Abstract
We identify a secreted chemokine inhibitor encoded by orf virus (ORFV), the prototypic poxvirus of the Parapoxvirus genus, and show that it is related to the poxvirus type II CC-chemokine-binding proteins (CBP-II) produced by members of the Orthopoxvirus and Leporipoxvirus genera. The ORFV chemokine-binding protein (CBP) is functionally similar to the CBP-II proteins in its ability to bind and inhibit many CC-chemokines with high affinity. However, unlike CBP-II, the ORFV CBP also binds with high affinity to lymphotactin, a member of the C-chemokine family, demonstrating that the ORFV CBP possesses an altered binding specificity. Interestingly, the amino acid sequence of ORFV CBP more closely resembles the granulocyte–macrophage colony-stimulating factor/IL-2 inhibitory factor also produced by ORFV, implicating the granulocyte–macrophage colony-stimulating factor/IL-2 inhibitory factor protein as a highly diverged, but related, member of the CBP-II protein family. Notably, these findings suggest that the genes that encode these proteins derive from a common poxvirus ancestral gene that has since been modified in binding specificity during speciation of the poxvirus genera. Overall, these findings illustrate the concept of evolution of viral proteins at the biophysical and molecular interface.
Modification of receptor binding specificity for a ligand is an essential evolutionary process for creating new signaling pathways and requires that one partner's binding surface be altered to complement changes in the other (1). The coevolution of viruses with their hosts also depends on alterations in protein–protein interaction specificity to enable mutual adaptation in binding recognition. Viral immune evasion proteins that possess receptor-like qualities (viroceptors) are no doubt subject to such adaptations in binding specificity for host-signaling proteins. Here, we report an interesting example of this phenomenon through the identification of a poxvirus-encoded chemokine-binding protein (CBP) with unique binding specificity. This finding also provides an unexpected familial link between a family of poxvirus CC-CBP and a previously described ORF virus (ORFV) granulocyte–macrophage colony-stimulating factor (GM-CSF)/IL-2 inhibitory factor (GIF) protein. Importantly, these data strongly support the notion that these poxvirus immune evasion proteins have undergone shifts in ligand specificity during the course of poxvirus evolution.
Chemokines comprise a family of small proteins that function to attract and activate leukocytes during processes of inflammation or infection (2, 3). Based on the arrangement of N-terminal cysteine residues, chemokines are partitioned into four classes: CXC, CC, CX3C, and C (where X is any residue). Discrete and overlapping residues on the surface of chemokines define the specificity for binding and signaling through the variety of chemokine G protein-coupled receptors (3). Interactions with cell-surface glycosaminoglycans are also thought to modulate chemokine activity in addition to providing a means for chemokines to form solid-phase gradients that help guide leukocytes along endothelial surfaces and into tissue (4–6). As critical coordinators of immune cell trafficking and activation, these distinct binding interactions for chemokines present two attractive targets for disruption by viruses (7, 8).
Poxviruses of the Leporipoxvirus and Orthopoxvirus genera express a secreted CBP that is capable of binding many, but not all, CC-chemokines (7, 9–15). Members of this type II CC-CBP (CBP-II, also called vCCI) family have no sequence or structural homology to any known G protein-coupled receptor or mammalian protein but are able to competitively bind and inhibit CC-chemokine interactions with cognate receptors and thus prevent chemokine signaling and chemokine-induced chemotaxis (11–16). Supporting these in vitro observations, M-T1 protein from myxoma virus (Leporipoxvirus) and the 35-kDa protein from rabbitpox virus (Orthopoxvirus) have been shown to prevent the acute infiltration of leukocytes into sites of virus infection, with no effect on overall lethality (15, 17), whereas the vaccinia virus 35-kDa (VV-35kDa) and the cowpox virus (CPV) p35 CBPs have been shown to reduce the recruitment of inflammatory cells in experimental models of inflammation (11, 18). Structurally, the CPV p35 CBP is composed of a β-sandwich with patches of conserved negatively charged residues that are thought to facilitate binding to positively charged chemokines (16). Interestingly, a recent cocrystal structure of the secreted murine γ-herpesvirus 68 M3 CBP, a protein that binds CX3C-, CXC-, CC-, and C-chemokines (19, 20), demonstrated that its N-terminal domain possesses a β-sandwich that remotely resembles the general structure of the CPV CBP, although its connecting topology is rather different (21). One member of the CBP-II family of proteins encoded by vaccinia virus, termed A41L, has immunomodulatory properties but thus far has no known binding partner (22). Despite the number of CBPs discovered among the Leporipoxvirus and Orthopoxvirus genera, no CBP has yet been identified among any other poxvirus genera, including the Parapoxvirus genus.
ORFV, also known as contagious ecthyma, is a prototypic Parapoxvirus that causes pustular dermatitis in sheep, goats, and humans with worldwide distribution and is therefore of clinical and agricultural importance (23). The ORFV host-modulating arsenal includes an IL-10 homolog (24, 25), a vascular endothelial growth factor homolog (26–29), an IFN resistance gene (30, 31), and GIF, a dual-specific cytokine-binding protein (32). Interestingly, it has been noted that GIF has low sequence homology to the orphan CBP-II member A41L, suggesting that it may be a related member of this family (22, 32).
Here, we identify an ORFV-encoded CBP that shares sequence similarities to the GIF protein from ORFV and a less conspicuous resemblance to the CBP-II proteins produced by the Orthopoxvirus and Leporipoxvirus genera. Thus, this ORFV CBP provides an evolutionary link to the ORFV GIF protein and confirms the GIF protein as a member of the CBP-II family. We describe the binding properties of the ORFV CBP, demonstrating that it not only interacts with various CC-chemokines similarly to the CBP-II proteins but, surprisingly, also binds the C-chemokine lymphotactin. Importantly, the unique binding profiles and diverged primary sequence structures of the ORFV CBP and GIF proteins compared with the poxvirus CBP-II proteins as well as the accumulating data on the binding specificities of other CBP-II family members suggest that distinct evolutionary pressures within the various genera of poxviruses have molded the unique binding specificities of each member of this family of proteins.
Materials and Methods
Viruses and DNA. The origin of ORFV strains NZ2 (ORFV NZ2) and NZ7 (ORFV NZ7) have been reported (33). The restriction fragment EcoRI-D from ORFV NZ2 was cloned into pBR328 (pVUI) and maintained in Escherichia coli HB101 (34). The restriction fragment HindIII-E from ORFV NZ7 was cloned into pUC8 (33). The ORFV NZ2 and NZ7 genes have been deposited in GenBank under accession nos. CAD99366 and AY453066, respectively.
DNA Cloning and Sequencing. The methods used for cloning have been described (35). Templates were prepared by cloning random fragments of EcoRI-D (ORFV NZ2) into pTZ as was recommended by Applied Biosystems. To obtain sequence across junctions showing ambiguities, primers were synthesized on the basis of emerging sequences. Reagents used for sequencing were supplied by Applied Biosystems, and the products of the sequencing reactions were analyzed with an Applied Biosystems model 373A sequencing system.
Expression and Purification of ORFV CBP. The coding region of the CBP ORFV NZ2 was amplified by PCR from the EcoRI-D fragment. The primers used for PCR amplification were 5′-AGCGCCCGGCGCGCCAGAAAGCGGTGTTGTTGCT and 5′-AGCGCCCGGCGCGCCGCATTGCCAGGGTTGAGGTTAA and were based on the 5′ and 3′ ends of the CBP coding region, respectively. Each primer possessed an AscI cleavage site to allow cloning into the plasmid pEFBOS-FLAG (a gift from Clare McFarlane, Walter and Eliza Hall Institute, Melbourne). This cloning step incorporated a Kozak sequence at the 5′ end of the gene and a FLAG sequence at the 3′ end of the coding sequence. To express the CBP–FLAG fusion protein, the gene was subcloned into the eukaryotic expression vector pAPEX-3 (a gift from Clare McFarlane). The gene was excised from pEFBOS with NheI and cloned into pAPEX-3 at the XbaI site. pAPEX-3 contains a simian virus 40 promoter sequence, a transcription termination sequence, and a gene for hygromycin resistance.
The ORFV protein was expressed in 293 cells expressing Epstein–Barr virus-encoded nuclear antigen. The cells were transfected with recombinant pAPEX-3 by using Lipofectamine (GIBCO/BRL) or the FuGENE reagent (Roche, Mannheim, Germany) and cells expressing the ORFV CBP were selected by using hygromycin. FLAG-tagged proteins were purified from cell culture supernatants by affinity chromatography with anti-FLAG M2 affinity gel (Sigma). Proteins were visualized and assessed for purity by Coomassie blue staining or silver staining after SDS/PAGE (data not shown). Polypeptides separated by SDS/PAGE were confirmed by N-terminal sequencing analysis (Edman degradation).
Chemokines and Preparation and Characterization of Human Monocyte Chemoattractant Protein (MCP)-1 Mutants. Chemokines were purchased from Peprotech (Rocky Hill, NJ) or were a generous gift from Chemocentryx (San Carlos, CA). MCP-1 mutants were generated as described (36, 37). All mutants were made in the context of MCP-1 M64I, which has been shown to behave identically to wild type in binding assays to CCR2b. With respect to binding to ORFV NZ2 CBP, the kinetic parameters of wild-type MCP-1 and M64I are indistinguishable (data not shown).
Biomolecular Interaction Analysis by Using Surface Plasmon Resonance (SPR). For ligand screening experiments, the ORFV NZ2 protein was immobilized at high density onto a CM5 chip by using standard amine-coupling chemistry to a level of ≈800 response units (RU) (800 pg/mm2) and a BiacoreX biosensor (Biacore, Uppsala, Sweden). For kinetic analysis, ORFV NZ2 CBP was immobilized at low density (400 pg/mm2). All experiments were performed at 25°C with HBS-EP (10 mM Hepes, pH 7.4/150 mM NaCl/3 mM EDTA/0.005% polysorbate 20) as the running buffer. Kinetic and affinity analysis by using BiacoreX has been described elsewhere (38).
Leukocyte Preparation and Intracellular Calcium Measurements. Cells at a concentration of 2.0 × 106 per ml were loaded with 2.0 μM Indo-1 acetoxymethyl ester (Molecular Probes) in conditioned DMEM for 30 min at 37°C. Cells were resuspended in Hepes-buffered MEM at a concentration of 1 × 106 cells per ml. For calcium flux experiments, 2 × 106 cells were resuspended into a 2-ml volume of Na+ solution (20 mM Hepes/135 mM NaCl/1mM MgCl2/10 mM glucose/5 mM KCl/1 mM CaCl2) and added to a 2-ml cuvette that was continuously stirred at 37°C in a model RF-M2004 dual-wavelength fluorimeter (Photon Technology International, Lawrenceville, NJ). Purified ORFV NZ2 CBP was injected into the cuvette containing cells ≈10 s before injections of human macrophage inflammatory protein (hMIP)-1α, hMCP-1, RANTES, and I-309. Calcium responses were monitored continuously as a relative fluorescence ratio by using an excitation wavelength of 355 nm and emission wavelengths of 405 nm and 485 nm.
Results
Identification of a Putative ORFV Binding Protein. Sequencing of a 10-kbp fragment of EcoRI-D (34) derived from the right terminus of the ORFV (strain NZ2) genome revealed an ORF with homology to ORFV GIF as well as low homology to the CBP-II proteins of the Orthopoxvirus and Leporipoxvirus genera. No sequence similarity has been found between the ORFV proteins and the γ-herpesvirus 68 M3 protein. The ORFV NZ2 gene possesses 858 base pairs (Fig. 5, which is published as supporting information on the PNAS web site) and encodes a 286-aa protein with a predicted mass of 31,182 Da. To determine whether other strains of ORFV possess a similar gene, the HindIII-E fragment of the ORFV NZ7 genome was subjected to PCR using primers based on the ORFV NZ2 gene sequence. The nucleotide sequence of the ORFV NZ7 gene is 27 bp longer than its NZ2 counterpart and has three insertions. The ORFV NZ7 protein is 295 aa with a predicted mass of 32,193 Da. The amino acid identity and similarity of the ORFV NZ2 and NZ7 proteins are 78% and 87.3%, respectively.
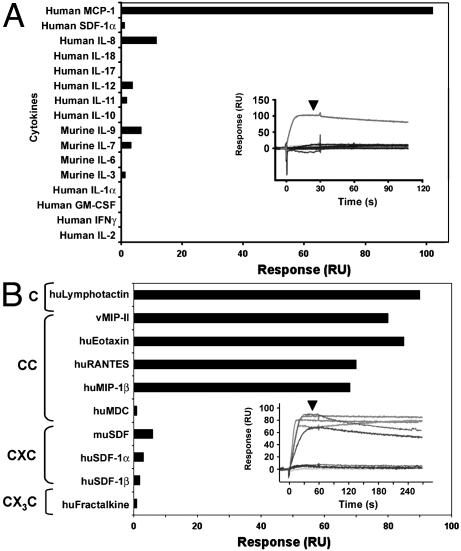
Ligand Screening by Using SPR. The similarity of the ORFV orphan protein to other cytokine-binding proteins and CBPs suggested that it may have a similar ability to interact with host cytokines. To screen for possible ligands, we immobilized the ORFV NZ2 protein at high density (Rmax = ≈100 RU) onto flow cell 2 of a Biacore CM5 chip. We injected various cytokines and chemokines over the control (flow cell 1) and ORFV NZ2 protein surface (flow cell 2) and monitored the response. Following an association period of 30 s, HBS-EP was injected over both surfaces to monitor the dissociation phase of binding (Inset of Fig. 1A). As summarized by the histogram shown in Fig. 1 A, we detected a specific interaction of the CC-chemokine MCP-1 binding the ORFV NZ2 protein, suggesting that the ORFV NZ2 protein is a CBP.
Fig. 1.
Screening of cytokines and chemokines binding to the ORFV NZ2 protein. Shown is a summary of binding data of various cytokines (A) or representative chemokines (B) generated from Biacore sensorgrams (Inset). Purified cytokines or chemokines (50 nM) were injected (at time 0) over the immobilized orphan ORFV NZ2 protein, and binding was monitored for 30 (A) or 60 (B) s. Sensorgrams (Inset) show the mass of protein binding in RU as a function of time. Arrowheads indicate point of reference for histograms. SDF, stromal cell-derived factor; hu-, human; mu-, murine; MDC, monocyte-derived chemokine.
Screening of Chemokine Classes Bound by ORFV CBP to Define Chemokine-Binding Specificity. To determine whether the ORFV CBP binds other classes of chemokines, we exploited SPR's realtime monitoring to assess affinity based on association and dissociation binding profiles. As shown in Fig. 1B, the ORFV CBP preferentially interacts with various CC-chemokines, including the Kaposi's sarcoma virus-encoded chemokine homolog vMIP-II, with binding profiles suggestive of high-affinity interactions (i.e., fast association rate and slow dissociation rate) but not with various CXC-chemokines and with the CX3C-chemokine fractalkine. Two CC-chemokines that did not bind the ORFV protein were monocyte-derived chemokine and thymus- and activation-regulated chemokine, a finding that was also observed in a screening of chemokines binding the VV-35kDa CBP (Fig. 1B and Table 1) (10). A notable high-affinity interaction occurred with the C-chemokine lymphotactin. Thus, the screening of various chemokine families by using SPR indicates that the ORFV CBP possesses a binding specificity for CC-chemokines similar to that of the CBP-II family of proteins but has an additional capacity to bind the C-chemokine lymphotactin.
Table 1. Kinetic binding parameters of ORFV NZ2 CBP to various human chemokines.
| Chemokine | kon × 107, M-1·s-1 | koff × 10-3, s-1 | Kd, nM |
|---|---|---|---|
| CC-chemokines | |||
| Eotaxin | 0.56 ± 0.02 | 0.05 ± 0.003 | 0.008 |
| MCP-3 | 0.71 ± 0.14 | 0.29 ± 0.08 | 0.043 |
| MCP-1 | 1.02 ± 0.23 | 1.86 ± 0.13 | 0.186 |
| MIP-1α | 0.64 ± 0.13 | 2.12 ± 0.36 | 0.032 |
| MIP-1β | 2.02 ± 0.39 | 11.99 ± 4.03 | 0.583 |
| I-309 | 0.23 ± 0.08 | 20.27 ± 5.69 | 9.25 |
| MDC | NB | ||
| TARC | NB | ||
| C-chemokine | |||
| Lymphotactin | 1.33 ± 0.48 | 8.01 ± 3.02 | 0.598 |
Values represent mean ± SD and were obtained from global fitting analysis of four different concentrations, each performed in triplicate. Sensorgrams were generated by observing the association and dissociation phases of chemokines binding immobilized ORFV NZ2 CBP. Chemokines that did not bind are indicated by NB. CX3C-chemokine (fractalkine) and CXC-chemokines [murine stromal cell-derived factor (SDF)-1, human SDF-1 α and β, and IL-8] did not bind and are not shown. MDC, monocyte-derived chemokine; TARC, thymus- and activation-regulated chemokine.
Kinetic and Affinity Analysis of the ORFV NZ2 CBP with Various Chemokines. To more accurately assess the affinity of the ORFV CBP interaction with chemokines, we performed kinetic binding analysis of various chemokines interacting with the ORFV CBP (Fig. 2). As summarized in Table 1, we demonstrate that the ORFV CBP interacts with the inflammatory CC-chemokines eotaxin, MCP-3, MIP-1α, MIP-1β, and MCP-1 with high affinity (Kd < 1 nM). The high-affinity interactions are the result of very fast association rates (kon > 106 M-1·s-1) and slow dissociation rates (koff < 102 s-1). A noteworthy interaction occurred between I-309 and the ORFV CBP, which exhibited a Kd of 9.3 nM (Table 1), contrasting the observed affinity (Kd > 850 nM) that was previously observed for I-309 binding the Orthopoxvirus VV-35kDa CBP (11). The most notable difference between the CBP-II binding profile and the ORFV CBP was observed with the interaction of the ORFV CBP with lymphotactin (Table 1). In contrast to the VV-35kDa protein, which was unable to interact with lymphotactin (12), ORFV CBP bound lymphotactin with high affinity (Kd = 598 pM) characterized by fast association kinetics (kon = 1.33 × 107 M-1·s-1) and slow dissociation kinetics (koff = 8.01 × 10-3 s-1). These binding parameters suggest that the ORFV CBP should be able to compete for the binding of these chemokines to their receptors, whose affinity for chemokines ranges from the low picomolar to low nanomolar range.
Fig. 2.
Sensorgrams of immobilized ORFV NZ2 CBP binding chemokines. Sensorgrams are plotted as the mass of protein binding (in RU) to immobilized ORFV NZ2 CBP as a function of time. Experimentally derived curves (black lines) from three repeat injections of MCP-1 (A), MIP-1α (B), MCP-3 (C), or eotaxin (D) at various concentrations (1, 3, 9, and 27 nM, bottom line to top line) are shown overlaid. Sensorgram on right in D shows the extended dissociation of eotaxin from the ORFV CBP. Triplicate curves were globally fitted with bia-evaluation 3.1 software using a 1:1 mass transport model (red lines) to determine the kinetic parameters presented in Table 1.
Identification of the MCP-1 Residues That Contribute to the ORFV CBP's Interaction. We previously demonstrated that the Orthopoxvirus VV-35kDa CBP-II contacts a set of conserved residues on MCP-1 that overlap with those used by the cellular MCP-1 receptor CCR2b (36–38). The results provided a structural basis for the ability of VV-35kDa to block CC-chemokines from binding their G protein-coupled receptors and revealed how the CBP-II proteins promiscuously recognize CC-chemokines (38). To assess whether the ORFV CBP binds similar residues on MCP-1 that are used by CCR2b, we performed a screen using SPR to monitor the association and dissociation profiles of single injections of each mutant (Fig. 3A). Using the wild-type MCP-1 interaction as a reference, we monitored the interaction of 32 MCP-1 point mutants (described in ref. 38) and identified at least four residues (Y13, R18, R24, and K49) that, when changed to alanine, alter the binding profile from that of the wild-type MCP-1 (Fig. 3A and Table 2, which is published as supporting information on the PNAS web site). These residues overlap with residues on MCP-1 that contribute to recognition by CCR2b (36) (Fig. 3B, Right vs. Left). Moreover, these residues coincide with the major contact residues that contribute to the interaction of VV-35kDa with MCP-1 (38, 39). Thus, the ORFV CBP binding mechanism occludes the receptor-binding site on chemokines in a manner similar to the CBP-II family of chemokine inhibitors.
Fig. 3.
ORFV CBP binds overlapping epitopes on MCP-1 that are used by the VV-35kDa protein and CCR2b and inhibits chemokine-induced signaling. (A) Overlaid sensorgrams of single injections of MCP-1 mutants at a single concentration (5 nM) by using the high-density chip (see Materials and Methods). Mutants are listed in Table 2 and are described elsewhere (36, 38). (B) Structures were generated in insight ii (Accelrys, San Diego) by using the MCP-1 NMR structure (47); the structures depict van der Waals surface representations with colors showing residues important for binding CCR2b (Left) or ORFV NZ2 CBP (Right). Light blue represents basic residues that have a minor effect on binding; purple represents basic residues that have a major effect on binding; green represents aromatic residues that have a major effect on binding; yellow represents the N terminus involved in CCR2b signaling. In each structure, arrow shows the hydrophobic groove identified as a CCR2 N-terminal peptide interaction site. (C) ORFV CBP inhibits chemokine-induced signaling in THP-1 monocytes. Shown are changes in relative fluorescence, which are proportional to levels of intracellular calcium in THP-1 cells in response to human RANTES (Left) and MIP-1α (Right). Cells were pretreated with 0, 10, 20, or 40 nM ORFV CBP (from top line to bottom line). Bar represents 5 s.
ORFV CBP Inhibits Signaling of Chemokines Through Cognate G Protein-Coupled Receptors. The observations that the ORFV CBP is capable of interacting with C- and CC-chemokines with high affinity and that the interaction is mediated by contacting residues that comprise chemokine receptor binding epitopes suggest that the ORFV CBP binds to these chemokines as a competitive inhibitor. We assessed whether the ORFV CBP is capable of blocking the binding and signaling events mediated by chemokines by testing the ability of the ORFV CBP to inhibit chemokine-induced signaling. Using a fluorometric assay to monitor the intracellular calcium levels in THP-1 cells, we demonstrated that the ORFV CBP can inhibit MIP-1α- and RANTES-induced signaling in a dose-dependent manner (Fig. 3C). Signaling in THP-1 cells in response to I-309 (CCR8) and MCP-1 (CCR2b) was also inhibited by the ORFV CBP (data not shown). Consistent with the SPR binding data, the ORFV CBP was unable to inhibit fractalkine (CX3C-chemokine) or IL-8 (CXC-chemokine) signaling in CX3CR-expressing 3T3 cells and CXCR1-expressing HEK 293 cells, respectively (data not shown). Together, these results demonstrate that ORFV CBP is a competitive inhibitor of chemokine binding and signaling that is mechanistically similar to the CBP-II family of proteins.
ORFV CBPs from Strain NZ2 and NZ7 Are Related to the ORFV GIF and the CC-CBP Family. Initial BLASTP (www.ncbi.nlm.nih.gov) searches using the amino acid sequence from the ORFV NZ2 CBP revealed that it had the highest resemblance to the ORFV GIF protein. Subsequently, we found that other poxvirus CBP-II proteins could be retrieved with BLASTP, but the homology scores were extremely low.
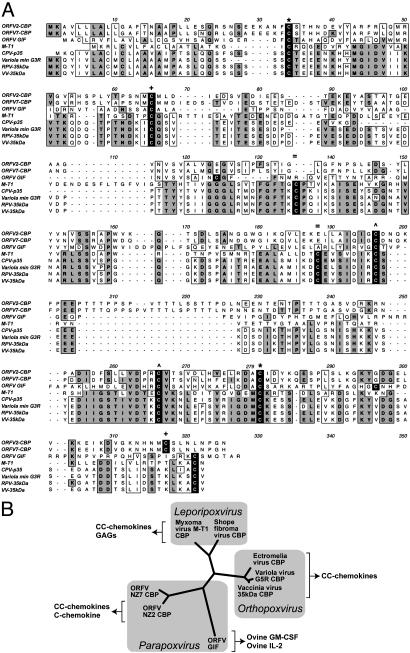
Based on the observation that the ORFV NZ2 CBP had sequence similarity to the ORFV GIF as well as limited sequence, but strong functional, similarity to known poxvirus CBPs, we performed an amino acid sequence alignment of the ORFV CBPs (from the NZ2 and NZ7 strains) with members of the CBP-II family of proteins and the ORFV GIF protein (Fig. 4A). The alignment of the ORFV CBP from the NZ2 and NZ7 strains gave the highest score against the ORFV GIF protein, displaying 19–21% identity and 33–36% similarity (Table 3, which is published as supporting information on the PNAS web site). The poxvirus CBP-II proteins exhibited 12–18% identity and 26–32% similarity with the ORFV NZ2 and NZ7 CBPs. Despite these relatively low values, inspection of the amino acid sequence alignment of these proteins revealed that these proteins do share notable regions of identity and similarity across the entire stretch of sequence (Fig. 4A). Importantly, six of the eight conserved cysteines of the CBP-II family are conserved in the ORFV CBP. Notably, these six cysteines correspond to six cysteines among the CBP-II family of proteins that form three disulfide bonds. The pair of cysteine residues absent from the ORFV CBP proteins corresponds to two residues that, among the CBP-II family of proteins, have been shown to be disulfide bonded (Fig. 4A) (16). Therefore, it is probable that the ORFV CBPs possess identical disulfide bond pairing between existing cysteine residues.
Fig. 4.
ORFV CBP and ORFV GIF are related to the CBP-II family of CC-chemokine inhibitors. (A) Sequence alignment between the ORFV CBP from strains NZ2 and NZ7, ORFV GIF, myxoma virus M-T1, CPV-p35, variola virus G3R, rabbitpox virus (RPV) 35-kDa, and the VV-35kDa. The cysteines are shown in black, and those that are involved in disulfide pairs are marked with the following symbols:*, +, =, and ^. Alignment was performed by using macvector 6.5.3 (Accelrys). (B) A phylogenetic tree, generated by using clustalw alignment (48) and phylip (49), shows the relatedness of the ORFV CBPs from strains NZ2 and NZ7, ORFV GIF (strain NZ2), and various Orthopoxvirus and Leporipoxvirus CBPs. Note that the function of the ORFV NZ7 CBP is as of yet uncharacterized and that vaccinia virus A41L, whose function is unknown, is not shown. GAGs, glycosaminoglycans.
Despite the difference in binding specificity of the ORFV GIF protein compared with the ORFV CBP, these proteins possess the highest alignment scores. Alignment of the GIF protein with the poxvirus CBPs demonstrated that GIF possesses some degree of similarity, including the positioning of cysteine residues. Although GIF possesses eight cysteines in total, only six correspond to those that are conserved among the CBP-II proteins. Two other cysteines are in nonconserved regions of the protein. Based on the sequence similarity between the CBPs from the Orthopoxviruses, Leporipoxviruses, and Parapoxvirus ORFVs and their strikingly similarity with the ORFV GIF protein, we suggest that the ORFV CBP and GIF can be classified as distant members of the CBP-II family of proteins.
Comparison of Binding Specificities Among an Expanded Family of Poxvirus CBP-II Viroceptors. As related members of a family, the genes that encode these proteins likely derive from a common ancestral poxvirus gene that has since diverged, not only in sequence, but also in binding specificity (Fig. 4B). Previous studies have characterized members of the CBP-II family of proteins and have observed several important similarities and distinctions. For instance, although the Orthopoxvirus and Leporipoxvirus CBPs have similar in vitro binding and inhibitory characteristics toward CC-chemokines (14), the Leporipoxvirus CBP-II protein from myxoma virus, termed M-T1, has an added ability to bind heparin-like molecules through a glycosaminoglycan-binding domain that is absent among the Orthopoxvirus CBP-II from vaccinia virus (40). Vaccinia virus A41L, in contrast, is a known CBP-II family member with immune-modulating activity but has no known ligand despite having been tested against various cytokines, including C5a and chemokines, suggesting it may also have an altered ligand specificity compared with the original CBP-II proteins (22). The data here demonstrate that the ORFV CBP has retained similar binding properties toward CC-chemokines as its CBP-II relatives but has the added ability to bind the CC-chemokine I-309 and the C-chemokine lymphotactin. The ORFV GIF, on the other hand, has a close sequence relationship to the ORFV CBP but has acquired a particularly distinct binding specificity toward ovine GM-CSF and ovine IL-2. GIF has shown no interaction with various chemokines (MCP-1, RANTES, IL-8, or MIP-1α), various cytokines (IL-3, IL-4, IL-5, IFN-γ, or TNF-α) or heparin (32). Importantly, the fact that members of this family of proteins possess a variety of different binding specificities (Fig. 4B) suggests that an ancestral poxvirus protein was used as a common structural scaffold that was modified over time to create binding proteins of various specificities for host molecules.
Discussion
The intimate relationship between virus and host places a special necessity on the coevolution of virus and host proteins to ensure productive infection in a host as well as efficient adaptation to a new host. This concept can be applied to viral proteins that are necessary for infection, including those proteins that dictate host range and virulence. We have described the identification of a member of the poxvirus CBP-II proteins encoded by ORFV that provides a link to a previously identified ORFV cytokine-binding protein, GIF. The sequence similarity of these proteins and the unique binding specificities that each engenders reveal that these proteins are likely the descendants of an ancestral poxvirus gene that has been altered in binding specificity during the course of poxvirus evolution.
ORFV is an epitheliotropic poxvirus able to reinfect its host after clearance of initial infection, suggesting that the immune response is dampened by viral factors (23). A number of ORFV proteins have been identified that target distinct host antiviral pathways (24, 26, 30–32). Our findings identify a CBP encoded by a member of the Parapoxvirus genus, ORFV. The ORFV CBP binds C- and CC-chemokines with high affinity and can inhibit chemokine function by occluding surface residues found on chemokines that generally overlap receptor-binding sites.
The ORFV CBP possesses characteristics similar to those of the CBP-II family of chemokine inhibitors encoded by the Orthopox-virus and Leporipoxvirus genera despite a low resemblance in their amino acid sequences. Similar to the CBP-II proteins, the ORFV CBP binds many inflammatory CC-chemokines, such as MCP-1, MIP-1α, and RANTES, that control monocyte, macrophage, and T cell recruitment to sites of infection. Interestingly, ORFV CBP does not bind monocyte-derived chemokines or thymus- and activation-regulated chemokines, suggesting the importance of inhibiting the inflammatory CC-chemokines rather than homeostatic CC-chemokines. In addition to binding CC-chemokines, ORFV CBP also interacts with the C-chemokine lymphotactin, which has been implicated in T cell chemotaxis and more recently in chemotaxis of neutrophils and B cells that express the lymphotactin receptor XCR1 (41–43). Moreover, MIP-1α, MIP-1β, RANTES, and lymphotactin can function in concert with IFN-γ as helper T cell type 1 (Th1) cytokines and can coactivate macrophages and promote natural killer and CD8+ T cells in driving a Th1 response (44). The targeting of these chemokines suggests that the ORFV CBP may function to reduce the Th1 antiviral responses mediated by chemokines in addition to inhibiting C- and CC-chemokine-induced chemotaxis. Moreover, the ORFV CBP binding specificity highlights an emerging pattern in which ORFV selectively inhibits Th1-mediated responses by having assembled a collection of Th1-dampening proteins, such as the ORFV homolog of IL-10 (25), an anti-IFN eIF2α homolog (30, 31), and the GIF protein, which binds and inhibits IL-2 and GM-CSF (32).
The regions of amino acid sequence similarity between the ORFV CBP compared with the CBP-II family of proteins strongly suggest that these proteins derive from a common poxvirus protein ancestor (Fig. 4). Moreover, the identification of the ORFV CBP and its close sequence relationship to the ORFV GIF protein provides an evolutionary link that bridges the CBP-II proteins of the Leporipoxvirus and Orthopoxvirus genera with the ORFV GIF protein, which may have been generated from a duplication of the ORFV CBP gene early after the divergence of the Parapoxvirus genus (Fig. 4B). One of the notable features that supports the familial relationship of these proteins is the conspicuous conservation of cysteine residues in the ORFV CBP and GIF proteins that possess six of the eight conserved cysteine residues found among all members of the CBP-II proteins (Fig. 4A) (16). A precedent for this type of loss of paired cysteine residues among poxvirus viroceptors has been observed in the IFN-γ receptor homolog protein (B8R) encoded by the vaccinia virus, which possesses six of eight cysteine residues that are typically conserved in mammalian IFN-γ receptors and other poxvirus IFN-γ receptor homologs. Not only is the function of B8R intact, but the binding specificity for IFN-γ is not species restricted (45, 46). The observation that the ORFV CBP has retained six cysteines able to form three of the four disulfide bonds that were observed in the CPV-p35 protein implies that the ORFV CBP likely adopts the overall structure of the CPV-p35 protein. The absent disulfide bond in ORFV CBP may provide additional conformational flexibility to accommodate the additional binding to lymphotactin and I-309. GIF has two additional cysteines in nonconserved positions that replace the two lost conserved cysteines. The structural consequences for these additional cysteines are unknown but may affect the conformation and specificity of GIF. Based on the sequence similarity of the ORFV CBP and GIF proteins with the Orthopoxvirus and Leporipoxvirus CBPs, we suggest that the ORFV GIF and the ORFV CBP proteins are bona fide members of the CBP-II family of proteins.
One of the surprising features of this family of viral proteins is that various members appear to have acquired a variety of unique adaptations that are notably reflected in their binding specificity (Fig. 4B). As noted, although the Orthopoxvirus and Leporipoxvirus CBPs have similar binding affinities toward CC-chemokines (14), the Leporipoxvirus CBP-II protein M-T1 has an additional glycosaminoglycan-binding domain that is absent from the Orthopoxvirus VV-35kDa protein (40). Our findings here reveal that ORFV CBP has the ability to bind similar CC-chemokines that the Leporipoxvirus and Orthopoxvirus CC-CBPs bind but is also capable of binding the CC-chemokine I-309 and the C-chemokine lymphotactin. Finally, the ORFV GIF, which most resembles the ORFV CBP, appears to have acquired a distinct binding specificity for ovine GM-CSF and ovine IL-2 (Fig. 4B) (32). The observation that GIF is specific for ovine, but not human, GM-CSF and IL-2 demonstrates the unique evolutionary history of the ORFV with sheep as its primary host and suggests that the GIF binding specificity was likely modified after speciation of ORFV. Together, these observations suggest that, despite common ancestral origins, these proteins have since diverged, thus providing a remarkable evolutionary example of how viral proteins may undergo shifts in ligand specificity likely driven by host-mediated selective pressure.
Overall, as we begin to accumulate data on the specificity of binding interactions among various protein families encoded by viruses, the full extent of the relationships between these viral proteins and host proteins will be revealed. Comparing and contrasting the biophysical interactions of viral proteins that interact with the host immune system highlights the propensity for adaptation by viruses to accommodate change associated with speciation, host adaptation, and evolving host immune responses. Furthermore, these observations underscore the need to characterize each member within a family of related viral proteins to assess their unique properties, because it cannot be assumed that all members are identical. Overall, the poxvirus CBP-II family of cytokine-/CBPs provides an impressive example of how viroceptors can undergo subtle or more profound shifts in ligand specificity and demonstrates the astonishing versatility and adaptability of viral immune evasion proteins.
Supplementary Material
Acknowledgments
This work was funded by the Canadian Institutes of Health Research, the National Cancer Institute (Canada), and the Health Research Council of New Zealand. S.B.F., A.M., and C.A.M. are supported by the Health Research Council of New Zealand. G.M. holds a Canada Research Chair in Molecular Virology. B.T.S. was funded by an Ontario Graduate Scholarship for Science and Technology. T.M.H. was supported by grants from the National Institutes of Health and the American Heart Association.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ORFV, orf virus; CBP-II, type II CC-chemokine-binding protein; CBP, chemokine-binding protein; GM-CSF, granulocyte–macrophage colony-stimulating factor; GIF, GM-CSF/IL-2 inhibitory factor; VV-35kDa, vaccinia virus 35-kDa; RU, response units; CPV, cowpox virus; SPR, surface plasmon resonance; MCP, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; Th1, type I helper T cells.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. CAD99366 (ORFV NZ2 CBP) and AY453066 (ORFV NZ7 CBP)].
References
- 1.Goh, C. S., Bogan, A. A., Joachimiak, M., Walther, D. & Cohen, F. E. (2000) J. Mol. Biol. 299, 283-293. [DOI] [PubMed] [Google Scholar]
- 2.Rossi, D. & Zlotnik, A. (2000) Annu. Rev. Immunol. 18, 217-242. [DOI] [PubMed] [Google Scholar]
- 3.Murphy, P. M., Baggiolini, M., Charo, I. F., Hebert, C. A., Horuk, R., Matsushima, K., Miller, L. H., Oppenheim, J. J. & Power, C. A. (2000) Pharmacol. Rev. 52, 145-176. [PubMed] [Google Scholar]
- 4.Hoogewerf, A., Kuschert, G. S. V., Proudfoot, A. E., Borlat, F., Clark-Lewis, I., Power, C. A. & Wells, T. N. C. (1997) Biochemistry 36, 13570-13578. [DOI] [PubMed] [Google Scholar]
- 5.Kuschert, G. S. V., Coulin, F., Power, C. A., Proudfoot, A. E. I., Hubbard, R. E., Hoogewerf, A. J. & Wells, T. N. C. (1999) Biochemistry 38, 12959-12968. [DOI] [PubMed] [Google Scholar]
- 6.Proudfoot, A. E., Handel, T. M., Johnson, Z., Lau, E. K., LiWang, P., Clark-Lewis, I., Borlat, F., Wells, T. N. & Kosco-Vilbois, M. H. (2003) Proc. Natl. Acad. Sci. USA 100, 1885-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seet, B. T. & McFadden, G. (2002) J. Leukocyte Biol. 72, 24-34. [PubMed] [Google Scholar]
- 8.Murphy, P. M. (2001) Nat. Immunol. 2, 116-122. [DOI] [PubMed] [Google Scholar]
- 9.Seet, B. T., Johnston, J., Brunetti, C., Barrett, J., Everett, H., Cameron, C., Sypula, J., Nazarian, S., Lucas, A. & McFadden, G. (2002) Annu. Rev. Immunol. 21, 377-423. [DOI] [PubMed] [Google Scholar]
- 10.Burns, J. M., Dairaghi, D. J. & Schall, T. J. (2001) J. Biol. Chem. 277, 2785-2789. [DOI] [PubMed] [Google Scholar]
- 11.Alcamí, A., Symons, J. A., Collins, P. D., Williams, T. J. & Smith, G. L. (1998) J. Immunol. 160, 624-633. [PubMed] [Google Scholar]
- 12.Smith, C. A., Smith, T. D., Smolak, P. J., Friend, D., Hagen, H., Gerhart, M., Park, L., Pickup, D. J., Torrance, D., Mohler, K., et al. (1997) Virology 236, 316-327. [DOI] [PubMed] [Google Scholar]
- 13.Smith, V. P. & Alcamí, A. (2000) J. Virol. 74, 8460-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalani, A. S., Ness, T. L., Singh, R., Harrison, J. K., Seet, B. T., Kelvin, D. J., McFadden, G. & Moyer, R. W. (1998) Virology 250, 173-184. [DOI] [PubMed] [Google Scholar]
- 15.Graham, K. A., Lalani, A. S., Macen, J. L., Ness, T. L., Barry, M., Liu, L.-Y., Lucas, A., Clark-Lewis, I., Moyer, R. W. & McFadden, G. (1997) Virology 229, 12-24. [DOI] [PubMed] [Google Scholar]
- 16.Carfi, A., Smith, C. A., Smolak, P. J., McGrew, J. & Wiley, D. C. (1999) Proc. Natl. Acad. Sci. USA 96, 12379-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalani, A. S., Masters, J., Graham, K., Liu, L., Lucas, A. & McFadden, G. (1999) Virology 256, 233-245. [DOI] [PubMed] [Google Scholar]
- 18.Dabbagh, K., Xiao, Y., Smith, C., Stepick-Biek, P., Kim, S. G., Lamm, W. J., Liggitt, D. H. & Lewis, D. B. (2000) J. Immunol. 165, 3418-3422. [DOI] [PubMed] [Google Scholar]
- 19.Parry, C. M., Simas, J. P., Smith, V. P., Stewart, C. A., Minson, A. C., Efstathiou, S. & Alcamí, A. (2000) J. Exp. Med. 191, 573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Berkel, V., Barrett, J., Tiffany, H. L., Fremont, D. H., Murphy, P. M., McFadden, G., Speck, S. H. & Virgin, H. W. (2000) J. Virol. 74, 6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander, J. M., Nelson, C. A., van Berkel, V., Lau, E. K., Studts, J. M., Brett, T. J., Speck, S. H., Handel, T. M., Virgin, H. W. & Fremont, D. H. (2002) Cell 111, 343-356. [DOI] [PubMed] [Google Scholar]
- 22.Ng, A., Tscharke, D. C., Reading, P. C. & Smith, G. L. (2001) J. Gen. Virol. 82, 2095-2105. [DOI] [PubMed] [Google Scholar]
- 23.Haig, D. M. & Mercer, A. A. (1998) Vet. Res. 29, 311-326. [PubMed] [Google Scholar]
- 24.Fleming, S. B., McCaughan, C. A., Andrews, A. E., Nash, A. D. & Mercer, A. A. (1997) J. Virol. 71, 4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming, S. B., Haig, D. M., Nettleton, P., Reid, H. W., McCaughan, C. A., Wise, L. M. & Mercer, A. (2000) Virus Genes 21, 85-95. [DOI] [PubMed] [Google Scholar]
- 26.Lyttle, D. J., Fraser, K. M., Fleming, S. B., Mercer, A. A. & Robinson, A. J. (1994) J. Virol. 68, 84-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, M., Clauss, M., Lepple-Wienhues, A., Waltenberger, J., Augustin, H. G., Ziche, M., Lanz, C., Buttner, M., Rziha, H. J. & Dehio, C. (1999) EMBO J. 18, 362-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savory, L. J., Stacker, S. A., Fleming, S. B., Niven, B. E. & Mercer, A. A. (2000) J. Virol. 74, 10699-10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise, L. M., Veikkola, T., Mercer, A. A., Savory, L. J., Fleming, S. B., Caesar, C., Vitali, A., Makinen, T., Alitalo, K. & Stacker, S. A. (1999) Proc. Natl. Acad. Sci. USA 96, 3071-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haig, D. M., McInnes, C. J., Thomson, J., Wood, A., Bunyan, K. & Mercer, A. (1998) Immunology 93, 335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McInnes, C. J., Wood, A. R. & Mercer, A. A. (1998) Virus Genes 17, 107-115. [DOI] [PubMed] [Google Scholar]
- 32.Deane, D., McInnes, C. J., Percival, A., Wood, A., Thomson, J., Lear, A., Gilray, J., Fleming, S., Mercer, A. & Haig, D. (2000) J. Virol. 74, 1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, A. J., Barns, G., Fraser, K., Carpenter, E. & Mercer, A. A. (1987) Virology 157, 13-23. [DOI] [PubMed] [Google Scholar]
- 34.Mercer, A. A., Fraser, K., Barns, G. & Robinson, A. J. (1987) Virology 157, 1-12. [DOI] [PubMed] [Google Scholar]
- 35.Fraser, K. M., Hill, D. F., Mercer, A. A. & Robinson, A. J. (1990) Virology 176, 379-389. [DOI] [PubMed] [Google Scholar]
- 36.Hemmerich, S., Paavola, C., Bloom, A., Bhakta, S., Freedman, R., Grunberger, D., Krstenansky, J., Lee, S., McCarley, D., Mulkins, M., et al. (1999) Biochemistry 38, 13013-13025. [DOI] [PubMed] [Google Scholar]
- 37.Jarnagin, K., Grunberger, D., Mulkins, M., Wong, B., Hemmerich, S., Paavola, C., Bloom, A., Bhakta, S., Diehl, F., Freedman, R., et al. (1999) Biochemistry 38, 16167-16177. [DOI] [PubMed] [Google Scholar]
- 38.Seet, B. T., Singh, R., Paavola, C., Lau, E. K., Handel, T. M. & McFadden, G. (2001) Proc. Natl. Acad. Sci. USA 98, 9008-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beck, C. G., Studer, C., Zuber, J. F., Jachez Demange, B., Manning, U. & Urfer, R. (2001) J. Biol. Chem. 276, 43270-43276. [DOI] [PubMed] [Google Scholar]
- 40.Seet, B. T., Barrett, J., Robichaud, J., Shilton, B., Singh, R. & McFadden, G. (2001) J. Biol. Chem. 276, 30504-30513. [DOI] [PubMed] [Google Scholar]
- 41.Huang, H., Li, F., Cairns, C. M., Gordon, J. R. & Xiang, J. (2001) Biochem. Biophys. Res. Commun. 281, 378-382. [DOI] [PubMed] [Google Scholar]
- 42.Kelner, G. S., Kennedy, J., Bacon, K. B., Kleyensteuber, S., Largaespada, D. A., Jenkins, N. A., Copeland, N. G., Bazan, J. F., Moore, K. W. & Schall, T. J. (1994) Science 266, 1395-1399. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, T., Imai, T., Kakizaki, M., Nishimura, M., Takagi, S. & Yoshie, O. (1998) J. Biol. Chem. 273, 16551-16554. [DOI] [PubMed] [Google Scholar]
- 44.Dorner, B. G., Scheffold, A., Rolph, M. S., Hüser, M. B., Kaufmann, S. H., Radbruch, A., Flesch, I. E. & Kroczek, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcamí, A. & Smith, G. L. (1995) J. Virol. 69, 4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossman, K., Upton, C., Buller, R. M. & McFadden, G. (1995) Virology 208, 762-769. [DOI] [PubMed] [Google Scholar]
- 47.Handel, T. M. & Domaille, P. J. (1996) Biochemistry 35, 6569-6584. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Felsenstein, J. (1989) Cladistics 5, 164-166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.