Abstract
Vast world reserves of methane gas are underutilized as a feedstock for production of liquid fuels and chemicals due to the lack of economical and sustainable strategies for selective oxidation to methanol1. Current processes to activate the strong C–H bond (104 kcal/mol) in methane require high temperatures, are costly and inefficient, and produce waste2. In nature, methanotrophic bacteria perform this reaction under ambient conditions using metalloenzymes called methane monooxygenases (MMOs). MMOs are thus the optimal inspiration for an efficient, green catalyst3. There are two types of MMOs. Soluble MMO (sMMO), which is expressed by several strains of methanotrophs under copper limited conditions, oxidizes methane with a well characterized catalytic diiron center4. Particulate methane monooxygenase (pMMO), an integral membrane metalloenzyme produced by all methanotrophs, is composed of three subunits, pmoA, pmoB, and pmoC, arranged in a trimeric α3β3γ3 complex5. Despite 20 years of research and the availability of two crystal structures, the metal composition and location of the pMMO metal active site are not known. Here we show that pMMO activity is dependent on copper, not iron, and that the copper active site is located in the soluble domains of the pmoB subunit rather than within the membrane. Recombinant soluble fragments of pmoB (spmoB) bind copper and exhibit propylene and methane oxidation activities. Disruption of each copper center in spmoB by mutagenesis indicates that the active site is a dicopper center. These findings resolve the pMMO controversy and provide a promising new approach to developing environmentally friendly C–H oxidation catalysts.
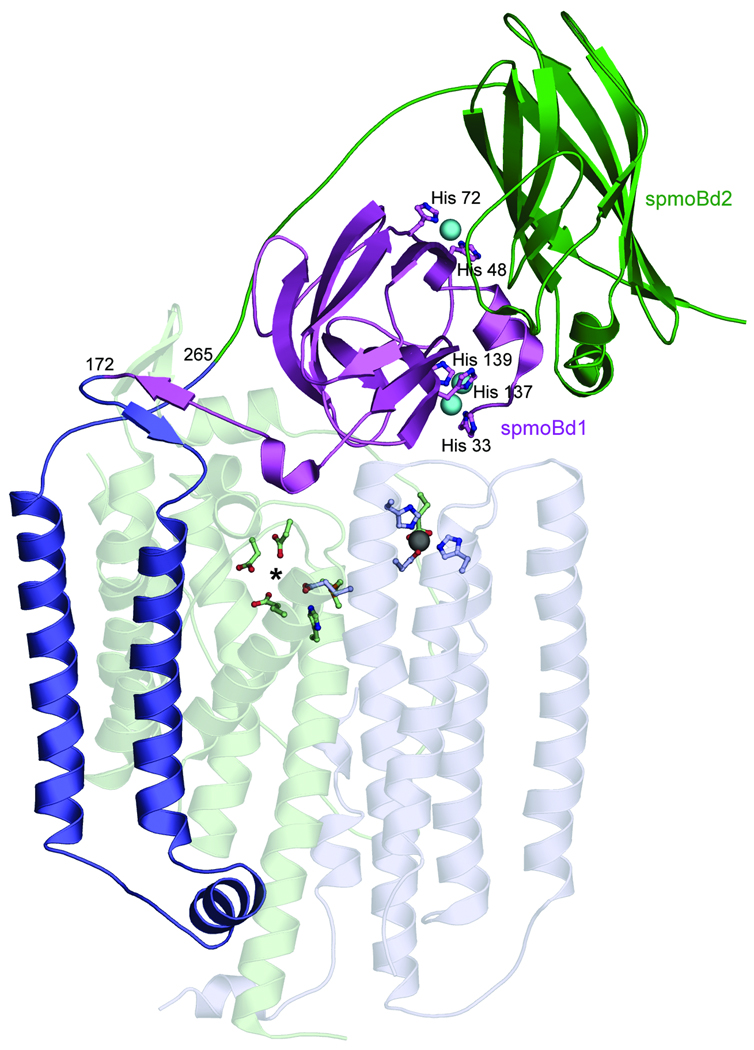
Three distinct metal centers were identified in the 2.8 Å resolution structure of pMMO from Methylococcus capsulatus (Bath)6, 7 (Fig. 1). Two copper centers are located in the soluble regions of the pmoB subunit, which form two cupredoxin domains. A dicopper center with a short Cu-Cu distance of 2.5–2.7 Å8, 9 is coordinated by highly conserved5 residues His 33, His 137, and His 139 and is also observed in pMMO from Methylosinus trichosporium OB3b10. The second site, a monocopper center coordinated by residues His 48 and His 72, is not conserved in M. trichosporium OB3b pMMO. A third metal center, occupied by zinc from the crystallization buffer, is located within the membrane and is ligated by Asp 156, His 160, and His 173 from pmoC and possibly Glu195 from pmoA6. This site houses a copper ion in the M. trichosporium OB3b pMMO structure10.
Figure 1. Structure of M. capsulatus (Bath) pMMO protomer.
(PDB accession code 1YEW). The N-terminal cupredoxin domain of pmoB (spmoBd1) is shown in purple, the C-terminal cupredoxin domain of pmoB (spmoBd2) is shown in green, and the two transmembrane helices are shown in blue. In the recombinant spmoB protein, spmoBd1 and spmoBd2 are connected by a GKLGGG sequence linking residues 172 and 265 instead of the two transmembrane helices. Copper ions are shown as cyan spheres and ligands are shown as ball-and-stick representations. The pmoA (transparent light green) and pmoC (transparent light blue) subunits are composed of transmembrane helices. The location of the zinc ion (gray sphere) has been proposed to house a diiron center. A hydrophilic patch of residues marked with an asterisk is the site of a proposed tricopper center.
The nature of the pMMO metal active site is not established and has been intensely controversial11, 12. On the basis of electron paramagnetic resonance (EPR) spectroscopic data, Chan and coworkers initially proposed the presence of multiple trinuclear copper clusters13, 14. In light of the crystallographic data, this model was revised to a single catalytic tricopper cluster located at an intramembrane site composed of conserved hydrophilic residues (Fig. 1)15. This model also includes the binding of an additional 10 copper ions to the C-terminal soluble domain of pmoB16. Both this intramembrane site and the C-terminal domain of pmoB are devoid of metal ions in the crystal structures. By contrast, DiSpirito, Münck, and coworkers have argued on the basis of Mössbauer spectroscopic data that the pMMO active site is a diiron center located at the intramembrane site occupied by zinc in the M. capsulatus (Bath) pMMO structure (Fig. 1) and copper in the M. trichosporium OB3b pMMO structure17.
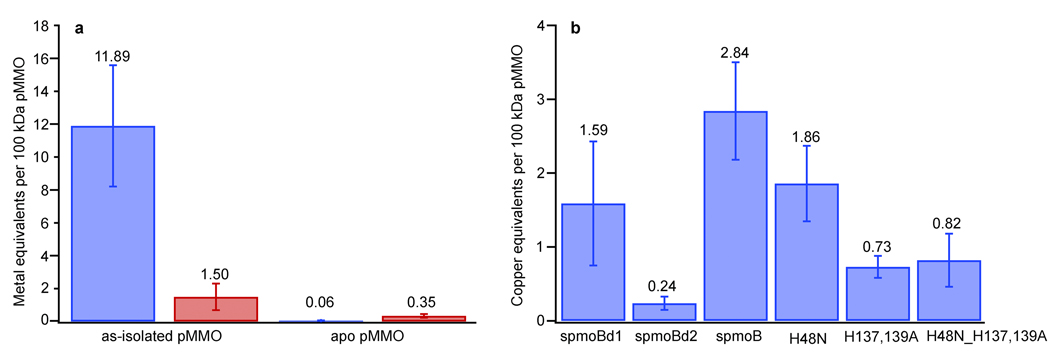
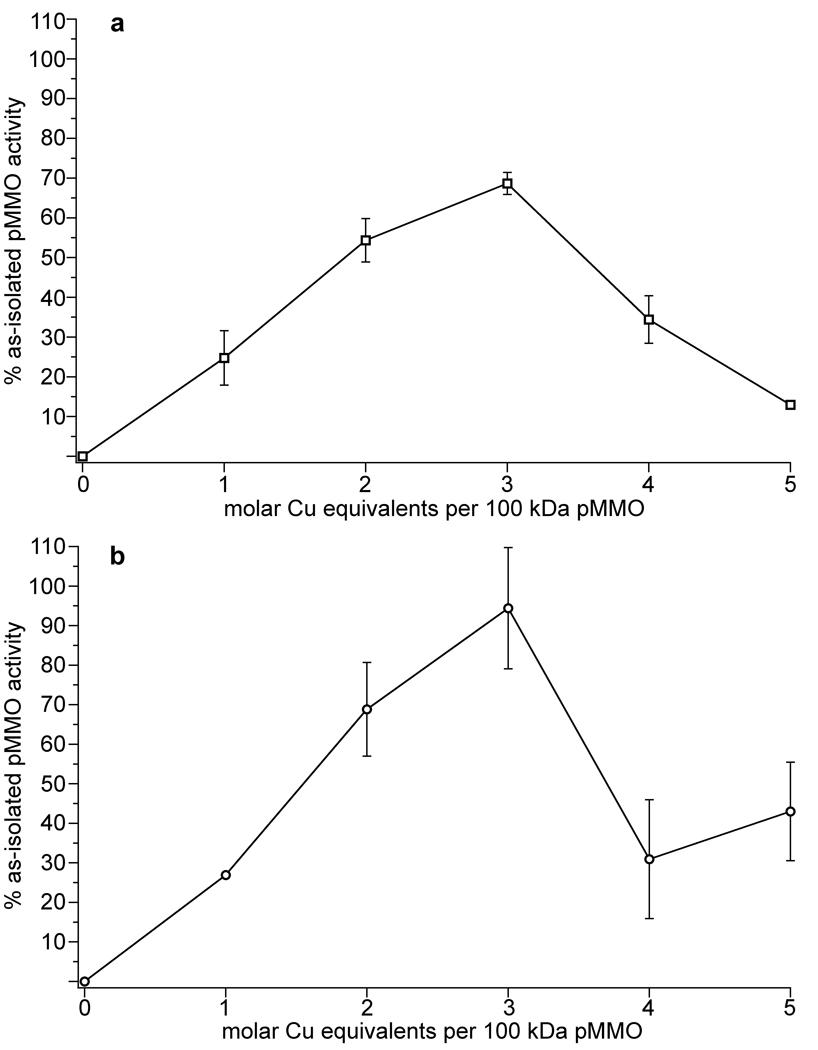
To determine the metal requirement for pMMO activity, we first removed the metals from as-isolated M. capsulatus (Bath) pMMO membranes by treatment with cyanide (Fig. 2a). Prior to cyanide treatment, as-isolated samples exhibited propylene epoxidation activities of 50–200 nmol propylene oxide min−1 mg−1. These values are comparable to or higher than values reported by other researchers for as-isolated membranes18–20 (Table S1). We used membranes rather than purified pMMO to more closely resemble the natural environment and to avoid activity loss typically observed upon solubilization and purification9. Cyanide treatment completely abolished pMMO activity as measured by the propylene epoxidation assay. We then titrated stoichiometric amounts of copper and/or iron into the apo pMMO samples and assessed the effects on activity. Addition of 2–3 equivalents of copper per 100 kDa pMMO protomer restores ~70% of the propylene epoxidation activity (Fig. 3a). We also tested the ability of pMMO reconstituted with copper to oxidize methane to methanol. As-isolated pMMO has a specific activity of 22.9 ± 6.1 nmol methanol mg−1 min−1 and apo pMMO reconstituted with 3 equivalents of copper exhibits an activity of 21.7 ± 3.5 nmol methanol mg−1 min−1, restoring greater than 90% of the methane oxidation activity (Fig. 3b). By contrast, addition of iron, aerobically or anaerobically, does not restore or improve activity if added alone or in combination with copper. These experiments demonstrate that copper, not iron17, is the metal at the active site of pMMO. The requirement for only 2–3 equivalents of copper indicates that large numbers of copper ions may not be necessary for activity as proposed previously12. Some of the copper associated with the as-isolated membranes may instead be bound adventitiously. Interestingly, addition of copper beyond three equivalents inhibits pMMO activity. Repeating the cyanide treatment and adding back 2–3 equivalents of copper can reverse this inhibition (Fig. S1a). The excess copper ions likely react with reductant to generate hydrogen peroxide, which reversibly inhibits pMMO. This phenomenon has been studied for pMMO from M. trichosporium OB3b21. In support of this explanation, catalase prevents inhibition upon addition of excess copper with maximal activity still observed at 2–3 equivalents (Fig. S1b).
Figure 2. Metal analysis.
a, Metal content of as-isolated pMMO and apo pMMO prepared by cyanide treatment. Metal content is expressed per 100 kDa pMMO protomer with copper in blue and iron in red. b, Copper content of refolded spmoB variants. Reported values are an average of at least four independent measurements for pMMO samples and at least six independent refolding experiments for each spmoB variant.
Figure 3. Restoration of activity to apo pMMO by the addition of copper.
Copper equivalents added are expressed per 100 kDa pMMO protomer. Representative titrations are shown. Addition of 2–3 equivalents of copper restored a, ~70% of the propylene epoxidation activity and b, ~90% of the methane oxidation activity.
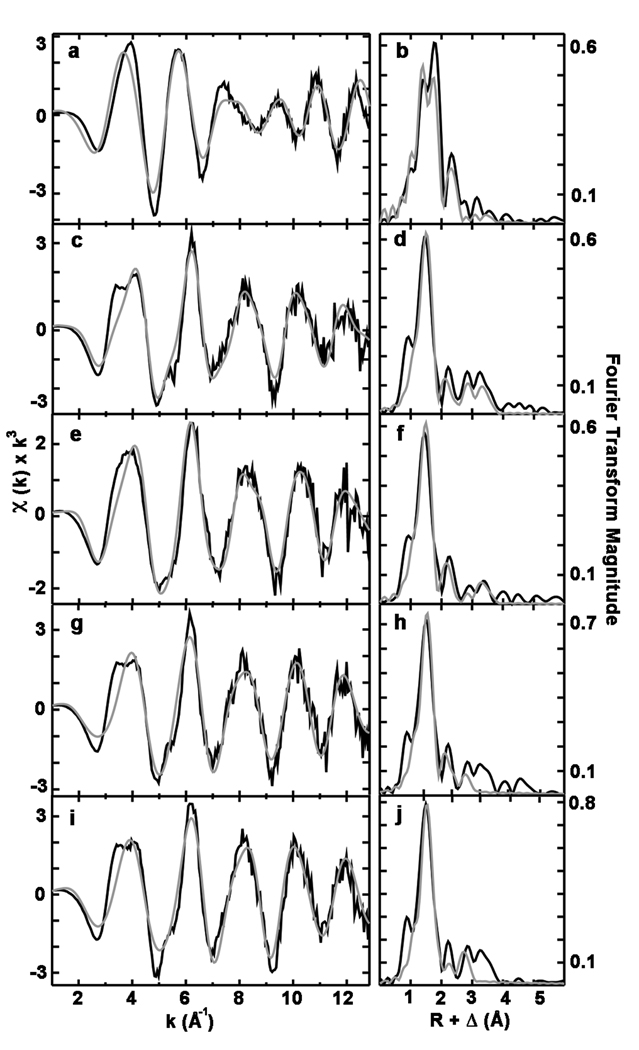
To probe the coordination environment of copper in pMMO, we collected X-ray absorption spectroscopic (XAS) data. As-isolated pMMO is a mixture of Cu(I) and Cu(II) as shown by both a 1s➔4p transition and a weak 1s➔3d feature22 in the near edge spectra (Fig. S2). Apo pMMO reconstituted with three equivalents of CuSO4 and treated with duroquinol to mimic the activity assay conditions has a stronger Cu(I) 1s➔4p transition consistent with the presence of reductant (Fig. S2). Fourier transforms of the extended X-ray absorption fine structure (EXAFS) data for both samples (Figs. 4b and 4d) show two scattering interactions corresponding to nearest neighbor ligands at approximately 2 and 2.5 Å. Long-range ligand scattering (> 3 Å) is also evident. The as-isolated pMMO EXAFS were best fit with two Cu-O/N environments at 1.92 and 2.09 Å and a Cu-Cu interaction centered at 2.66 Å. The reconstituted pMMO EXAFS were best fit with a single set of O/N ligands at 1.95 Å. Most important, there was a significant improvement in fit when a Cu-Cu scattering environment centered at 2.53 Å was included (Table S2). Debye-Waller factors, a measure of the metal-ligand bond disorder, were high in both Cu-Cu interaction simulations (> 5.5 × 10−3 Å2) while the coordination numbers were consistently low (≤ 0.25). These results suggest either a near stoichiometric population of distinct, but non-resolvable, Cu-Cu interactions with scattering that destructively overlaps centered at 2.66 Å and 2.53 Å or low populations (less than 25%) of bound copper coordinated in a multinuclear cluster. Regardless, restoration of activity upon copper addition is accompanied by formation of a copper cluster, which is most likely the dinuclear copper center observed in the crystal structures6, 10.
Figure 4. Cu EXAFS data and simulations for pMMO and spmoB variants.
Raw EXAFS and phase shifted Fourier transforms are shown for as-isolated pMMO (a and b), copper reconstituted pMMO (c and d), spmoB (e and f), spmoB_H48N (g and h), and spmoB_H137,139A (i and j). Raw unfiltered data are shown in black and best fit simulations are shown in gray.
Possible locations for the M. capsulatus (Bath) pMMO active site include the two copper centers in the soluble regions of pmoB, the intramembrane zinc site, and the intramembrane hydrophilic patch proposed to house a trinuclear copper site (Fig. 1)11. We cloned and expressed in E. coli several proteins corresponding to the soluble cupredoxin domains of the pmoB subunit. These proteins include the N-terminal cupredoxin domain (spmoBd1, residues 33–172), the C-terminal cupredoxin domain (spmoBd2, residues 265–414), and both domains tethered via a GKLGGG linker, which replaces the two transmembrane helices (spmoB) (Fig. 1). All ligands to the two copper centers are contained within spmoBd1, although the mononuclear site lies at the domain interface. These recombinant proteins were refolded from solubilized inclusion bodies in the presence of CuSO4. Copper analysis by the bicinchoninic acid (BCA) method23 indicates the presence of 2.84 ± 0.66 copper ions per protein for spmoB, 1.59 ± 0.84 copper ions per protein for spmoBd1, and 0.24 ± 0.09 copper ions per protein for spmoBd2 (Fig. 2b). These values, which are an average of at least six independent refolding experiments, are consistent with the M. capsulatus (Bath) pMMO crystal structure6 in which spmoB binds three copper ions and spmoBd2 binds no copper ions. Despite the excess of copper present during the refolding procedure, we do not observe the binding of approximately 10 copper ions to spmoBd2 as reported by Chan and coworkers16. Refolding of spmoB in the presence of iron yields 0.17 ± 0.1 iron ions per protein. The spmoBd1 protein is extremely unstable, probably due to the removal of spmoBd2. In the M. capsulatus (Bath) structure,6 the interface between the two soluble domains is extensive with ~1400 Å2 buried surface area for each chain. The loop region connecting dicopper ligand His 33 with monocopper ligand His 48 is heavily involved in this interface with a number of hydrogen bonds (Fig. S3a) and hydrophobic contacts with residues from the C-terminal cupredoxin domain. Increased flexibility upon disruption of all these interactions likely affects both copper centers and accounts for the reduced copper binding by spmoBd1.
In spite of loading with Cu(II) and possibly due to photoreduction, the XANES spectrum of spmoB (Fig. S2) is typical of three- to four-coordinate Cu(I) based on the low intensity of the 1s➔4p transition, the overall edge structure, and the lack of any discernible 1s➔3d feature. The EXAFS for spmoB (Figs. 4e and f) were best fit with a Cu-O/N environment at 1.95 Å and a Cu-Cu interaction at 2.53 Å, the inclusion of which significantly improved the fit (Table S2). The combined copper binding and XAS data indicate that spmoB can assemble copper centers similar to those in pMMO.
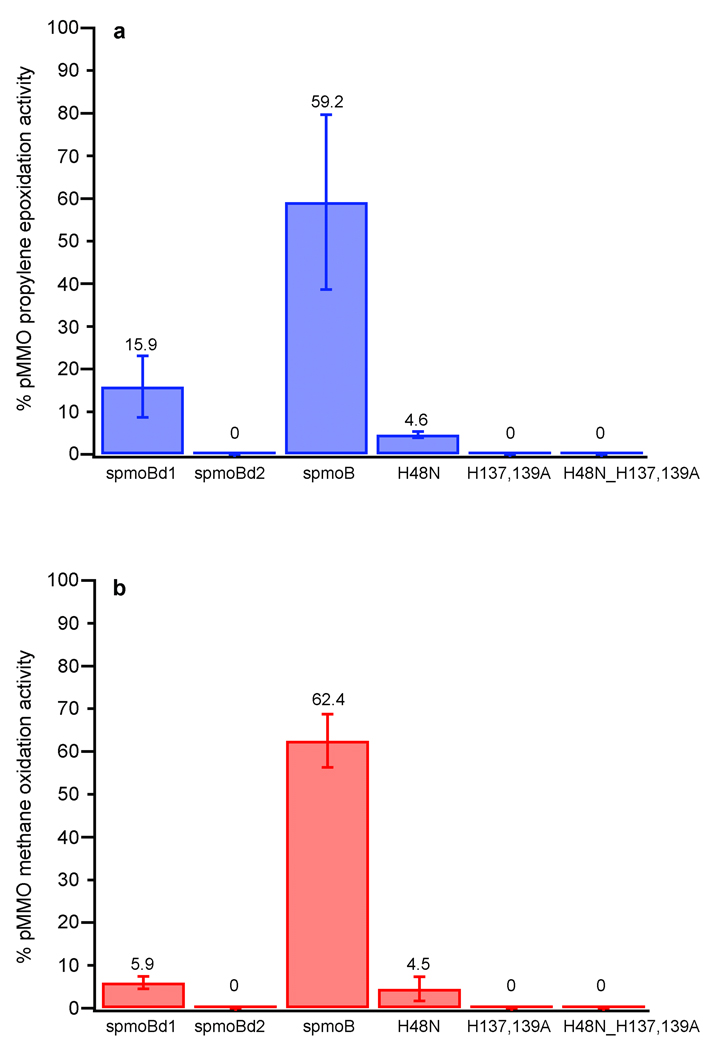
To determine whether the copper active site is located within spmoB, we performed activity assays. The spmoB protein is indeed active and can oxidize propylene with a specific activity of 30.2 ± 10.5 nmol propylene oxide µmoles−1 min−1 as compared to 51.1 ± 11.3 nmol propylene oxide µmoles−1 min−1 for as-isolated, membrane-bound pMMO measured under similar experimental conditions (Fig. 5a). Some activity is also detected for spmoBd1, 8.1 ± 3.7 nmol propylene oxide µmoles−1 min−1, whereas spmoBd2 is inactive. No activity is detected for spmoB refolded in the presence of iron. Although propylene epoxidation is routinely used for measuring MMO activity, the bond dissociation energies of the C=C bond in propylene (63 kcal/mol) and the C–H bond in methane (104 kcal/mol) differ significantly. Therefore, we also tested the ability of spmoB to oxidize methane to methanol. Under similar experimental conditions, the specific activity of as-isolated pMMO is 325.1 nmol methanol µmoles−1 min−1 and that of spmoB is 203.1 ± 20.2 nmol methanol µmoles−1 min−1 (Fig. 5b). Consistent with the trend observed for propylene epoxidation activity, spmoBd1 exhibits a specific activity of 19.3 ± 4.7 nmol methanol µmoles−1 min−1 and spmoBd2 is inactive. The reduced activity for spmoBd1 is consistent with increased lability of the copper sites upon removal of spmoBd2. These combined activity data indicate that the pMMO copper active site is located within spmoB and rule out the possibility of the active site being a diiron center located at the crystallographic zinc site17 or a trinuclear copper center located at the intramembrane hydrophilic patch12. In addition, the active site is not located in the pmoA subunit as suggested by radiolabeling experiments with the suicide substrate acetylene24, 25.
Figure 5. Catalytic activity of spmoB proteins.
a, Epoxidation of propylene to propylene oxide expressed as a percentage of the activity of M. capsulatus (Bath) as-isolated, membrane-bound pMMO. b, Oxidation of methane to methanol expressed as a percentage of the activity of M. capsulatus (Bath) as-isolated, membrane-bound pMMO. All values are an average of at least two independent refolding preparations. The activities of the spmoB proteins were compared to activities of membrane-bound pMMO measured under the same experimental conditions.
To pinpoint the active site location within spmoB, we generated site-specific variants. These variants were designed to disrupt binding at the mononuclear copper site (spmoB_H48N), the dinuclear copper site (spmoB_H137,139A) and both copper sites (spmoB_H48N_H137,139A). Proteins were refolded and the copper content was measured using the same procedures as for spmoB (Fig. 2b). The spmoB_H48N variant (asparagine was selected to mimic the site in M. trichosporium OB3b pMMO10) binds 1.86 ± 0.52 copper ions per protein, consistent with elimination of the mononuclear site. The spmoB_H137,139A variant binds 0.73 ± 0.15 copper ions per protein, suggesting that the dinuclear copper site has been abolished and the mononuclear copper site remains intact. The spmoB_H48N_H137,139A variant still binds some copper, 0.81 ± 0.36 copper ions per protein, as does a quadruple mutant in which His 33 is also changed to alanine. The spmoB_H48N_H137,139A variant refolded in the presence of iron binds 0.51 ± 0.14 iron per protein. This metal binding by spmoB_H48N_H137,139A is likely nonspecific and raises the possibility that some fraction of the copper in spmoB_H48N and spmoB_H137,139A is also bound adventitiously. The copper coordination environments in spmoB_H48N and spmoB_H137,139A were further investigated by XAS. These two samples contain a mixture of Cu(I) and Cu(II) as shown by both a discernible 1s➔4p transition and a weak 1s➔3d feature (Fig. S2). The EXAFS for both variants were best fit with a Cu-O/N ligand environment at 1.96 Å. For spmoB_H48N, inclusion of a Cu-Cu environment at 2.52 Å improved the fit significantly (Figs. 4g and h, Table S2). However, second shell fits for spmoB_H137,139A were better using C as a ligand rather than Cu, as suggested by lower F’ value for this fit (Figs. 4i and j, Table S2). Thus, it is probable that this double mutant does not contain a dinuclear copper center.
Each variant was then tested for propylene and methane oxidation activities (Fig. 5). Only the spmoB_H48N variant is active with a propylene epoxidation activity of 2.3 ± 0.4 nmol propylene oxide µmoles−1 min−1 and a methane oxidation activity of 14.8 ± 9.2 nmol methanol µmoles−1 min−1. Since removal of the mononuclear copper site does not abolish activity, it cannot be the active site. By contrast, spmoB_H137,139A and spmoB_H48N_H137,139A show no activity (Fig. 5). These activity results combined with the EXAFS data indicate that the dinuclear copper center in the pmoB subunit is required for substrate oxidation by pMMO. The dicopper site may not be fully occupied in spmoB_H48N due to the presence of adventitious copper, resulting in decreased activity. The activity may be further impaired by the mutation at the monocopper site, which likely imposes some reorganization on the domain interface (Fig. S3a). These two considerations could reasonably account for the ~5% activity measured for the spmoB_H48N variant (Fig. 5). The location of the dicopper center in the pMMO structure (Fig. 1) is consistent with some reduction in activity upon isolation of spmoB. A number of pmoB residues adjacent to the dicopper center interact with residues from the pmoC subunit (Fig. S3b). Removing these interactions likely confers additional flexibility at the dicopper center.
Our work shows that pMMO is a copper enzyme and that the active site is located in the N-terminal soluble domain of the pmoB subunit at the site of the crystallographic dicopper center. Neither the proposed intramembrane diiron center17 nor the proposed intramembrane tricopper center12 is the active site. The copper stoichiometry, XAS, and activity data are most consistent with a dicopper, rather than a tricopper, active site. Additional support comes from computational studies of methane activation by bis(µ-oxo) dicopper species26 and reports of direct methane oxidation by mono-(µ-oxo) dicopper cores in zeolites27. If spmoB can do the chemistry, why is pMMO a membrane protein? Substrate entry may occur through the lipid bilayer. The increased solubility of methane in the membrane28, 29 would provide a high local concentration. The methanol product likely exits through the soluble regions since the next metabolic enzyme, methanol dehydrogenase, is periplasmic30. The transmembrane regions could also shuttle electrons to the active site and might have other undiscovered activities. Given that the spmoB proteins have less activity than pMMO and that as-isolated pMMO has lower activity than whole cells17, understanding how the additional subunits and trimeric structure enhance the function is an important issue.
METHODS SUMMARY
M. capsulatus (Bath) was cultivated as described previously9 with minor modifications. Cells were lysed by sonication and ultracentrifuged with multiple washing/homogenization steps to obtain pMMO-containing membranes. Metals were removed from as-isolated pMMO by treatment with KCN. Genes encoding spmoBd1 (residues 33–172), spmoBd2 (residues 265–414), and spmoB (residues 33–172 and residues 265–414 connected via a GKLGGG linker instead of the two transmembrane helices) were amplified by PCR and cloned into a pET21b(+) expression vector. Site-directed variants were generated using commercial kits. All proteins expressed as inclusion bodies, which were centrifuged and treated with Triton X-100 to obtain >90% pure samples. Purified inclusion bodies were solubilized with urea and refolded by a stepwise dialysis procedure in the presence of CuSO4. Folding was assessed by circular dichroism (CD) spectroscopy and size exclusion chromatography. Metal content was determined by the bicinchoninic acid (BCA) method23 or by inductively coupled plasma-optical emission spectroscopy (ICP-OES). Propylene epoxidation and methane oxidation activities were measured by gas chromatography using Porapak Q packed and Rt-Q-BOND capillary columns, respectively. XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamline 9–38.
METHODS
Growth of M. capsulatus (Bath)
M. capsulatus (Bath) was cultivated as described previously9 with a few modifications. Briefly, 12 l of nitrate mineral salts (NMS) media was supplemented with a trace element solution, 50 µM CuSO4, and 80 µM FeEDTA. The pH of the culture was maintained at 6.8 using a 100 mM phosphate buffer. Adjustments to the pH during growth were made using NaOH and H2SO4. Prior to growth, methane gas was bubbled through the media for ~30 min. Approximately 5–10 g of highly active cell paste stock were used as the inoculum after resuspending in sterile NMS at 45 °C. Fermentations were conducted at 45 °C with an air:methane gas ratio of 4:1 and a constant agitation of 300 rpm. Cells were harvested at mid exponential phase (OD600 nm of 5–7) and centrifuged for 10 min at 8000 × g. Pelleted cells were washed by resuspension in 25 mM PIPES, pH 6.8, re-centrifuged, flash frozen in liquid nitrogen, and stored at −80 °C.
Isolation of pMMO-containing membranes
Frozen cells were resuspended in lysis buffer (25 mM PIPES, pH 6.8, 250 mM NaCl) and sonicated on ice. Cell debris was removed by centrifugation at 20,000 × g for 1.5 hrs. The lysate was then ultracentrifuged at 160,000 × g for 1 hr to pellet intracytoplasmic membranes. These membranes were resuspended in fresh lysis buffer using a Dounce homogenizer. Ultracentrifugation and resuspension were repeated three times to remove contaminating soluble proteins. SDS-PAGE analysis of the as-isolated membranes show that the predominant component is pMMO. The homogenized crude membranes at ~10–20 mg/ml were flash frozen in liquid nitrogen and stored at −80 °C.
Preparation of apo pMMO
Active as-isolated pMMO membranes were incubated with 50 mM MOPS, pH 8.0, 250 mM NaCl supplemented with a 10-fold molar excess (based on copper concentration) of KCN and freshly prepared sodium ascorbate. The addition of ascorbate did not affect the pH of the solution. After a 1 hr incubation at room temperature with stirring, the solution was ultracentrifuged at 160,000 × g for 30 min to pellet the pMMO membranes. The apo pMMO membranes were resuspended and washed three times with fresh lysis buffer to remove excess KCN and ascorbate. Cyanide treatment does not alter the integrity of the subunits according to SDS-PAGE. Moreover, apo pMMO stored at 4 °C exhibits higher activity upon copper reactivation than as-isolated pMMO stored at 4 °C for the same time period. This observation suggests that the strict anaerobic protocols used by other researchers17 might serve to protect against oxidative damage caused by excess metal ions.
Cloning of spmoB, spmoBd1, and spmoBd2
Three constructs of the soluble domain of pmoB (spmoB) were generated: spmoBd1 (residues 33–172), spmoBd2 (residues 265–414), and spmoB (residues 33–172 and residues 265–414 connected via a GKLGGG linker instead of the two transmembrane helices) (Fig. 1). This linker was designed by inspection of the pMMO structure, and the glycine residues were selected to impart flexibility. Primers spmoBd1F (5’-gga att cca tat gca cgg tga gaa atc gca gg-3’) and spmoBd1R (5’-gtg atc caa gct ttc cgg tgg tga cgg ggt tgc gaa-3’) were used to amplify spmoBd1 from M. capsulatus (Bath) genomic DNA. The NdeI/HindIII digested PCR product encoding spmoBd1 was then ligated into a NdeI/HindIII digested pET21b(+) vector (Novagen). Primers spmoBd2F (5’-gag aag caa gct tgg agg agg aca ggc cgc cgg cac cat gcg tgg-3’) and spmoBd2R1 (5’-gag atc cca agc tta cat gaa cga cgg gat cag cgg-3’) were used to amplify spmoBd2. The pET21b(+)-spmoBd2 vector was generated by ligation of the PCR product encoding spmoBd2 at the NdeI/HindIII sites. To generate the spmoB construct, HindIII digested PCR product encoding spmoBd2 amplified using primers spmoBd2F (5’-gag aag caa gct tgg agg agg aca ggc cgc cgg cac cat gcg tgg-3’) and spmoBd2R2 (5’-gag atc cca agc tta cat gaa cga cgg gat cag cgg-3’) was inserted into the HindIII digested pET21b(+)-spmoBd1 vector. Authenticity of the coding sequences for all constructs was verified by DNA sequencing. There is a silent mutation in the spmoB DNA sequence at position 1076 that changes C to G (ACC to ACG coding for Thr). The site-directed variants spmoB_H48N, spmoB_H137,139A, and spmoB_H48N_H137,139A were generated using either a traditional single mutagenesis kit or a multi-site directed mutagenesis kit from Stratagene and verified by DNA sequencing.
Protein expression, purification, and refolding
For protein expression, plasmids were transformed into either BL21(DE3) or Rosetta (DE3) strains of E. coli. Ten ml of an overnight culture of freshly transformed cells were transferred to 1 l of LB supplemented with 50 µg/ml ampicillin. After reaching an OD600 of 0.6 at 37 °C, 0.5 mM IPTG was added followed by 4 hrs of post induction growth. All the expression constructs tested produced proteins that formed inclusion bodies (Fig. S4). Alterations to the growth temperature, incubation times, inducer concentrations, and growth media had no effect on the protein solubility. Cells from 4 l of growth were collected by centrifugation at 5000 × g for 10 min, washed and resuspended in 200 ml of 20 mM TrisCl, pH 8.0, 50 mM NaCl, flash frozen in liquid nitrogen, and stored at −20 °C until further processing.
Frozen cells were thawed in lukewarm water and lysed by a 10 min sonication cycle of 10 s on, 30 s off pulses on ice at 50% output power. Inclusion bodies were separated from cell debris and other soluble proteins by centrifuging for 30 min at 3000 × g. The inclusion bodies were then washed 3 times with 20 mM TrisCl, pH 8.0, 50 mM NaCl, 1.0% Triton X-100 and once more with the same buffer lacking Triton X-100. Inclusion bodies isolated using this procedure were >90% pure and required no additional purification steps (Fig. S4). Typically, 1–3 g of inclusion bodies can be isolated from 1 l of cell culture. Purified inclusion bodies were solubilized in 8 M urea (20 ml urea per 1 g). This mixture was left stirring at room temperature for 1 hr and then centrifuged at 15000 × g for 30 min at room temperature. Urea solubilized inclusion bodies were aliquoted and stored at −80 °C until further use.
All attempts to express even partially soluble protein were unsuccessful. Therefore, a refolding procedure was developed. Urea solubilized inclusion bodies were diluted to ~5 mg/ml protein concentration. Protein refolding was performed using a stepwise dialysis procedure against buffers containing a stepwise reduction in the urea concentration. Each dialysis was performed for at least 3 hr. After a 3 hr dialysis against 7 M urea, the protein was dialyzed against 6 M urea for 3 hr followed by 5 M urea and so forth. After dialysis against a buffer containing 0.5 M urea, at least two rounds of dialysis were performed against a buffer containing either 20 mM TrisCl, pH 8.0 or 20 mM PIPES, pH 7.0, 250 mM NaCl (no urea). Based on SDS-PAGE band intensities, we estimate that about 0.2–5% of the total protein was folded after this procedure. Precipitates were removed by centrifugation at 20,000 × g for 10 min at 4 °C using a tabletop centrifuge. The spmoBd1 variant was generally less stable and more susceptible to precipitation during these procedures.
Circular dichroism (CD) spectra were recorded on a JASCO J715 spectrometer using a 2 mm path length quartz cuvette. Proteins were diluted to ~1–2 µM concentration in a 20 mM potassium phosphate, pH 7.5, 20 mM potassium fluoride buffer. An average of 5 scans was collected at 20 °C with 1 nm resolution. Laccase from Trametes versicolor (Sigma Aldrich) was used as a model cupredoxin. All the spmoB variants appear folded and exhibit similar secondary structure properties, suggesting that the mutations do not perturb the overall structure (Fig. S5).
Folding was further assessed by analytical size exclusion chromatography on 21 ml prepacked Superdex G75 column equilibrated with 20 mM PIPES, pH 7.0, 150 mM NaCl for at least three column volumes. Recombinant proteins were injected in 0.5 ml aliquots and the molecular masses (Table S3) were determined using the following standards: blue dextran (void volume), 2 MDa; aldolase, 158 kDa; concanavalin, 75 kDa; albumin, 66 kDa; ovalbumin, 43 kDa; chymotrypsinogen, 25 kDa; RNAse, 13.7 kDa; aprotinin, 6.5 kDa. None of the proteins appear to form trimers, suggesting that the transmembrane regions of pMMO drive oligomerization.
Copper loading of recombinant proteins
The spmoB protein contains the ligands to the two copper sites observed in the M. capsulatus (Bath) crystal structure (Fig. 1). Complete precipitation occurred upon copper addition to refolded spmoB. Therefore, a refolding procedure in the presence of copper was developed and used for all spmoB variants. The 6 M urea dialysis buffer included 1 mM CuSO4, and the stepwise reduction in urea concentration was accompanied by a reduction in copper concentration. Excess copper was then removed from Cu(II)-refolded proteins by dialysis against a buffer containing either 20 mM TrisCl, pH 8.0, or 20 mM PIPES, pH 7.0, 250 mM NaCl with no CuSO4 or urea. Proteins refolded by this procedure contained no detectable iron. For refolding of spmoB and its variants in the presence of iron, 1 mM Fe(NH4)2(SO4)2 was added during dialysis instead of CuSO4. All other procedures were identical to those used for refolding in the presence of copper.
Determination of metal content
The copper contents of spmoB variants were measured using the bicinchoninic acid (BCA) method23. Standards of 0–60 µM copper were prepared by diluting commercially available copper atomic absorption spectroscopy standard solutions (Sigma Aldrich) in water. The Cu(I)-BCA complex exhibits absorbance peaks at 360 and 562 nm. The iron contents of spmoB variants were determined using the ferrozine assay31. A solution containing 150 µl 1N HCl, 150 µl of freshly prepared 1.7 mM ascorbate, and 250 µl 0.1% ferrozine (Acros Organics) prepared in 50% ammonium acetate was added to 150 µl protein. The reaction mixture was incubated at 30 °C for 15 min, centrifuged, and the absorbance at 570 nm of the supernatant measured. A standard curve using iron atomic absorption standard (Aldrich) was generated for a 0–40 µM iron concentration range. The copper, iron, and zinc contents of as-isolated pMMO membranes and apo pMMO membranes were determined by inductively coupled plasma-optical emission spectroscopy (ICP-OES) using a Varian Vista-MPX CCD simultaneous ICP-OES instrument in the Northwestern IMSERC. Samples were digested in 5% trace-metal grade (TMG) nitric acid prior to analysis. Standard curves were generated from atomic absorption standards diluted in 5% nitric acid (TMG). All samples were measured in triplicate. Protein concentrations were determined using either Bradford reagent or detergent compatible Bio-Rad Dc protein assays. Alternatively, estimated extinction coefficients for the absorbance at 280 nm were used for the spmoB variants.
Propylene epoxidation assay
pMMO activity was assessed by the propylene epoxidation assay using duroquinol as the reductant9. In a typical assay, solid duroquinol is added to 50 µl as-isolated pMMO in a septa sealed 3 ml reaction vial such that the final concentration of duroquinol is 0.9–1.1 M. The assay is then initiated by replacing 2 ml of the headspace gas with propylene and the vial is shaken in a 45 °C water bath for 3 min. The amount of propylene oxide produced is measured by injecting 250 µl of headspace gas onto a Porapak Q column (Supelco) using a Hewlett Packard 5890A gas chromatograph. Quantitation is performed by comparison with pure propylene oxide standards (Sigma Aldrich). Typical activity values were between 50–200 nmol min−1 mg pMMO−1 (Table S1). For metal reconstituted activity assays, apo pMMO was incubated with varying molar equivalents of CuSO4 and/or Fe(NH4)2(SO4)2 for 30 min and measurements performed as above. The CuSO4 solution (280 ppm) contained < 0.02 ppm iron so addition of one equivalent of copper would correspond to ~ 10−7 equivalents of iron. The iron content of apo pMMO reconstituted with copper did not increase. The titration results shown in Fig. 3 were reproduced more than four times. For experiments with catalase, commercial enzyme (Sigma Aldrich) was added at a concentration of ~1 mg/ml to samples reconstituted with more than 2–3 equivalents of copper (Fig. S1b). For the spmoB variants, the propylene epoxidation assay was performed with several modifications. Protein sample volumes were 350 µl and 2.5 ml of headspace air were replaced with 2.0 ml propylene and 0.5 ml air. The reaction mixture was incubated in the 45 °C shaking water bath for at least 1 hr. A longer incubation time for spmoB was required to accurately measure product formation. For comparisons of the spmoB activity to that of as-isolated pMMO, the pMMO assay was also carried out for 1 hr (Fig. 5). All activity assays were performed in duplicate. To account for the differences in molecular masses when comparing activities of pMMO (~100 kDa) to that of spmoB and its variants (~16 kDa-32 kDa), the activities of spmoB and its variants are reported per mole instead of per mg.
Methane oxidation assay
Methanol production by both pMMO and the spmoB variants was measured by gas chromatography using a Rt-Q-BOND capillary column (Restek). The procedure for the propylene epoxidation assay was used with the following modifications. Methane (2 ml) was added to the headspace instead of propylene as the substrate. The reaction was performed for 3 min for pMMO at 45 °C (Fig. 3) and for at least 1 hr at 45 °C for spmoB and its variants (Fig. 5). For comparisons of the spmoB activity to that of as-isolated pMMO, the pMMO assay was also carried out for 1 hr (Fig. 5). After this procedure, the samples were heated to 85 °C for 10 min to stop the reaction and cooled on ice. Samples were then centrifuged briefly and 3 µl of the clear supernatant injected onto the capillary column at a constant oven temperature of 75 °C. Quantitation was performed using standard curves generated by analyzing methanol standards (Sigma Aldrich, spectrophotometric grade 99+%). For both propylene epoxidation and methane oxidation assays, numerous control assays in which duroquinol, substrate, or enzyme were eliminated systematically from the reaction mixture were performed.
X-ray absorption spectroscopy
XAS data were collected on the following five samples: as-isolated pMMO (sample 1), apo pMMO reconstituted with CuSO4 in the presence of duroquinol (sample 2), spmoB (sample 3), spmoB_H48N (sample 4) and spmoB_H137,139A (sample 5). As-isolated and apo pMMO membranes were ultracentrifuged and resuspended in 25 mM PIPES, pH 6.8, 250 mM NaCl, 50% glycerol before loading into Kapton tape wrapped Lucite cells and flash freezing in liquid nitrogen. The samples were stored in liquid nitrogen until data collection was performed. For reconstituted pMMO membranes, apo samples were incubated with 3 equivalents of CuSO4 and excess duroquinol for 30 min prior to ultracentrifugation, resuspension, cell loading, and freezing. The final copper concentrations were 0.7–2 mM. For spmoB and its variants, all samples were prepared in 10 mM TrisCl, pH 8.0, 250 mM NaCl, 50% glycerol. The final copper concentrations were 0.3–0.9 mM. All XAS results were reproduced using two independent samples.
XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamline 9–3 using a Si(220) double crystal monochromator equipped with a harmonic rejection mirror. Samples were maintained at 10 K using an Oxford Instruments continuous-flow liquid helium cryostat. Protein fluorescence excitation spectra were collected using a 30-element Ge solid-state array detector. A nickel filter (0.3 µm wide) was placed between the cryostat and detector to filter scattering fluorescence not associated with protein Cu signals. XAS spectra were measured using 5 eV steps in the pre-edge region (8750–8960 eV), 0.25 eV steps in the edge region (8986–9050 eV) and 0.05 Å−1 increments in the EXAFS region (to k = 13.0 Å−1), integrating from 1 s to 20 s in a k3 weighted manner for a total scan length of approximately 45 min. X-ray energies within the protein spectra were internally calibrated by simultaneously collecting Cu foil absorption spectra; the first inflection point in the Cu foil spectra was assigned as 8980 eV. Each fluorescence channel of each scan was examined for spectral anomalies prior to averaging and spectra were closely monitored for photoreduction. The represented data are the average of 7 to 8 scans.
XAS data were processed using the Macintosh OS X version of the EXAFSPAK program suite32 integrated with the Feff v7 software33 for theoretical model generation. Data reduction followed a previously published protocol for a spectral resolution in bond lengths of 0.13 Å8. EXAFS fitting analysis was performed on raw/unfiltered data. Protein EXAFS data were fit either using single scattering Feff v7 theoretical models, calculated for carbon, oxygen, sulfur and copper coordination to simulate copper-ligand environments, or a multiple scattering imidazole model, and values for the scale factors (Sc) and Eo calibrated by fitting crystallographically characterized copper model compounds, as outlined previously8. Criteria for judging the best-fit EXAFS simulations, utilized both the lowest mean square deviation between data and fit corrected for the number of degrees of freedom (F’)34 and reasonable Debye-Waller factors (σ2 < 0.006 Å2). First shell fits with a single set of O/N ligands gave dramatically better F' values than fits with a single Cu-S or Cu-Cu environment for all samples. Long range scattering (> 3 Å) evident for all five samples could not be fit with a multiple scattering Cu-imidazole model, as shown by the large F’ values for these fits (Table S2, fits 1.3, 2.3, 3.3, 4.3, and 5.3).
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM070473 (A. C. R.) and DK068139 (T. L. S.). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL). SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions R. B., S. M. S., S. R., and L. A. Y. performed experiments. R. B., S. M. S., S. R., T. L. S., and A. C. R. contributed to experimental design, data analysis, and manuscript preparation.
References
- 1.Hermans I, Spier ES, Neuenschwander U, Turra N, Baiker A. Selective oxidation catalysis: opportunities and challenges. Topics Catal. 2009;52:1162–1174. [Google Scholar]
- 2.Arakawa H, et al. Catalysis research of relevance to carbon management: progress, challenges, and opportunities. Chem. Rev. 2001;101:953–996. doi: 10.1021/cr000018s. [DOI] [PubMed] [Google Scholar]
- 3.Que L, Tolman WB. Biologically inspired oxidation catalysis. Nature. 2008;455:333–340. doi: 10.1038/nature07371. [DOI] [PubMed] [Google Scholar]
- 4.Merkx M, et al. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew. Chem. Int. Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Ann. Rev. Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 7.Balasubramanian R, Rosenzweig AC. Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc. Chem. Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman RL, et al. Characterization of the particulate methane monooxygenase metal centers in multiple redox states by X-ray absorption spectroscopy. Inorg. Chem. 2006;45:8372–8381. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman RL, et al. Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc. Natl. Acad. Sci. USA. 2003;100:3820–3825. doi: 10.1073/pnas.0536703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakemian AS, et al. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry. 2008;47:6793–6801. doi: 10.1021/bi800598h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenzweig AC. The metal centres of particulate methane monooxygenase. Biochem. Soc. Trans. 2008;36:1134–1137. doi: 10.1042/BST0361134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SI, Yu SSF. Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc. Chem. Res. 2008;41:969–979. doi: 10.1021/ar700277n. [DOI] [PubMed] [Google Scholar]
- 13.Chen KH-C, et al. The copper clusters in the particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath) J. Chin. Chem. Soc. 2004;51:1081–1098. [Google Scholar]
- 14.Nguyen H-HT, et al. X-ray absorption and EPR studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Cu(I) ions and their implications. J. Am. Chem. Soc. 1996;118:12766–12776. [Google Scholar]
- 15.Chan SI, et al. Redox potentiometry studies of particulate methane monooxygenase: support for a trinuclear copper cluster active site. Angew. Chem. Int. Ed. 2007;46:1992–1994. doi: 10.1002/anie.200604647. [DOI] [PubMed] [Google Scholar]
- 16.Yu SSF, et al. The C-terminal aqueous-exposed domain of the 45 kDa subunit of the particulate methane monooxygenase in Methylococcus capsulatus (Bath) is a Cu(I) sponge. Biochemistry. 2007;46:13762–13774. doi: 10.1021/bi700883g. [DOI] [PubMed] [Google Scholar]
- 17.Martinho M, et al. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J. Am. Chem. Soc. 2007;129:15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi DW, et al. The membrane-associated methane monooxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitmitto A, Myronova N, Basu P, Dalton H. Characterization and structural analysis of an active particulate methane monooxygenase trimer from Methylococcus capsulatus (Bath) Biochemistry. 2005;44:10954–10965. doi: 10.1021/bi050820u. [DOI] [PubMed] [Google Scholar]
- 20.Yu SS-F, et al. Production of high-quality particulate methane monooxygenase in high yields from Methylococcus capsulatus (Bath) with a hollow-fiber membrane bioreactor. J. Bacteriol. 2003;185:5915–5924. doi: 10.1128/JB.185.20.5915-5924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyaji A, Suzuki M, Baba T, Kamachi T, Okura I. Hydrogen peroxide as an effecter on the inactivation of particulate methane monooxygenase under aerobic conditions. J. Mol. Catal. B. 2009;57:211–215. [Google Scholar]
- 22.Kau L-S, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. X-ray absorption edge determination of the oxidation state and coordination number of copper. Application to the type 3 site in Rhus vernicifera laccase and its reaction with oxygen. J. Am. Chem. Soc. 1987;109:6433–6442. [Google Scholar]
- 23.Yatsunyk LA, Rosenzweig AC. Copper(I) binding and transfer by the N-terminus of the Wilson disease protein. J. Biol. Chem. 2007;282:8622–8631. doi: 10.1074/jbc.M609533200. [DOI] [PubMed] [Google Scholar]
- 24.Prior SD, Dalton H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath) FEMS Microbiol. Lett. 1985;29:105–109. [Google Scholar]
- 25.Zahn JA, DiSpirito AA. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J. Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiota Y, Yoshizawa K. Comparison of the reactivity of bis(µ-oxo)CuIICuIII and CuIIICuIII species to methane. Inorg. Chem. 2009;48:838–845. doi: 10.1021/ic8003933. [DOI] [PubMed] [Google Scholar]
- 27.Woertink JS, et al. A [Cu2O]2+ core in Cu-ZSM-5, the active site in the oxidation of methane to methanol. Proc. Natl. Acad. Sci. USA. 2009;106:18908–18913. doi: 10.1073/pnas.0910461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller KW, Hammond L, Porter EG. The solubility of hydrocarbon gases in lipid bilayers. Chem. Phys. Lipids. 1977;20:229–241. [Google Scholar]
- 29.Batliwala H, Somasundaram T, Uzgiris EE, Makowski L. Methane-induced hemolysis of human erythrocytes. Biochemical J. 1995;307:433–438. doi: 10.1042/bj3070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myronova N, Kitmitto A, Collins RF, Miyaji A, Dalton H. Three-dimensional structure determination of a protein supercomplex that oxidizes methane to formaldehyde in Methylococcus capsulatus (Bath) Biochemistry. 2006;45:11905–11914. doi: 10.1021/bi061294p. [DOI] [PubMed] [Google Scholar]
- 31.Stookey LL. Ferrozine - a new spectrophotometric reagent for iron. Anal. Chem. 1970;42:779–781. [Google Scholar]
- 32.George GN, George SJ, Pickering IJ. 2001. http://www-ssrl.slac.stanford.edu/~george/exafspak/exafs.htm.
- 33.Ankudinov AL, Rehr JJ. Relativistic calculations of spin-dependent X-ray absorption spectra. Phys. Rev. B. 1997;56:R1712–R1715. [Google Scholar]
- 34.Riggs-Gelasco PJ, Stemmler TL, Penner-Hahn JE. XAFS of dinuclear metal sites in proteins and model compounds. Coord. Chem. Rev. 1995;144:245–286. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.