carrying heterologous 3′ NTR sequences. Interestingly, insertion of the PV1(S) 3′ NTR into CBV-RICO drastically decreased viral gene expression and replication rates in Sk-N-Mc cells (compare Fig. 3 A to B). The replication profile and protein synthesis rate in Sk-N-Mc cells of CBV-RICO carrying a PV 3′ NTR corresponded to PV-RIPO (compare Fig. 1B to Fig. 3B). Insertion of the PV1(S) 3′ NTR had no effect on virus propagation in HeLa cells (Fig. 3B).
Fig. 3.
Replication profiles of CBV-RICO (shown on top) containing chimeric 3′ NTRs in HeLa (solid diamonds) and Sk-N-Mc neuroblastoma (open squares) cells. We analyzed constructs containing the cognate CBV3 3′ NTR (A), the PV1(S) 3′ NTR (B), the PV1(S) 3′ NTR containing the SLD Z (C), the CBV3 3′ NTR with the SLD Z deleted (D), the HRV2 3′ NTR (E), or the HRV2 3′ NTR replacing the CBV3 SLD Z (F). (Right) Western blot analyses of the kinetics of viral gene expression in Sk-N-Mc neuroblastoma cells for the corresponding constructs.
Next, we questioned whether neuron-specific modulation of IRES function indeed correlated with the structural differences of the viral 3′ NTRs. For that purpose, we constructed a PV 3′ NTR carrying a CBV3 Z domain insert in the authentic position (Fig. 2C) and a CBV3 3′ NTR, lacking the Z domain (Fig. 2D). These manipulated 3′ NTRs were used to replace the cognate CBV3 3′ NTR in the CBV-RICO background (Fig. 3 C and D). All chimeric viruses retained replication kinetics in HeLa cells at WT levels but differed dramatically with regard to viral gene expression in cells of neuronal origin (Fig. 3 C and D).
Neuronal propagation deficits mediated by incompatibility of the HRV2 IRES with the PV1(S) 3′ NTR could be completely reversed by inserting the CBV3 SLD Z, restoring replication rates and translation efficiency in neuronal cells to WT levels (compare Fig. 3 A to C). Inversely, truncating the CBV3 3′ NTR by deletion of the SLD Z diminished viral protein synthesis in Sk-N-Mc cells to the levels of PV-RIPO (Fig. 3D).
Replacement of the cognate CBV3 3′ NTR in CBV-RICO with its counterpart from HRV2 yielded a chimeric virus with WT level replication and IRES-mediated translation in Sk-N-Mc cells (Fig. 3E). Given the substantial structural diversity between the CBV3 and HRV2 3′ NTRs (Fig. 2 A and E), this result was surprising. However, SLD Z of the CBV3 3′ NTR and the HRV2 3′ NTR share significant structural similarity (Fig. 2 E and F). Reflecting this similarity, exchange of the CBV3 SLD Z with the entire HRV2 3′ NTR (Fig. 2G) in CBV-RICO produced WT IRES activity and particle propagation in Sk-N-Mc neuroblastoma cells (Fig. 3F).
Our observations suggested that both the CBV3 SLD Z as well as the HRV2 3′ NTR feature structural characteristics required for efficient HRV2 IRES function. To exclude any effects of SLD Z deletion on neuronal virus replication itself independent of IRES sequence, we constructed a WT CBV3 variant containing the CBV3 ΔZ 3′ NTR (Fig. 4). Deletion of the SLD Z in WT CBV3 had a minor effect on virus growth in Sk-N-Mc cells but failed to induce the drastic inhibition of viral propagation seen in the presence of the HRV2 IRES (Fig. 4B, compare to Fig. 3D). Because virus propagation driven by the HRV2 IRES in CBV3 covaried with the presence of the 3′-terminal Z domain, we speculate that IRES and 3′ NTR sequences cooperatively influence IRES-controlled gene expression.
Fig. 4.
Effect of 3′ NTR manipulation on IRES activity of WT CBV3 (on top). Particle propagation and viral gene expression of WT CBV3 (A) and CBV3 ΔZ (B) in HeLa (solid diamonds) and Sk-N-Mc neuroblastoma (open squares) cells.
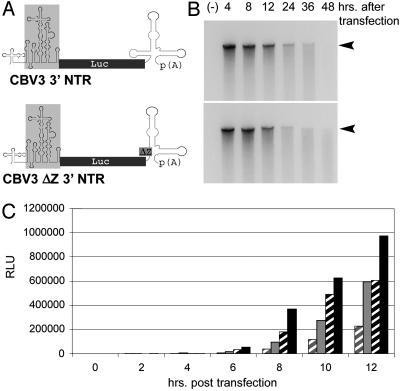
To demonstrate that the deletion of CBV3 SLD Z directly affected IRES-mediated translation, we generated nonreplicating reporter constructs (Fig. 5A) in which the HRV2 IRES drives translation of an rLuc gene. In vitro transcripts of two reporter constructs containing either the complete CBV3 3′ NTR or the CBV3 ΔZ variant were used for transfection of Sk-N-Mc and HeLa cells (Fig. 5A).
Fig. 5.
(A) Genetic structure of two HRV2 IRES (boxed in gray) driven rLuc reporter constructs with a full-length CBV3 (Upper) or CBV3 ΔZ3′ NTR (Lower), respectively. (B) Stability of reporter RNAs containing the complete CBV3 (Upper) or a CBV3 ΔZ 3′ NTR (Lower) in Sk-N-MC cells at different intervals posttransfection. Negative control (-) represents “mock” transfection without transfection reagent. (C) The kinetics of rLuc expression in Sk-N-Mc (gray) and HeLa (black) cells transfected with in vitro transcript RNA containing the CBV3 ΔZ3′ NTR (hatched bars) or the full-length CBV3 3′ NTR (solid bars). The data represent the averages of three independent experiments.
To exclude an effect of 3′ NTR structure on RNA stability, we analyzed the integrity of viral RNAs and the in vitro transcript reporter RNAs after transfection into Sk-N-Mc cells. Both, viral genomic (data not shown) as well as in vitro transcript RNAs (Fig. 5B) displayed equal rates of turnover after transfection into Sk-N-Mc cells, independent of the presence of the CBV3 SLD Z.
rLuc expression rates covaried with the presence of SLD Z in the 3′ NTR of the reporters (Fig. 5C). This effect was evident in Sk-N-Mc cells and, to a lesser extent, in HeLa cells. Translation levels of the ΔZ construct in HeLa cells were consistently higher than with the WT CBV3 3′ NTR in Sk-N-Mc cells and significantly exceeded the performance of the ΔZ construct in the latter (Fig. 5C). These results reflect the effects of SLD Z deletion on neuronal propagation of CBV-RICO (see Fig. 3).
Discussion
Regulation of gene expression through cap-dependent translation involves several cis-acting elements that mediate long-range interactions within mRNAs and affect translation rates and mRNA stability. These include sequence elements within the ORF (coding region determinants; refs. 7, 35) and AU-rich elements within the 3′ NTR (36) and the poly(A) tail (37). The influence of cis-acting elements on protein synthesis is believed to reflect the action of RNA-binding proteins that establish contact with the initiation complex (38), which may determine translation initiation rate and mRNA stability (35).
The role of 5′–3′ NTR interactions in cap-independent translation initiation at the IRES is less clear. Formation of ribonucleoprotein complexes at the ends of genome RNA has been suggested to play a role in polioviral genome replication (39, 40). Our findings provide evidence that, similar to eukaryotic mRNAs, 3′ NTR cis elements can also control translation rate at the IRES.
A precedent for such a scenario has been proposed for hepatitis C virus (HCV), although an involvement of the 3′ NTR in translation control at the HCV IRES is controversial. The HCV genome contains a highly conserved 3′ NTR stem–loop structure (the X region), which has been reported to either stimulate IRES function in cis (41), not influence translation at all (42), or even down-regulate IRES translation (43).
Our observations primarily reflect the cell type-specific effect of IRES–3′ NTR interactions on virus propagation. Interestingly, Merkle et al. (44) demonstrated that deletion of SLD Z from WT CBV3 resulted in reduced cardiovirulence in BALB/c mice. Our findings indicate that the incompatibility of the HRV2 IRES and 3′ NTRs lacking SLD Z produced the defective growth phenotype in neuronal cells (Figs. 3 and 4). Modulation of IRES-mediated gene expression by 3′ NTR sequences could be recapitulated with nonreplicating expression constructs in living cells, suggesting that these interactions influence virus propagation at the level of translation control. However, gene expression and genome replication of picornaviruses are intertwined processes involving overlapping regulatory sequences that are difficult to dissect experimentally. Thus, influences of IRES–3′ NTR interactions on cell type-specific virus growth defects beyond translation control cannot be excluded categorically.
Our experiments were carried out by measuring gene expression by viral genomes or monocistronic expression constructs with authentic 5′ and 3′ regulatory regions in living host cells. Experimental systems that detect translation rates of IRES elements outside their native context [e.g., dicistronic reporter constructs (30)] may miss the critical influence of regulatory elements residing outside the IRES proper. Furthermore, the basis for cell type-specific differences in IRES function may involve not only transactivating factors binding to the IRES itself [e.g., as proposed by Pilipenko et al. (29)] but also RNA–protein interactions with sequence elements outside the 5′ NTR.
We have described a regulatory mechanism of controlling IRES function. This mechanism involves 3′-terminal sequence elements that affect IRES translation and results in a cell type-specific viral growth defect. Our findings may explain the pathogenic properties of IRES-containing viruses, because IRES–3′ NTR interactions may favor viral gene expression and virus propagation at specific sites. Furthermore, we have provided evidence for fundamentally different conditions for IRES-mediated translation in neuronal cells from rapidly growing malignant cells. Oncolytic PV recombinants to be used against CNS malignancies (32) are the first example of targeting tumor-specific mechanisms of translational control for therapeutic purposes.
Acknowledgments
We are grateful to K. Klingel (University of Tuebingen, Tuebingen, Germany); E. Wimmer (State University of New York, Stony Brook); and N. Chapman (University of Nebraska, Omaha) for supplying valuable materials. We thank M. Garcia-Blanco and J. Keene for critical reading of the manuscript. This work was supported by Public Health Service Grant CA87537 (to M.G.) and the Accelerate Brain Cancer Cure Foundation. M.G. is a recipient of a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Abbreviations: IRES, internal ribosomal entry site; PABP, poly(A)-binding protein; eIF, eukaryotic initiation factor; PV, poliovirus; HRV2, human rhinovirus type 2; NTR, nontranslated region; CBV3, coxsackievirus B3; PV1(S), PV type 1 Sabin; rLuc, Renilla luciferase; SLD, stem–loop domain.
References
- 1.Le, H., Tanguay, R. L., Balasta, M. L., Wei, C. C., Browning, K. S., Metz, A. M., Goss, D. J. & Gallie, D. R. (1997) J. Biol. Chem. 272, 16247-16255. [DOI] [PubMed] [Google Scholar]
- 2.Tarun, S. Z. & Sachs, A. B. (1996) EMBO J. 15, 7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 3.Wells, S. E., Hillner, P. E., Vale, R. D. & Sachs, A. B. (1998) Mol. Cell 2, 135-140. [DOI] [PubMed] [Google Scholar]
- 4.Preiss, T. & Hentze, M. W. (1999) Curr. Opin. Genet. Dev. 9, 515-521. [DOI] [PubMed] [Google Scholar]
- 5.Sachs, A. B. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Matthews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 447-466.
- 6.Jang, S. K., Kräusslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C. & Wimmer, E. (1988) J. Virol. 62, 2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier, J., Kaplan, G., Racaniello, V. R. & Sonenberg, N. (1988) Mol. Cell. Biol. 8, 1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukiyama-Kohara, K., Iizuka, N., Kohara, M. & Nomoto, A. (1992) J. Virol. 6, 1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etchison, D., Milburn, S. C., Edery, I., Sonenberg, N. & Hershey J. W. B. (1982) J. Biol. Chem. 257, 14806-14810. [PubMed] [Google Scholar]
- 10.Belsham, G. J. & Jackson, R. J. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Matthews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 869-900.
- 11.Joachims, M., Van Breugel, P. C. & Lloyd, R. (1999) J. Virol. 73, 718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerekatte, V., Keiper, B. D., Badorff, C., Cai, A., Knowlton, K. U. & Rhoads, R. E. (1999) J. Virol. 73, 709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergamini, G., Preiss, T. & Hentze, M. W. (2000) RNA 6, 1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hruby, D. E. & Roberts, W. K. (1977) J. Virol. 23, 338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel, Y. M., Borman, A. M., Paulous, S. & Kean, K. (2001) Mol. Cell. Biol. 21, 4097-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svitkin, Y. V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H. D. & Sonenberg, N. (2001) RNA 7, 1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 17.Gromeier, M., Alexander, L. & Wimmer, E. (1996) Proc. Natl. Acad. Sci. USA 93, 2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaMonica, N. & Racaniello, V. R. (1989) J. Virol. 63, 2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agol, V. A., Drozdov, S. G., Ivannikova, T. A., Kolesnikova, M. S., Korolev, M. B. & Tolskaya, E. A. (1989) J. Virol. 63, 4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gromeier, M., Bossert, B., Arita, M., Nomoto, A. & Wimmer, E. (1999) J. Virol. 73, 958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macadam, A. J., Ferguson, G., Burlison, J., Stone D., Skuce, R., Almond, J. W. & Minor, P. D. (1992) Virology 189, 415-422. [DOI] [PubMed] [Google Scholar]
- 22.Skinner, M. A., Racaniello, V. R., Dunn, G., Cooper, J., Minor, P. D. & Almond, J. W. (1989) J. Mol. Biol. 207, 379-392. [DOI] [PubMed] [Google Scholar]
- 23.Hellen, C. U. T., Witherell, G. W., Schmid, M., Shin, S. H., Pestova, T. V., Gil, A. & Wimmer, E. (1993) Proc. Natl. Acad. Sci. USA 90, 7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meerovitch, K., Svitkin, Y. V., Lee, H. S., Lejbkowicz, F., Kenan, D. J., Chan, E. K. L., Agol, V. I., Keene, J. D. & Sonenberg, N. (1993) J. Virol. 67, 3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blyn, L. B., Swiderek, K. M., Richards, O., Stahl, D. C., Semler, B. L. & Ehrenfeld, E. (1996) Proc. Natl. Acad. Sci. USA 93, 11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blyn, L. B., Towner, J., Semler, B. L. & Ehrenfeld, E. (1997) J. Virol. 7, 6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt, S. L., Hsuan, J. J., Totty, N. & Jackson, R. J. (1999) Genes Dev. 13, 437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilipenko, E. V., Pestova, T. V., Kolupaeva, V. G., Khitrina, E. V., Poperechnaya, A. N., Agol, V. I. & Hellen, C. U. (2000) Genes Dev. 14, 2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 29.Pilipenko, E. V., Viktorova, E. G., Guest, S. T., Agol, V. I. & Roos, R. P. (2001) EMBO J. 20, 6899-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creancier, L., Morello, D., Mercier, P. & Prats, A. C. (2000) J. Cell Biol. 150, 275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creancier, L., Mercier P., Prats, A. C. & Morello, D. (2001) Mol. Cell. Biol. 21, 1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gromeier, M., Lachmann, S., Rosenfeld, M., Gutin, P. H. & Wimmer, E. (2000) Proc. Natl. Acad. Sci. USA 97, 6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray, N. & Wickens, M. (1998) Annu. Rev. Cell Dev. Biol. 14, 399-458. [DOI] [PubMed] [Google Scholar]
- 34.Rohll, J. B., Moon, D. H., Evans, D. J. & Almond, J. W. (1995) J. Virol. 69, 7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wisdom, R. & Lee, W. (1991) Genes Dev. 5, 232-243. [DOI] [PubMed] [Google Scholar]
- 36.Meijlink, F., Curran, T., Miller, D. A. & Verma, I. (1985) Proc. Natl. Acad. Sci. USA 8, 4987-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallie, D. R. (1991) Genes Dev. 5, 2108-2116. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein, P. Peltz, S. & Ross, J. (1989) Mol. Cell. Biol. 9, 659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herold, J. & Andino, R. (2001) Mol. Cell 7, 581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barton, D. J., O'Donnell, B. J. & Flanegan, J. B. (2001) EMBO J. 20, 1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito, T., Tahara, S. M. & Lai, M. M. (1998) J. Virol. 72, 8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong, L. K. & Sarnow, P. (2002) J. Virol. 76, 12457-12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami, K., Abe, M., Kageyama, T., Kamoshita, N. & Nomoto, A. (2001) Arch. Virol. 146, 729-741. [DOI] [PubMed] [Google Scholar]
- 44.Merkle, I., van Ooij, M. J., van Kuppeveld, F. J., Glaudemans, D. H., Galama, J. M., Henke, A., Zell, R. & Melchers, W. J. G. (2002) J. Virol. 76, 9900-9909. [DOI] [PMC free article] [PubMed] [Google Scholar]