Abstract
Human cells resist viral infections by a variety of mechanisms. Viruses must overcome host cell restrictions to successfully reproduce their genetic material. Here, we identify a host restriction to viral replication that acts at the stage of particle assembly. Viral protein U (Vpu) is an HIV-1 accessory protein that enhances particle assembly and release in most human cells, but not in simian cells. By using human-simian cell heterokaryons, we show that the inhibition of assembly in human cells is dominant. Vpu overcomes the block to assembly in human cells and in human-simian heterokaryons. The HIV-1 vpu gene may have evolved to counteract an assembly restriction that is present in human cells.
Mammalian cells have developed a number of nonimmune mechanisms to inhibit the replication of retroviruses. Fv-1 is a murine host cell gene whose product inhibits replication of murine leukemia viruses (1). Restrictions in human cells inhibit replication of murine retroviruses (Ref1), while restrictions in nonhuman primate cells block infection by multiple divergent retroviruses (Lv-1) (2). A subset of humans exhibit genetic polymorphisms that restrict HIV replication, such as the CCR5-Δ32 allele that provides protection from macrophage-tropic HIV strains in the homozygous state by preventing entry (3). Recently, a cellular cytidine deaminase, CEM15/APOBEC3G, was found to restrict HIV infection in human T lymphocytes by rendering progeny virions noninfectious (4). The activity of this host cell restriction factor is counteracted by the HIV accessory protein virion infectivity factor (Vif). Here, we describe a host cell restriction to retroviral replication that acts at the stage of particle assembly and release. The restriction is present in most human cells but lacking in simian cells, and is overcome by the HIV-1 accessory protein viral protein U (Vpu).
Vpu is a 16-kDa phosphoprotein that is expressed late in virus replication from a bicistronic mRNA that also codes for the HIV-1 envelope glycoprotein (5, 6). Vpu is a type I integral membrane protein that forms homooligomers in cellular membranes (7). Vpu is not incorporated into HIV-1 particles, and, therefore, must perform its functions within the infected cell in which it is expressed. Two distinct functions have been associated with Vpu. Vpu interacts with CD4 within the endoplasmic reticulum of infected cells, where it targets CD4 for degradation through interactions with the WD domain protein h-βTrCP (8, 9). Vpu is also known to enhance HIV particle assembly through an as-yet-undefined mechanism. These two functions of Vpu are associated with distinct functional domains and occur in separate subcellular locations (10, 11). Curiously, the effect of Vpu on particle release is cell-type-dependent (12–14). A strong effect on particle assembly has been shown in human epithelial cell lines, T cell lines, primary T lymphocytes, and primary macrophages (12, 13, 15). In contrast, simian cells such as CV-1, Cos-1, and Cos-7 allow efficient particle assembly and release in the absence of Vpu, and enhancement of particle production by Vpu is not seen in these cells. Whereas HIV requires an intact vpu gene for efficient viral particle release, HIV-2 and most SIVs do not encode a vpu homolog. Thus, while HIV requires Vpu to efficiently replicate in human cells, most SIV species do not require Vpu for replication in their host primate species. However, several HIV-2 isolates have been shown to encode a Vpu-like activity in their env gene that facilitates viral particle release (16, 17).
Here we demonstrate that human cells express a cellular restrictive factor that remarkably impedes HIV-1 particle production. The restrictive factor is dominant in simian-human heterokaryons, resulting in significant inhibition of viral particle release. The cellular block to particle production in these simian-human heterokaryons is relieved by the expression of Vpu. These results point to a new host cell restriction of HIV replication that is present in human cells and overcome by Vpu.
Materials and Methods
Cells and Plasmids. HeLa (CCL-2), Hep-2 (CCL-23), Cos-7 (CRL 1651), and Vero (CCL-81) cells were obtained from the American Type Culture Collection; BSC-40 were obtained from B. Moss (National Institutes of Health). These cells were maintained in DMEM with 10% FBS and antibiotics at 37°C in 5% CO2. Jurkat clone E6–1 cells were obtained from A. Weiss (National Institutes of Health) through the National Institutes of Health AIDS Reference and Reagent Repository, and were maintained in RPMI medium 1640 supplemented with 2 mM glutamine and 10% FBS. HeLa-CD4-LTR-βGal (p4 cells; ref. 18) were cultured in DMEM (plus 10% FBS) supplemented with G418 at 200 μg/ml. The full-length infectious HIV-1 molecular clone pNL4–3 and the Vpu deletion mutant, pNL4–3/Udel, have been described (19). The HIV-1 Gag/protease expression vector, 3-CCCC, was obtained from H.-G. Krausslich (Universitätsklinikum, Heidelberg) (20). This vector expresses both HIV-1 gag and protease genes under the control of the cytomegalovirus (CMV) immediate-early promoter. pcDNA-Vphu is an expression plasmid bearing a codon-optimized Vpu sequence obtained by PCR cloning into the EcoRI–HindIII sites of pcDNA 3.1 (Invitrogen). pBABE-Vphu was constructed by inserting the codon-optimized vpu gene into the BamHI–EcoRI sites of pBABE-puro (21). The amino acid sequence of the protein encoded by pcDNA-Vphu is identical to that of NL4–3 Vpu (S.P.B., unpublished work).
Virus Production and Infections. HIV-1 virus stocks were generated by calcium phosphate transfection of 293T cells with the infectious molecular clone, pNL4–3 or pNL4–3/Udel, with or without the vesicular stomatitis virus envelope glycoprotein G (VSV-G) expression plasmid pHCMV-G (22). After an overnight transfection, fresh growth medium was added. Virus stocks were harvested after 48 h, were filter sterilized, and assessed for infectivity by using p4 cells (18). For quantitation of HIV-1 release, Cos-7 and HeLa cells were seeded in a six-well tissue culture plate at 1 × 106 cells per well and cultured overnight in complete growth medium. Infections were performed in triplicate the following day by using a multiplicity of infection of 1 for NL4–3 or NL4–3/Udel viruses. Twelve to 16 h after infection, the cells were washed and fresh growth medium was added. Virion content in the supernatants was quantified at the indicated time points by using a p24 capture ELISA as described (23). A retroviral stock encoding Vphu was prepared by transfecting 293T cells with plasmids pBABE-Vphu, pCL-Ampho (24), and pHCMV-G (22). Cellular supernatants were harvested 2 days posttransfection, filtered through 0.45-μm filters, and aliquoted for use.
Transfection/Transduction. Cos-7 cells were transfected with 3-CCCC plasmid DNA by using a commercial liposome preparation, DMRIE-C (Invitrogen) with protocols provided by the manufacturer. After an overnight transfection, medium was replaced and cells were processed further for the cell fusion assays. Similarly, HeLa cells or Hep-2 cells were transfected with pcDNA-Vphu before cell fusion for particle release rescue. Jurkat E6–1 T-cells were transduced with a retroviral vector (pBABE-Vphu) for expression of Vpu before fusion with Cos-7 cells. In the HIV Gag particle release assays shown in Fig. 1, HeLa or Cos-7 cells (50,000 cells per well) were transfected with 1 μg of 3-cccc DNA together with 500 ng of pcDNA 3.1 or pCDNA-Vphu per well with DMRIE-C. Cells and supernatants were harvested at the indicated time points for p24 quantitation, and, in some experiments, the efficiency of release was expressed as the percentage of released p24 (p24 in supernatant/p24 in supernatant plus p24 in cells).
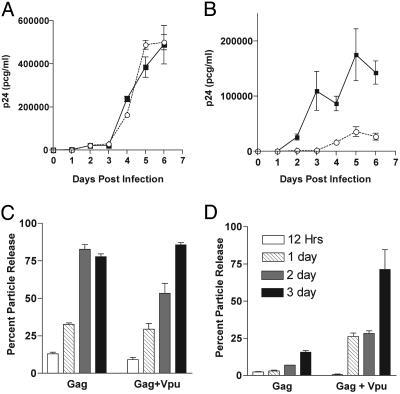
Fig. 1.
Vpu-mediated enhancement of particle production is cell-type-dependent. (A) Cos-7 cells were infected with VSV-G-pseudotyped NL4–3(▪) or NL4–3/Udel (○). p24 release was followed over time. (B) HeLa cells were infected with pseudotyped viruses and monitored for p24 release as in A. Error bars indicate SD from triplicate wells. (C) Cos-7 cells were transfected with Gag-pro alone or Gag-pro plus Vpu expression plasmids, and p24 in cells and supernatants were measured at the time points listed in D. Percent particle release was calculated as p24 in supernatant/(p24 in supernatant plus p24 in cells). (D) HeLa cells were transfected with Gag-pro alone or Gag-pro plus Vpu expression plasmids and p24 monitored as in C. The key indicating the number of days posttransfection is the same for C and D.
Staining of Cells with CellTracker Probes. Cos-7 cells were stained with CellTracker green CMFDA (5-chloromethylfluorescein diacetate; Molecular Probes) and HeLa, Hep-2, Vero, or Jurkat cells were stained with CellTracker orange CMTMR (5-(and- 6)-{[(4-chloromethyl)benzoyl]amino} tetramethylrhodamine; Molecular Probes) before fusion. Dyes were dissolved in DMSO and stored as 10-mM stock solutions. Cells were stained for 15–45 min at 37°C in culture medium containing 5–10 μM of the respective dye. After staining, the cells were washed with Dulbecco's PBS and further incubated for at least 30 min at 37°C in growth medium.
Cell–Cell Fusion with Polyethylene Glycol (PEG). Cos-7 and HeLa cells stained with CellTracker dyes were fused by PEG, with a molecular mass of 3,000–3,700 Da (Sigma). Briefly, 10 × 106 each of HeLa and Cos-7 cells were mixed. The cells were subjected to low-speed centrifugation (1,000 × g), the supernatant was removed, and the pellets were resuspended by gentle tapping. One milliliter of 50% PEG in PBS plus 2% glucose was added dropwise with intermittent gentle mixing. The cell mixture was incubated for 90 s at room temperature. After the incubation step, PBS (1 ml) was added slowly and the cells were incubated for an additional 60 s. Three milliliters of PBS plus 2% FBS was next added slowly, and the cells were pelleted by low-speed centrifugation. The cells were washed twice with PBS plus 2% FBS to remove PEG, suspended in growth medium with 20% FBS, and incubated overnight or at least 4 h at 37°C before sorting. Cell sorting was performed in the Nashville Veteran's Administration Hospital Flow Cytometry Special Resource Center with a FACStar Plus FACsorter (Beckton Dickinson) with an excitation laser frequency of 488 nm and emission detected at 525 nm. Data acquisition and analysis was performed with cellquest software. Sorted cells plated in replicates of 2 × 104 cells per well in a 12-well tissue culture plate and the supernatants were assayed periodically for p24 release. In the experiments where results are presented as percent particle release, cells and supernatants were harvested at each time point and assayed for p24 content by ELISA. We noted that the majority of the heterokaryons remained viable as judged by Trypan blue exclusion at 5 days postsorting, and that their morphology and continued adherence to plastic suggested that they remained viable beyond 7 days after cell sorting.
Results
Enhancement of HIV Particle Release by Vpu Is Cell-Type-Dependent. We compared particle release between a human epithelial cell line (HeLa) and African green monkey epithelial cell line (Cos-7). Viral infections of these CD4-negative cells were facilitated by pseudotyping with VSV-G. Cells were infected with NL4–3 or with the Vpu deletion mutant NL4–3/Udel at a multiplicity of infection of 1. We then monitored these cells for virus release from a single round replication in culture (Fig. 1). Cos-7 cells supported the efficient production and release of viral particles in the presence or absence of Vpu (Fig. 1 A). In contrast, HeLa cells supported efficient particle production only when Vpu was expressed (Fig. 1B, filled squares versus open circles). We next sought to confirm that the observed cell-type-specific differences reflect an effect of Vpu on the efficiency of Gag protein release from cells in the absence of other viral proteins. To confirm this hypothesis, Cos-7 and HeLa cells were transfected with the Gag-protease expression construct, 3-CCCC, with or without expression of Vpu. Cells and supernatants were harvested at 12, 24, 48, and 72 h after transfection for determination of p24 content. Results are reported as percent Gag particle release (Fig. 1 C and D). Gag alone in the absence of Vpu was released efficiently from Cos-7 cells. Thirteen percent of the protein produced in the transfected cells was able to exit from cells at 12 h, and the efficiency of release increased gradually, reaching 30% by day 1 and a maximum of 80% by days 2 and 3 (Fig. 1C). In HeLa cells. the release of Gag in the absence of Vpu was remarkably restricted. Only 2% of Gag was released at 12 h, and the particle release efficiency remained low, reaching a maximum of only 15% by day 3 (Fig. 1D). The defect was overcome when Vpu was transiently expressed in HeLa cells along with Gag, with >70% particle release by day 3. These data are consistent with published reports indicating a cell-type-specific activity of Vpu (12–14) and provide the basis for further experiments designed to examine the nature of the cell-specific defect in particle assembly. We term the particle production phenotype of Cos-7 cells “permissive/Vpu-unresponsive,” and that of HeLa cells “restrictive/Vpu-responsive.”
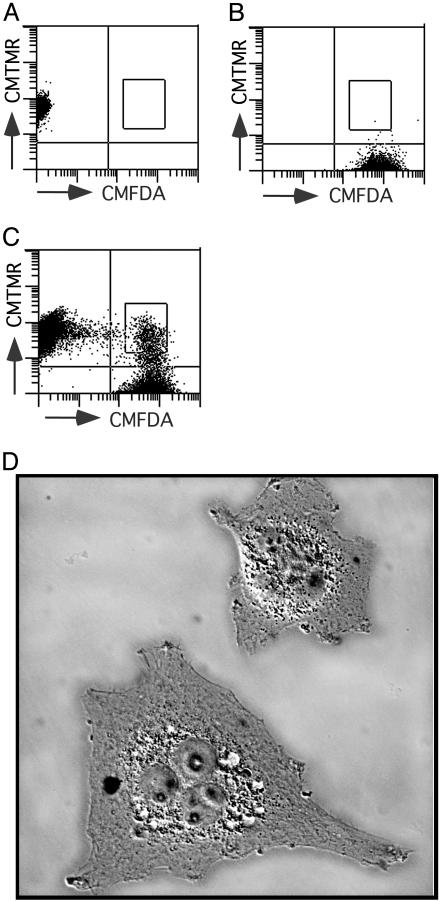
Defining the Nature of Cellular Factor Responsible for Vpu-Dependent Particle Release. The differences in particle release reported above could be explained in two ways: (i) Cos-7 cells express a Vpu-like factor that enables virus release in the absence of viral Vpu, and this Vpu-like factor is lacking or diminished in HeLa cells; or (ii) HeLa cells contain a cellular factor that inhibits HIV-1 particle release, and Vpu overcomes this inhibitory factor. The experimental strategy we used to differentiate between the two possibilities was to analyze particle production from heterokaryons produced from permissive Cos-7 cells and restrictive HeLa cells. Fig. 2 illustrates the technique used for cell–cell fusion and subsequent sorting of the heterokaryons for analysis of Vpu-mediated particle release. Before fusion, Cos-7 cells were transfected with the Gag/protease expression construct, 3-CCCC. Gag-expressing permissive Cos-7 cells were stained green with CMFDA (Fig. 2B), while HeLa cells were stained orange with CMTMR (Fig. 2 A). Cell–cell fusions were performed with PEG, and fused HeLa-Cos-7 cells were sorted, based on the double staining with CMFDA and CMTMR (Fig. 2C). Staining with these fluorescent probes allowed isolation of heterokaryons generated only between HeLa and Cos-7, but not HeLa-HeLa or Cos-Cos, which was critical for the study. Heterokaryons generated between Gag-expressing Cos-7 cells stained green with CMFDA and mock-transfected Cos-7 cells stained orange with CMTMR were also sorted to assess the impact of fusion process itself on the viral particle release phenotype. By microscopy, the sorted double-positive cells were large and multinucleated (Fig. 2D).
Fig. 2.
Sorting for purification of Cos-HeLa heterokaryons. (A) HeLa cells stained with CMTMR and examined by flow cytometry. (B) Cos-7 cells stained with CMFDA and examined by flow cytometry. (C) Dual-staining Cos-HeLa heterokaryons were sorted as indicated by the box in the right upper quadrant. (D) Heterokaryons were examined by differential interference contrast microscopy on a Nikon TE2000 inverted microscope at ×400 magnification. Intensity of background (coverslip) has been adjusted downward to emphasize heterokaryons.
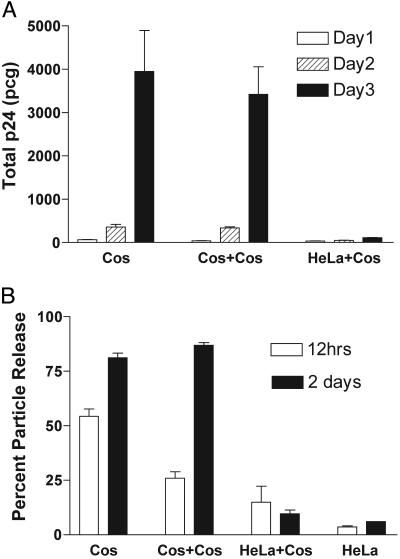
Cos-HeLa Heterokaryons Demonstrate a Dominant-Restrictive Particle Release Phenotype. Cos-HeLa and Cos-Cos heterokaryons were cultured in equal numbers and supernatants were monitored for p24 release over time. Results are presented in Fig. 3A as total p24 release in picograms. Cos-Cos heterokaryons demonstrated particle production that was equal to that of transfected, non-fused Cos cells (Fig. 3A). In marked contrast, the same Cos cells when fused with HeLa cells formed heterokaryons that released extremely low levels of Gag into the supernatant (Fig. 3A). These results indicate that Cos-HeLa heterokaryons are restrictive for particle production and that the process of fusion itself does not account for the observed restriction.
Fig. 3.
Cos-HeLa heterokaryons are restricted for particle production. Fusions between Cos cells expressing Gag-pro and untransfected Cos or HeLa cells were performed and purified by fluorescence-activated cell sorting as described in the text. (A) An equal number of transfected Cos cells, Cos-Cos heterokaryons, or Cos-HeLa heterokaryons were plated in parallel and analyzed for particle output by p24 ELISA. Experiments were performed in triplicate wells and error bars representing SD are shown. (B) In a separate heterokaryon experiment, cellular and supernatant p24 were determined to define the efficiency of particle release. Cos-Cos and Cos-HeLa heterokaryons expressing Gag-pro were prepared as described in the text. An equal number of HeLa cells transfected with the same Gag-pro expression construct were included as controls (bars on the right). The p24 antigen content of the cells and supernatants was determined by ELISA, with results presented as percent particle release.
To demonstrate that the block observed in Cos-HeLa heterokaryons occurs at the level of assembly, and was not due to decreased total amounts of protein production, we repeated the Cos-HeLa fusion experiment and harvested cells as well as supernatants for measurement of p24. For comparison, we included unfused HeLa cells transfected with the identical Gag-protease expression vector. Results are expressed as percent particle release from duplicate experiments (Fig. 3B). Approximately 50% of the Gag protein produced in Cos-7 cells was present in supernatants at 12 h, and by 2 days in culture cells, this level increased to 80%. Cos-Cos heterokaryons revealed similar levels of protein release. In contrast, the transfected HeLa control cells revealed a maximum of only 6% Gag release at the 2-day time point. Cos-HeLa heterokaryons demonstrated a remarkably similar restriction to that observed in HeLa cells, resulting in only 9% Gag release at 2 days. These results indicate that the HeLa cells possess a cellular factor that dominantly inhibits HIV Gag release from HeLa-Cos heterokaryon and argue against the hypothesis that Cos cells express a Vpu-like factor that enhances particle release.
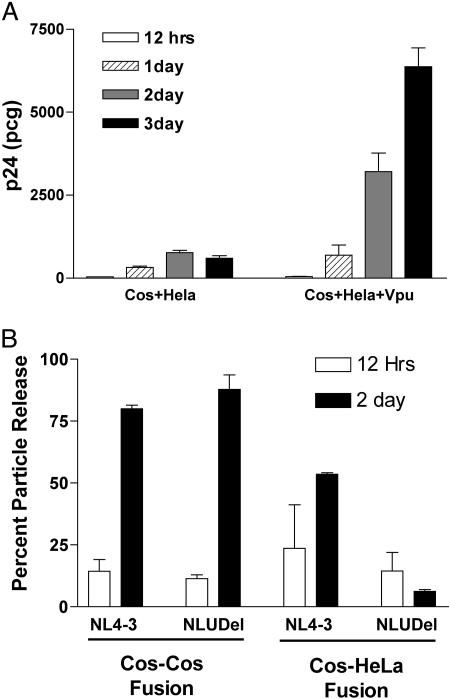
The Cellular Inhibitory Factor in Cos-HeLa Heterokaryons Is Vpu-Responsive. We next examined the role of Vpu in overcoming the potent cellular block to assembly observed in Cos-HeLa heterokaryons. We performed cell–cell fusions between Cos-7 cells expressing Gag and HeLa cells transfected with pcDNA3.1 (control) or with a Vpu expression construct, pcDNA-Vphu. The biological activity of the Vpu protein expressed by this construct has been shown in an earlier experiment (Fig. 1). Cos-HeLa and Cos-HeLa+Vpu heterokaryons were monitored for Gag release in culture. Results are expressed as means of total p24 concentration in supernatants derived from triplicate experiments. The Cos-HeLa heterokaryons resulting from fusion with control HeLa cells revealed a potent cellular block as already described (Fig. 4A). Remarkably, transient expression of Vpu in HeLa cells restored particle release from Cos-HeLa heterokaryons. At 12 h, the amount of Gag in the supernatants was comparable to that released from Cos-HeLa heterokaryons without Vpu, but, as time progressed, the Gag release from Cos-HeLa+Vpu heterokaryons increased steadily to ≈700 pg by day 1, 3,200 pg by day 2, and a maximum of 6.3 ng after 3 days in culture.
Fig. 4.
Vpu rescues the host cell restriction in Cos-HeLa heterokaryons. (A) Cos cells expressing Gag-pro were fused with HeLa cells (Left) or with HeLa cells expressing Vpu (Right). p24 output over time is indicated. Experiments were performed in triplicate and SD are indicated. (B) Vpu rescues particle output in Cos-HeLa heterokaryons in the setting of viral infection. Cos-HeLa heterokaryons were infected with VSV-G-pseudotyped NL4–3 or NL4–3/Udel (NLUDel), and p24 was measured in supernatant and cells to calculate the percent particle release.
We demonstrated above that transient expression of Vpu in HeLa cells rescued the Gag release defect present in Cos-HeLa heterokaryons. We next sought to determine whether this cellular block to replication was relieved with Vpu in the context of infectious virus. In this experiment, Cos-Cos and Cos-HeLa heterokaryons were generated as described. An equal number of these heterokaryons were infected with VSV-G-pseudotyped NL4–3 or NL4–3/Udel viruses at a multiplicity of infection of 1. Cells and supernatants were harvested at 12 h and 2 days postinfection and assayed for virion release by p24 ELISA. Results are expressed as percent particle release from duplicate experiments (Fig. 4B). Two days after the infection, the efficiency of release of both NL4–3 and NL4–3/Udel viruses was very similar in Cos-Cos heterokaryons (≈80% and 87%). In Cos-HeLa heterokaryons, 54% of the wild-type NL4–3 virus produced within the cells was found in the supernatant, whereas the release of Vpu deletion mutant, NL4–3/Udel, was severely impaired (6% at 2 days postinfection). These results demonstrate that Vpu reverses the host cell restriction of particle release that is present in Cos-HeLa heterokaryons in the context of viral infection.
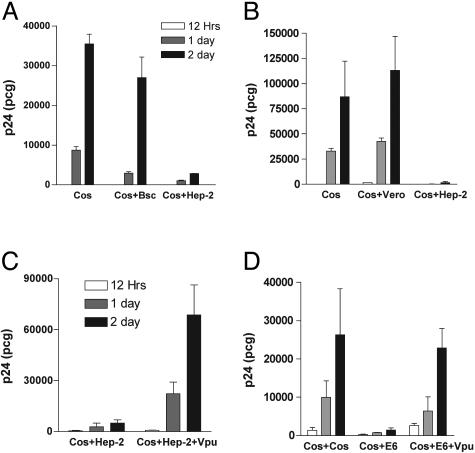
The Cellular Restrictive Factor Is Present in Human Cells and Is Absent in African Green Monkey Cells. The data presented thus far argue for the presence of a potent cellular restrictive factor for particle release in HeLa cells. To determine whether this restrictive factor is present in other human cells, we generated heterokaryons between Gag-expressing Cos-7 and mock-transfected Hep-2 (human laryngeal carcinoma epithelial cell line) or BSC-40 (African green monkey kidney epithelial cell line). Heterokaryons were generated, sorted, and cultured in replicates of equal numbers. An identical number of Gag-expressing Cos-7 cells were included as a reference for the amount of Gag release from the transfected, permissive cells in the experiment. Cos-BSC-40 heterokaryons released a significant amount of Gag into the culture medium, suggesting that these simian cells also lack the host restrictive factor (Fig. 5A). Remarkably, Gag release was very low from Cos-Hep-2 heterokaryons, indicating a dominant-restrictive phenotype similar to that of Cos-HeLa heterokaryons. This experiment was then repeated with the substitution of Vero cells as a fusion partner for Cos-7 cells, to extend the number of simian cells examined. Vero cells exhibited a permissive phenotype very similar to that of Cos-7 and Bsc-40 cells, whereas Hep-2 cells again demonstrated a strong restriction to particle release (Fig. 5B). We then tested the ability of Vpu to overcome the restriction present in Hep-2 cells. Cos cells were fused with Hep-2 cells that were transfected with pcDNA 3.1 (control) or with pcDNA-Vphu. Results are presented as total p24 release from triplicate cultures of equal number of Cos-Hep-2 and Cos-Hep-2+Vpu heterokaryons (Fig. 5C). Vpu expression in Hep-2 cells resulted in a dramatic rescue of particle release in the context of Cos-Hep-2 heterokaryons. These results strongly support the presence of a cellular restrictive factor that is common to human cell lines and indicate that Vpu overcomes this inhibitor to promote efficient particle release.
Fig. 5.
Restrictive versus permissive phenotype of additional human and simian cells. (A) Cos cells transfected with Gag/protease expression plasmid were plated (bars on the left) or fused to BSC-40 cells or Hep-2 cells. Output is reported as p24 released in supernatant at 12 h, 1 day, and 2 days. Error bars indicate SD from mean of triplicate samples. (B) Cos cells expressing Gag/protease were plated alone or after fusion with Vero or Hep-2 cells. p24 output was measured as described. (C) Vpu rescues the host cell restriction of Hep-2 cells. Cos cells expressing Gag and protease were fused with Hep-2 cells (Left) or with Hep-2 cells expressing Vpu (Right). Results reported as p24 output in supernatant, mean, and SD of triplicate samples. (D) Cos cells expressing Gag and protease were fused with untransfected Cos (Left), with Jurkat E6–1 cells (Center), or with Jurkat E6–1 cells expressing Vpu through retrovirus-mediated transduction (Right).
The Restrictive Factor Is Present in Human T Cells. HeLa and Hep-2 cells are human epithelial cell lines and may not be representative of cells that are normally infected with HIV in vivo.Totest for the presence of the Vpu-responsive cellular inhibitor in additional human cells, we generated heterokaryons between Cos-7 cells and Jurkat T cells (E6–1 clone). As shown in Fig. 5D, Cos–Cos fusions were permissive for particle release, whereas Cos-Jurkat heterokaryons were highly restricted. To rescue this restriction in the heterokaryon assay, Vpu was expressed in the Jurkat fusion partner by using retroviral transduction. Vpu successfully restored particle release from the Cos-Jurkat cell heterokaryons (Fig. 5D). These results indicate that relevant human cells express a cellular inhibitor of particle assembly or release that can be overcome by Vpu.
Discussion
The identification of host cell restrictions to viral replication can provide fundamental new knowledge that is useful in understanding viral pathogenesis. For example, the discovery of CCR5 as the coreceptor for macrophage-tropic strains of HIV prompted a search for polymorphisms in the CCR5 gene that might correlate with infection or disease progression. The discovery that CCR5-Δ32 is protective for HIV infection when present in both alleles and partially protective for disease in heterozygous individuals, provided evidence of the central importance of CCR5 in HIV transmission (3). Such fundamental knowledge can also lead to the development of novel antiviral therapies, as illustrated by small molecule inhibitors of CCR5 that are presently under development (25, 26). Cell-type-specific restrictions may initially be recognized by the observation of differential viral replication kinetics in specific cell types. Recently (4), a unique host cell restrictive factor that inhibits HIV-1 particle infectivity, CEM15/APOBEC3G, was identified by a subtractive hybridization approach. The search for this inhibitory molecule began with the observation that some cells were restricted for infectious particle production in the absence of the vif gene, whereas others were permissive. By using cell–cell fusion techniques, the existence of a dominant host restriction that is overcome by Vif was confirmed (27). By using a similar approach, we now provide evidence for a distinct host restriction that acts at the stage of particle assembly and is overcome by HIV-1 Vpu.
It has been recognized for some time that Vpu enhances particle assembly in human cells, such as HeLa cells, and not in simian cells, such as CV-1 and Cos (12, 13). The basis of this difference, however, had remained obscure. Gottlinger et al. (12) demonstrated that the Vpu-mediated enhancement of particle release in HeLa cells was not specific for HIV-1, but was also effective for HIV-2, visna virus, and Moloney murine leukemia virus. Vpu also has been shown to enhance particle release by the pathogenic SIVmac239 isolate (17). These results suggest that Vpu acts to alter a cellular pathway that is used by diverse retroviruses. Our results now clarify and extend these previous findings. The fact that the restrictive phenotype of HeLa, Hep-2 cells, and Jurkat E6–1 cells was dominant in Cos-HeLa, Cos-Hep-2, and Cos-Jurkat heterokaryons indicates that a cellular inhibitor of retroviral particle assembly exists in restrictive cells. It is important to note that primary human lymphocytes and macrophages depend on Vpu for efficient particle production, suggesting that they also express the Vpu-responsive inhibitor of assembly (15). However, the expression of the inhibitor may not be universal in human cells, because some human cell lines have been characterized as Vpu-unresponsive (14). In addition, SIVagm, which lacks a vpu gene, can efficiently replicate in a subset of human cells (28). It is also not yet known whether all simian cells lack this host restriction to viral replication. It is tempting to speculate that the acquisition of the vpu gene by SIV species was a key step to overcoming a restriction to replication in human cells, and that this acquisition was important in the origins of the HIV-1 epidemic. HIV-2 may have used a distinct mechanism to overcome the same host cell restriction, because the HIV-2 envelope glycoprotein has been shown to contain Vpu-like activity (16, 17, 29). The Vpu-like activity of the HIV-2 ROD10 isolate enhances HIV-1 particle output in a manner indistinguishable from that of Vpu (17). Thus, both human immunodeficiency viruses can overcome the common host restriction described in this work, but have evolved genetically distinct strategies to do so.
An inhibitor of particle assembly may act at one of several steps in the assembly process. Retroviral Gag proteins are known to interact very rapidly with cellular membranes and to form homomultimers that are resistant to detergent extraction (30). The precise pathway taken by Gag in reaching the plasma membrane assembly site is not certain. However, clues have come from the recent observation that HIV Gag interacts with TSG101, a member of the ESCRT complex involved in vesicular trafficking within the cell (31). Other retroviruses such as MLV also use the endosomal sorting machinery, although the MLV late domain does not interact with TSG101 (31). There is presently intense interest in defining the interactions of Gag with endosomal trafficking pathways. We speculate that the inhibitor of assembly present in restrictive cells blocks the normal trafficking of Gag through vesicular sorting pathways, and that Vpu releases this block to allow Gag to proceed along this pathway.
In summary, we provide evidence for the existence of a host cell restriction to retroviral particle assembly in human cells. HIV-1 Vpu overcomes this restriction through a mechanism that remains to be defined. A complete understanding of the Vpu-responsive restriction in human cells will require elucidation of the molecular basis of this host cell restriction.
Acknowledgments
We thank H-G. Krausslich for the gift of the 3-CCCC plasmid and J. Burns (University of California at San Diego, La Jolla, CA) for pHCMV-G. This work was supported by National Institutes of Health (NIH) Grant R01 AI40338, NIH Grants AI52007 and AI47985 (to P.S.), a grant from the Elizabeth Glaser Pediatric AIDS Foundation, and by NIH Grant T32 AI07474 (to V.V.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Vpu, viral protein U; PEG, polyethylene glycol; SIV, simian immunodeficiency virus; VSV-G, vesicular stomatitis virus glycoprotein G; CMFDA, 5-chloromethylfluorescein diacetate; CMTMR, (5-(and-6)-{[(4-chloromethyl)benzoyl]amino} tetramethylrhodamine.
References
- 1.Bishop, K. N., Bock, M., Towers, G. & Stoye, J. P. (2001) J. Virol. 75, 5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatziioannou, T., Cowan, S., Goff, S. P., Bieniasz, P. D. & Towers, G. J. (2003) EMBO J. 22, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang, Y., Paxton, W. A., Wolinsky, S. M., Neumann, A. U., Zhang, L., He, T., Kang, S., Ceradini, D., Jin, Z., Yazdanbakhsh, K., et al. (1996) Nat. Med. 2, 1240-1243. [DOI] [PubMed] [Google Scholar]
- 4.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646-650. [DOI] [PubMed] [Google Scholar]
- 5.Arrigo, S. J. & Chen, I. S. (1991) Genes Dev. 5, 808-819. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz, S., Felber, B. K., Fenyo, E. M. & Pavlakis, G. N. (1990) J. Virol. 64, 5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strebel, K., Klimkait, T., Maldarelli, F. & Martin, M. A. (1989) J. Virol. 63, 3784-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bour, S., Schubert, U. & Strebel, K. (1995) J. Virol. 69, 1510-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margottin, F., Bour, S. P., Durand, H., Selig, L., Benichou, S., Richard, V., Thomas, D., Strebel, K. & Benarous, R. (1998) Mol. Cell 1, 565-574. [DOI] [PubMed] [Google Scholar]
- 10.Schubert, U. & Strebel, K. (1994) J. Virol. 68, 2260-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert, U., Bour, S., Ferrer-Montiel, A. V., Montal, M., Maldarell, F. & Strebel, K. (1996) J. Virol. 70, 809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlinger, H. G., Dorfman, T., Cohen, E. A. & Haseltine, W. A. (1993) Proc. Natl. Acad. Sci. USA 90, 7381-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geraghty, R. J., Talbot, K. J., Callahan, M., Harper, W. & Panganiban, A. T. (1994) J. Med. Primatol. 23, 146-150. [DOI] [PubMed] [Google Scholar]
- 14.Sakai, H., Tokunaga, K., Kawamura, M. & Adachi, A. (1995) J. Gen. Virol. 76, 2717-2722. [DOI] [PubMed] [Google Scholar]
- 15.Schubert, U., Clouse, K. A. & Strebel, K. (1995) J. Virol. 69, 7699-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bour, S., Akari, H., Miyagi, E. & Strebel, K. (2003) Virology 309, 85-98. [DOI] [PubMed] [Google Scholar]
- 17.Bour, S. & Strebel, K. (1996) J. Virol. 70, 8285-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragic, T., Charneau, P., Clavel, F. & Alizon, M. (1992) J. Virol. 66, 4794-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimkait, T., Strebel, K., Hoggan, M. D., Martin, M. A. & Orenstein, J. M. (1990) J. Virol. 64, 621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wodrich, H., Schambach, A. & Krausslich, H. G. (2000) Nucleic Acids Res. 28, 901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgenstern, J. P. & Land, H. (1990) Nucleic Acids Res. 18, 3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yee, J. K., Friedmann, T. & Burns, J. C. (1994) Methods Cell Biol. 43, 99-112. [DOI] [PubMed] [Google Scholar]
- 23.Wehrly, K. & Chesebro, B. (1997) Methods 12, 288-293. [DOI] [PubMed] [Google Scholar]
- 24.Naviaux, R. K., Costanzi, E., Haas, M. & Verma, I. M. (1996) J. Virol. 70, 5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strizki, J. M., Xu, S., Wagner, N. E., Wojcik, L., Liu, J., Hou, Y., Endres, M., Palani, A., Shapiro, S., Clader, J. W., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dragic, T., Trkola, A., Thompson, D. A., Cormier, E. G., Kajumo, F. A., Maxwell, E., Lin, S. W., Ying, W., Smith, S. O., Sakmar, T. P. & Moore, J. P. (2000) Proc. Natl. Acad. Sci. USA 97, 5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon, J. H., Gaddis, N. C., Fouchier, R. A. & Malim, M. H. (1998) Nat. Med. 4, 1397-1400. [DOI] [PubMed] [Google Scholar]
- 28.Shimano, R., Inubushi, R., Amano, K., Ogasawa, T. & Adachi, A. (1998) Virus Genes 16, 137-139. [DOI] [PubMed] [Google Scholar]
- 29.Bour, S., Schubert, U., Peden, K. & Strebel, K. (1996) J. Virol. 70, 820-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding, L., Derdowski, A., Wang, J. J. & Spearman, P. (2003) J. Virol. 77, 1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, 55-65. [DOI] [PubMed] [Google Scholar]