Abstract
Dax1 (Nr0b1) is an atypical orphan nuclear receptor that has recently been shown to play a role in mouse embryonic stem (mES) cell pluripotency. Here we describe a mechanism by which Dax1 maintains pluripotency. In steroidogenic cells, Dax1 protein interacts with the NR5A nuclear receptor steroidogenic factor 1 (Nr5a1) to inhibit transcription of target genes. In mES cells, liver receptor homolog 1 (LRH-1, Nr5a2), the other NR5A family member, is expressed, and LRH-1 has been shown to interact with Dax1. We demonstrate by coimmunoprecipitation that Dax1 is, indeed, able to form a complex with LRH-1 in mES cells. Because Dax1 was historically characterized as an inhibitor of steroidogenic factor 1-mediated transcriptional activation, we hypothesized that Dax1 would inhibit LRH-1 action in mES cells. Therefore, we examined the effect of Dax1 on the LRH-1-mediated activation of the critical ES cell factor Oct4 (Pou5f1). Chromatin immunoprecipitation localized Dax1 to the Oct4 promoter at the LRH-1 binding site, and luciferase assays together with Dax1 overexpression and knockdown experiments revealed that, rather than repress, Dax1 accentuated LRH-1-mediated activation of the Oct4 gene. Similar to our previously published studies that defined the RNA coactivator steroid receptor RNA activator as the critical mediator of Dax1 coactivation function, Dax1 augmentation of LRH-1-mediated Oct4 activation is dependent upon steroid receptor RNA activator. Finally, utilizing published chromatin immunoprecipitation data of whole-genome binding sites of LRH-1 and Dax1, we show that LRH-1 and Dax1 commonly colocalize at 288 genes (43% of LRH-1 target genes), many of which are involved in mES cell pluripotency. Thus, our results indicate that Dax1 plays an important role in the maintenance of pluripotency in mES cells through interaction with LRH-1 and transcriptional activation of Oct4 and other genes.
Dax1 coactivates LRH-1-mediated Oct4 transcription in mES cells. Additionally, Dax1 and LRH-1 localize to common genomic sites and activate target genes.
Self-renewal and pluripotency are essential properties that allow embryonic stem (ES) cells to form every cell of an organism. In the case of mouse embryonic stem (mES) cells, the network circuitry that maintains the undifferentiated state is complex and involves several transcription factors, including Oct4, Nanog, Sox2, and signal transducer and activator of transcription 3 (for review, see Ref. 1). The regulation of expression of these factors is marked by a high degree of redundancy, likely due to their critical roles in maintaining pluripotency. For example, Oct4 levels in mES cells must remain within an exquisitely tight window, because only 2-fold increase in expression of Oct4 results in endoderm/mesoderm differentiation (2). Indeed, there have been reports that at least eight factors, including Oct4 itself, regulate the proximal Oct4 promoter (reviewed in Ref. 1).
Dax1 is an atypical orphan nuclear receptor that was cloned more than a decade ago as the gene mutated in patients with X-linked congenital adrenal hypoplasia (3). Since that time, Dax1 has been characterized as a transcriptional repressor that binds the nuclear receptor steroidogenic factor 1 (Sf1) to inhibit Sf1-dependent transcription of steroidogenic genes (reviewed in Ref. 4). Recently, our understanding of the roles of Dax1 has expanded enormously. The observation that Dax1 is expressed in mES cells suggested that Dax1 participates in cellular processes in addition to the inhibition of steroidogenesis (5,6). Indeed, deletion or knockdown of Dax1 results in differentiation of mES cells to multiple lineages, showing that Dax1 is required for the maintenance of pluripotency (6,7,8). Additionally, microarray and whole-genome binding studies in mES cells have shown that Dax1 binds to and regulates expression of thousands of genes (7,9). These reports illustrate that Dax1 plays a significant, but largely undefined, role in the maintenance of pluripotency of mES cells.
Although an early study suggested that Dax1 can bind to a hairpin loop structure in the promoter of the steroidogenic acute regulatory protein, minimal evidence exists for direct DNA binding by Dax1 (10). Therefore, we began to explore a role for Dax1 in mES cells by hypothesizing a protein-protein interaction to provide clues to its targets. Indeed, a recent publication (11) showed that Dax1 could bind directly to Oct4 protein and inhibit its activation of target genes, indicating one way in which Dax1 regulates gene transcription in mES cells. In steroidogenic cells, Dax1 interacts with Sf1 protein and through this interaction alters target gene activation (12,13). Sf1 is not expressed in mES cells, but liver receptor homolog 1 (LRH-1), the closest nuclear receptor family member, is expressed in these cells, and furthermore is required for expression of the critical ES cell factor Oct4 (14). We therefore hypothesized that Dax1 may interact with LRH-1 in mES cells to regulate mES cell pluripotency. In this report, we set out to define molecular interactions between LRH-1 and Dax1 and resultant target gene regulation in mES cells. As detailed below, we uncover novel mechanisms of Dax1-mediated transcriptional control of Oct4 and other genes that contribute to mES cell pluripotency.
Results
Dax1 and LRH-1 interact in mES cells
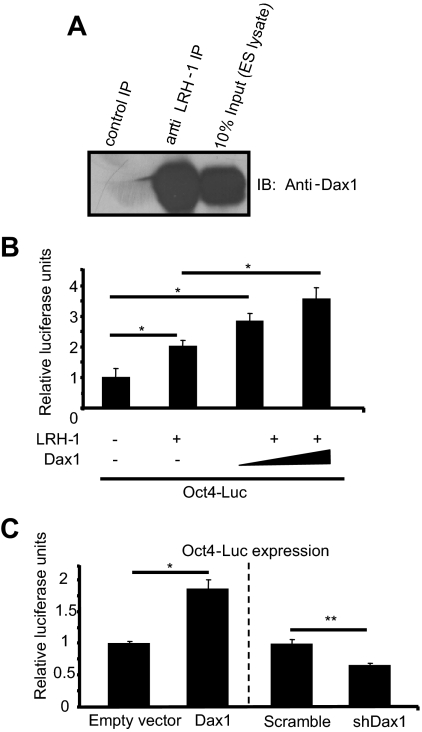
To further our understanding of the role for Dax1 in mES cells, we began by looking for a protein-protein interaction. Dax1 is an atypical nuclear receptor with a well-conserved ligand-binding domain, but no conserved DNA-binding domain (3). Although there is a report suggesting that Dax1 can bind specific DNA structures within a promoter, the predominant role of Dax1 has been to interact with the nuclear receptor steroidogenic factor 1 (Sf1) to inhibit transactivation of target genes (10,12,15,16,17). Although Sf1 is not expressed in mES cells, the closest nuclear receptor family member liver receptor homolog 1 (LRH-1), is highly expressed. Because Sf1 and LRH-1 share similar protein structures, we hypothesized that LRH-1 and Dax1 may interact in mES cells. Indeed, the crystal structure of a LRH-1/Dax1 protein complex has recently been published (18). To determine whether this interaction occurs in mES cells, we performed coimmunoprecipitation (co-IP) experiments. When we immunoprecipitated with anti-LRH-1 antibody, immunoblotting with an anti-Dax1 antibody shows enrichment of Dax1 protein over control immunoprecipitation with normal serum (Fig. 1A). These results demonstrate that LRH-1 and Dax1 are present in a protein-protein complex in mES cells.
Figure 1.
Dax1 and LRH-1 cooperate to up-regulate Oct4 in mES cells. A, Dax1 and LRH-1 interact in mES cells. mES cell lysates were immunoprecipitated (IP) with anti-LRH-1 antibody and Protein A agarose beads. Bound proteins were subjected to SDS-PAGE, and immunoblotting (IB) was performed with anti-Dax1 antibody as described in Materials and Methods. B, Dax1 up-regulates LRH-1-mediated Oct4-Luc activation in F9 cells. F9 embryonal carcinoma cells in 24-well plates were transfected with 200 ng Oct4-Luc and cotransfected with 200 ng empty vector, pcDNA3.1 LRH-1, and/or empty vector or 150–300 ng Dax1. Luciferase assays were carried out on lysates 48 h after transfection, and values were normalized to Renilla luciferase internal control. Data are presented as relative to empty vector control. *, P < 0.03; **, P < 0.002. C, Overexpression or knockdown of Dax1 in mES cells alters Oct4-Luc reporter expression. mES cells in 24-well plates were transfected with 200 ng Oct4-Luc and cotransfected with 200 ng empty vector, pcDNA3 Dax1, scramble, or short hairpin Dax 1 (shDax1) vector. For Dax1 and empty vector transfection, cells were harvested 48 h after transfection, and lysates were subjected to luciferase assay. shDax1 and scramble transfected cells were harvested 24 h after transfection, and the luciferase assay was carried out as described above. Data are presented as relative to empty vector and scramble controls.
Dax1 coactivates LRH-1-mediated Oct4 activation
Because Dax1 is known to inhibit Sf1-mediated activation of transcription, we hypothesized that Dax1 similarly inhibits LRH-1 activity. LRH-1 has been shown to be critical in regulating the expression of Oct4 (2,14). As such, we predicted that Dax1 would regulate LRH-1-mediated activation of the Oct4 promoter to maintain the appropriate expression level of Oct4 in mES cells. To investigate this possibility, we carried out luciferase reporter assays utilizing a cell line that lacks endogenous LRH-1, F9 embryonal carcinoma cells. When we transfected an Oct4-luciferase reporter along with the LRH-1 expression vector, we saw the expected LRH-1-dependent increase in reporter expression (Fig. 1B). However, when we cotransfected increasing amounts of pcDNA3 Dax1, we found that Oct4 promoter activity increased above the level with LRH-1 alone, indicating that Dax1 coactivated LRH-1-mediated Oct4 promoter activity.
To confirm that Dax1 activated Oct4 promoter activity, we carried out luciferase assays in mES cells. When we transfected cells with the luciferase reporter driven by the Oct4 promoter and cotransfected a Dax1 expression vector, we observed a modest, but statistically significant, increase in Oct4 promoter activity (Fig. 1C). Conversely, we transfected the Oct4-luciferase reporter and cotransfected a vector that codes for a short hairpin RNA (shRNA) against Dax1, and harvested cells for luciferase assay only 24 h after transfection to examine direct effects of Dax1 knockdown and not differentiation. Knockdown of endogenous Dax1 inhibited Oct4 promoter activity when compared with scramble control (Fig. 1C). These data confirm that Dax1 acts as an activator of Oct4 promoter activity in mES cells.
Dax1 overexpression does not drive differentiation
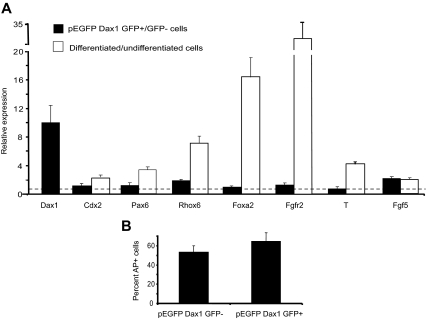
Stable overexpression of Dax1 has been shown to result in differentiation of mES cells (7,11). Because both Oct4 and Dax1 have been shown to induce mES cell differentiation when stably overexpressed, it is possible that any changes observed in our studies reflect secondary effects of the Dax1-mediated differentiation and not primary effects of Dax1 on LRH-1-mediated activation of Oct4. To rule out this possibility, we overexpressed a Dax1-green fluorescent protein (GFP) fusion protein in mES cells and sorted for Dax1 overexpressing cells by GFP. Quantitative PCR analyses of these cells indicate that markers of differentiation to the germ layers and trophectoderm are not up-regulated, relative to their levels in cells differentiated by retinoic acid or withdrawal of leukemia-inhibitory factor (LIF) (Fig. 2A). Additionally, to confirm that overexpression of Dax1 does not change the number of undifferentiated cell colonies, we replated the pEGFP Dax1 transfected/sorted cells and counted the percentage of alkaline phosphatase (AP) staining cells in GFP-positive vs. negative cells. Overexpression of Dax1 did not change the percentage of undifferentiated colonies (Fig. 2B). Therefore, in D3 mES cells, after transient overexpression of Dax1, there is no loss of pluripotency.
Figure 2.
Transient overexpression of Dax1 does not drive differentiation. A, mES cells in 10-cm plates were transfected with 10 μg pEGFP Dax1 and 48 h later sorted for GFP-positive and -negative cells. Cells were harvested for QPCR analysis. As positive controls, cells differentiated by retinoic acid (RA) treatment or withdrawal of LIF (−LIF) were also subjected to QPCR analysis (as described in Materials and Methods; RA: Cdx2, Pax6, Rhox6, Foxa2, Fgfr2; -LIF: T, Fgf5). B, Sorted cells were replated and AP staining was performed. Percent of AP-positive cells was determined by Metamorph software.
Dax1 activates the Oct4 promoter through the LRH-1 proximal promoter site
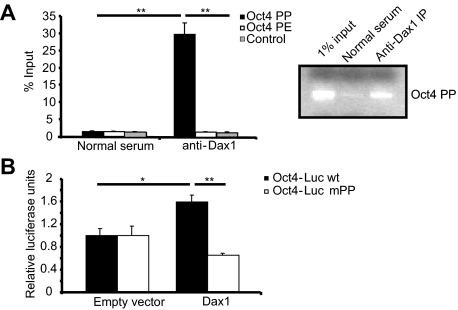
Based on our data that Dax1 activates Oct4 promoter activity through interaction with LRH-1, we investigated whether Dax1 is localized to the Oct4 promoter at a previously characterized site of LRH-1 binding (14). To this end, we performed chromatin immunoprecipitation (ChIP) assays that detect protein bound to DNA either directly or through a complex. Immunoprecipitation with anti-Dax1 antibody, and not with normal serum, enriched for the LRH-1 site within the proximal promoter (PP) of Oct4 (Fig. 3A) (14). These data, together with the lack of enrichment of sequence in the proximal enhancer or control sites, are consistent with Dax1 localization to the Oct4 promoter at the LRH-1 binding site within the PP. These results were observed by both quantitative PCR (QPCR) and PCR followed by agarose gel electrophoresis (Fig. 3A, right).
Figure 3.
Dax1 binds to and up-regulates Oct4 transcription through the LRH-1 proximal promoter site. A, ChIP was performed in mES cells as described in Materials and Methods. Immunoprecipitation (IP) was performed with normal serum or anti-Dax1 antibody, and QPCR was carried out with primer sets flanking the Oct4 PP LRH-1 site, proximal enhancer site (PE), or control site. Results are shown from independent experiments quantitated by either QPCR or PCR and agarose gel analysis. *, P < 0.02; **, P < 0.006. B, mES cells were transfected with either 200 ng wild-type (wt) Oct4-Luc reporter or reporter in which the LRH-1 PP site is mutated. Cells were cotransfected with 200 ng empty vector or Dax1 and 48 h later were harvested and luciferase assay was carried out.
The binding of Dax1 to the Oct4 promoter at the LRH-1 PP binding site suggested that the activation of the Oct4 promoter by Dax1 was mediated by the LRH-1 PP site. To test this hypothesis, an Oct4 promoter-luciferase construct harboring a previously characterized mutation in the LRH-1 PP site (14) was tested in luciferase assays in mES cells. When mES cells were transfected with the wild-type reporter along with either empty vector or pcDNA3 Dax1, a Dax1-dependent up-regulation of the reporter was observed, but when cells were transfected with the mutated reporter (Oct4-Luc mPP), there was no increase in promoter activity upon overexpression of Dax1 (Fig. 3B). The slight decrease in expression of Oct4-Luc mPP with Dax1 overexpression was not statistically significant. These data indicate that Dax1 up-regulates Oct4 promoter activity through interaction with the LRH-1 PP site.
Dax1 overexpression and knockdown result in changes in Oct4 mRNA and protein
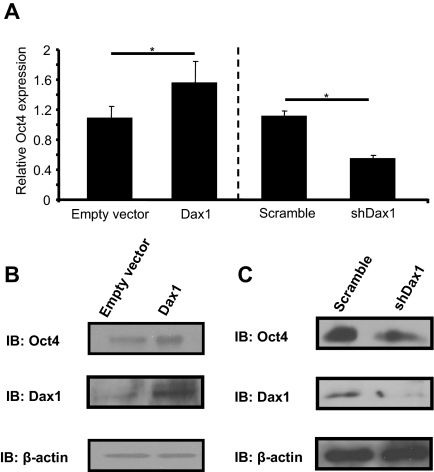
To examine the effect of Dax1 expression on endogenous Oct4 levels, mES cells were transiently transfected with pcDNA3 Dax1 and 48 h later harvested for RNA, and gene expression studies were performed. QPCR revealed that overexpression of Dax1 resulted in modest, but statistically significant, up-regulation of Oct4 mRNA levels (Fig. 4A). Indeed, analysis of Oct4 mRNA levels from Dax1-GFP-positive cells in Fig. 2 also showed an up-regulation, with Oct mRNA levels 1.9-fold higher than in GFP-negative cells. To examine direct changes in Oct4 mRNA levels with Dax1 knockdown, as opposed to secondary effects due to differentiation, mES cells were transfected with a vector containing an shRNA against Dax1 or scrambled and a GFP cassette, and sorted for GFP-positive cells 24 h after transfection. Before any potential differentiation phenotype, a greater than 50% loss in Oct4 levels was observed (Fig. 4A). These data indicate overexpression and knockdown of Dax1 result in up-regulation and down-regulation of Oct4 mRNA, respectively.
Figure 4.
Overexpression or knockdown of Dax1 results in alteration of endogenous Oct4. A, For overexpression, mES cells were transiently transfected in a six-well plate with 3 μg empty vector or pcDNA3 Dax1, and 48 h later cells were harvested and RNA was isolated. For knockdown, mES cells were transfected in a 10-cm plate with 10 μg of a vector containing shRNA against Dax1 or scrambled control, and a GFP expression cassette. Cells were harvested 24 h after transfection, and GFP-positive cells were sorted and then RNA isolated. For both, cDNA synthesis and QPCR were carried out as described in Materials and Methods. Oct4 values were normalized to glyceraldehyde-3-phosphate dehydrogenase and data presented as fold over empty vector control. *, P < 0.005. B, mES cells were transfected in 10-cm dishes with 10 μg empty vector or pcDNA3 Dax1, and 48 h later cells were harvested and protein was isolated. Western blot analysis was performed as described in Materials and Methods with anti-Dax1, anti-Oct4, and anti-β-actin antibodies. C, mES cells were transfected in a 10-cm plate with 10 μg of a vector containing shRNA against Dax1 or scrambled control, and a GFP expression cassette. Cells were harvested 24 h after transfection and GFP-positive cells were sorted after which cells were harvested and protein was isolated. Western blot analysis was performed as described in Materials and Methods with anti-Dax1, anti-Oct4, and anti-β-actin antibodies. IB, Immunoblotting; shDax1, short hairpin Dax1.
To determine whether Oct4 protein levels are altered with the mRNA levels, mES cells were transiently transfected with pcDNA3 Dax1 and 48 h later harvested for protein. Western blot analysis determined that when Dax1 is overexpressed, Oct4 protein levels are concomitantly increased (Fig. 4B). Conversely, to examine Oct4 protein changes with Dax1 knockdown, mES cells were transfected with a vector containing an shRNA against Dax1 or scrambled and a GFP cassette, and sorted for GFP-positive cells 24 h after transfection. Western blot analysis determined that when Dax1 is knocked down, Oct4 protein levels are likewise decreased (Fig. 4C). These data show that Dax1 overexpression and knockdown result in an increase or decrease in Oct4 protein in mES cells, respectively.
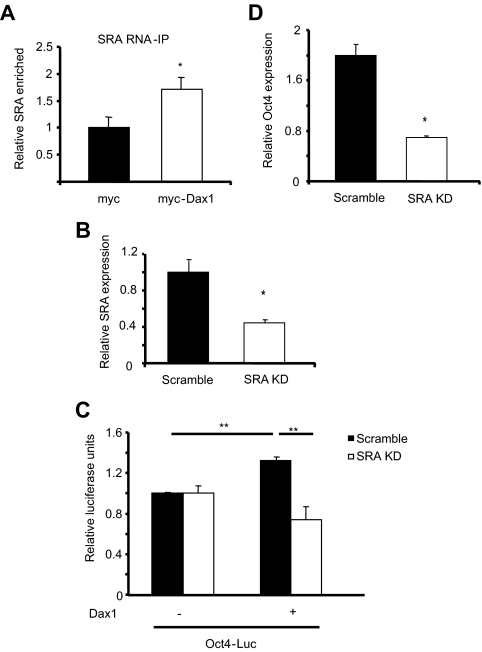
Dax1 interacts with steroid receptor RNA activator (SRA) in mES cells
The steroid receptor RNA activator (SRA), an RNA that interacts with nuclear receptors and forms a scaffold for a p160 family coactivator complex to activate target gene transcription, has recently been shown to bind directly to Dax1 to facilitate Dax1 coactivator (and not corepressor) function (13,19). Therefore, we hypothesized that SRA may mediate the ability of Dax1 to activate Oct4 expression in mES cells. RNA-immunoprecipitation experiments using a myc-tagged Dax1 revealed that immunoprecipitation with anti-myc antibody significantly enriched for SRA in myc-Dax1 transfected mES cell lysates over the empty myc vector transfection (Fig. 5A), indicating that Dax1 interacts with SRA in mES cells.
Figure 5.
Dax1 regulation of Oct4 is mediated by SRA. A, mES cells were transfected in three 10-cm dishes with 10 μg pDax-1-Myc or pCMV-3tag-4A vector per dish, and RNA-immunoprecipitation was performed as described in Materials and Methods. The Final RNA immunoprecipitate (IP) was used to synthesize cDNA and QPCR was performed for SRA and glyceraldehyde-3-phosphate dehydrogenase (as a nonspecific normalization control). *, P < 0.02. B, mES cells stably expressing shRNA against SRA or scrambled were generated as described in Materials and Methods. RNA was harvested, cDNA was synthesized, and QPCR was carried out to analyze the amount of SRA knocked down. Results are presented as relative to scramble control. C, Scramble or SRA knockdown (KD) mES cells were transfected with 200 ng Oct4-Luc reporter and cotransfected with empty vector or 300 ng pcDNA3 Dax1. Cells were harvested 48 h after transfection and lysates were analyzed for luciferase activity. Reporter alone for each cell line was set as 1 luciferase unit. **, P < 0.006. D, Scramble or SRA KD mES cells were transfected in six-well plates with 3 μg empty vector or Dax1. Cells were harvested 48 h after transfection for RNA, cDNA was synthesized, and QPCR was carried out. Change in Oct4 was normalized, and results are presented as fold change over empty vector control.
Loss of SRA results in attenuation of the Dax1 effect on Oct4
Having determined that Dax1 interacts with SRA in mES cells, we set out to determine whether SRA mediates the Dax1-enhanced activation of Oct4. Therefore, a mES cell line with stable shRNA mediated knockdown of SRA was generated. An approximately 60% knockdown of SRA compared with the scramble control was achieved (Fig. 5B). To determine whether SRA was responsible for the Dax1-activating effect on the Oct4 promoter, we carried out luciferase assays in the SRA knockdown or scramble mES cells. When scramble control cells were transfected with the luciferase reporter driven by the Oct4 promoter and cotransfected with pcDNA3 Dax1, a statistically significant increase in reporter activity was observed. In SRA knockdown cells; in contrast, overexpression of Dax1 did not induce reporter activity (Fig. 5C). Similarly, when we overexpress Dax1 in SRA knockdown and scramble control cells, the SRA knockdown cells have at least 2-fold less up-regulation of endogenous Oct4 as determined by QPCR (Fig. 5D). These data indicate that interaction between Dax1 and SRA is necessary for the ability of Dax1 to coactivate Oct4 transcription.
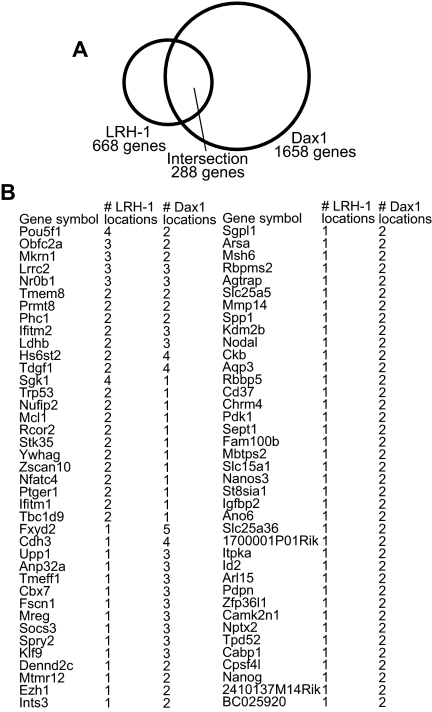
LRH-1 and Dax1 are localized to 288 common genes in mES cells
To determine whether LRH-1 and Dax1 together activate other genes in mES cells, we interrogated published ChIP data sets from whole-genome binding studies in mES cells; 288 genes were shared by the LRH-1 and Dax1 genome binding-site lists, which is 5.6 times as many as expected by chance, indicating a significant overlap in promoter occupancy for LRH-1 and Dax1 (P = 1 × 10−6) (Fig. 6A and Supplemental Table 1 published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Dax1 was found at 43% of the predicted LRH-1 target genes. Of these 288 genes, 78 were identified as having more than one binding location in one of the data sets (Fig. 6B). Literature searches for these 78 genes revealed that 15 of these genes have been shown to be involved in mES cell pluripotency (Table 1). We therefore examined changes in gene expression of these 15 genes in Dax1-overexpressing and knockdown mES cells. As predicted for Dax1 serving a primary coactivation role for LRH-1-mediated gene activation, 11 of the genes were significantly up-regulated when Dax1 was overexpressed, and nine were down-regulated when Dax1 expression was knocked down. Interestingly, the expression of two genes (Mcl1 and Nodal) increased in both the Dax1-overexpressing and knockdown mES cells, consistent with our earlier observations of dose dependence of Dax-1 corepression (low dose) and coactivation (high dose) (13). The expression of Nanog was decreased with both overexpression and knockdown, whereas Fscn1 did not change significantly with Dax1 overexpression or knockdown. These data support a model in which Dax1 coregulates a variety of LRH-1 target genes with most genes being activated and others repressed or being regulated in a dose-dependent manner by Dax1. Such complex regulatory mechanisms are predicted to play a major role in the maintenance of ES cell pluripotency.
Figure 6.
LRH-1 and Dax1 localize to common gene targets in mES cells. A, Venn diagram showing the intersection between ChIP-determined whole-genome binding sites of LRH-1 and Dax1. From these data sets, 288 genes overlap as common targets. B, List of 78 gene targets that have more than one location indicated by the ChIP data sets and the number of locations for LRH-1 and Dax1.
Table 1.
Target gene expression in Dax1-overexpressing and knockdown mES cells
| Gene | RefSeq identification no. | Fold change (mRNA) Dax1 OE mES cells | Fold change (mRNA) Dax1 KD mES cells | Reference |
|---|---|---|---|---|
| Socs3 | NM_007707 | 1.12 (0.17) | 0.18 (0.0005) | 29 |
| Mmp14 | NM_008608 | 1.86 (0.00003) | 0.29 (0.00002) | 30 |
| Pdk1 | NM_172665 | 1.40 (0.001) | 0.44 (0.0006) | 31 |
| Spp1 | NM_009263 | 1.14 (0.33) | 0.52 (0.003) | 32 |
| Tdgf1 | NM_011562 | 1.58 (0.00003) | 0.55 (0.003) | 33 |
| Phc1 | NM_007905 | 1.31 (0.0003) | 0.59 (0.022) | 34 |
| Ezh1 | NM_007970 | 1.32 (0.005) | 0.61 (0.002) | 35 |
| Trp53 | NM_011640 | 1.37 (0.0002) | 0.65 (0.043) | 36 |
| Nanog | NM_028016 | 0.71 (0.0005) | 0.67 (0.027) | 21 |
| Msh6 | NM_010830 | 2.33 (0.00003) | 0.80 (0.11) | 37 |
| Fscn1 | NM_007984 | 1.10 (0.25) | 0.88 (0.14) | 38 |
| Zfp36l1 | NM_007564 | 3.11 (0.0003) | 0.96 (0.75) | 39 |
| Aqp3 | NM_016689 | 1.16 (0.030) | 1.58 (0.055) | 40 |
| Mcl1 | NM_008562 | 3.11 (0.00001) | 1.95 (0.0001) | 41 |
| Nodal | NM_013611 | 1.48 (0.0003) | 2.08 (0.006) | 42 |
Whole-genome binding regions corresponding to promoters of 78 distinct genes were found to contain more than one binding site for LRH-1 or Dax1 and at least one site for both. For further validation studies, we selected the 15 genes from this set that have known roles in mES cell biology and examined gene expression levels by QPCR in mES cells that overexpressed Dax1 vs. mES cells with Dax1 knocked down (as described in Materials and Methods). Shown are fold changes defined as mRNA level in Dax1-overexpressing cells relative to empty vector, or mRNA in Dax1 knockdown cells relative to scramble control, from triplicate experiments. Values were normalized to GAPDH. P values for the changes are given in parentheses. KD, Knockdown; OE, overexpression.
Discussion
Although evidence that Dax1 plays a significant role in the maintenance of pluripotency in mES cells is abundant, few studies have examined a specific mechanism of Dax1 action in these cells. In this study, we elucidate a mechanism by which Dax1 up-regulates Oct4 levels that is predicted to be important in the maintenance of Oct4 levels within the window of pluripotency. Our data show that Dax1 interacts with LRH-1 and is localized to the Oct4 promoter at the site of LRH-1 binding in the proximal promoter, and it is through this site that Dax1 exerts its activating effects. Dax1 is typically known as a corepressor; however, we found that Dax1 coactivates Oct4 transcription mediated by LRH-1. We recently reported that interaction of Dax1 with the RNA activator, SRA, in steroidogenic cells resulted in the ability of Dax1 to act as an activator in certain contexts in a dose-dependent manner (13). In the current study, we demonstrate that Dax1 interacts with SRA in mES cells, and stable knockdown of SRA results in loss of the ability of Dax1 to coactivate Oct4. In addition, we show that LRH-1 and Dax1 localize to a significant number of common genes by ChIP, suggesting that the LRH-1/Dax1 protein complex is important in gene regulation in mES cells. We provide data that Dax1, through interaction with LRH-1 and SRA on the proximal promoter, up-regulates Oct4 expression. These are likely important mechanisms by which Dax1 maintains pluripotency in mES cells.
A previous study reported that Dax1 overexpression caused mES cell differentiation (11). Because high levels of Oct4 cause differentiation and our study shows that Dax1 overexpression results in up-regulation of Oct4, we carefully evaluated the mES cells for any differentiation phenotype to ensure that our results were not simply secondary effects of differentiation rather than primary effects of Dax1 on LRH-1-mediated activation of Oct4. By sorting for a pure population of Dax1-overexpressing mES cells, QPCR revealed that there was no up-regulation of differentiation factors. Additionally, we did not see a decrease in percentage of cells expressing a marker of pluripotency (AP). Perhaps these results are different than the previous report because experiments described herein were not performed in stable Dax1-overexpressing lines and used traditional, as opposed to episomal, plasmids. The well-characterized dose and time dependence of Dax1 coactivation vs. corepression function on transcription may also play a role in the different results observed in these two studies. In humans, duplication of Dax1 is associated with gonadal sex reversal, and our recent work indicates that different concentrations of Dax1 determine whether Dax1 acts as a transcriptional repressor or activator (13,20). Thus, our finding that Dax1 overexpression does not cause differentiation but does up-regulate Oct4 levels may reflect temporal and concentration-dependent actions of Dax1 in the transcriptional control of mES cell pluripotency.
Our results demonstrate that Dax1 interacts with both LRH-1 and SRA in mES cells. Whether SRA may bind to LRH-1 as well, in the absence of Dax1, remains an open question. This interaction is most likely not restricted to the Oct4 promoter but is an important mechanism of transcriptional regulation of other gene promoters in mES cells. Indeed, the intersection of published ChIP results shows that LRH-1 and Dax1 localize to 288 common genes. Changes in expression of a substantial subset of these genes in Dax1-overexpressing vs. knockdown mES cells support a model whereby Dax1 serves as both an activator and a repressor in different contexts, perhaps in a dose-dependent manner. It is interesting to hypothesize that Dax1 may act on many different promoters with varied protein or RNA partners, each resulting in a context-specific activation or repression.
Oct4 expression levels must be kept within a tight window to maintain pluripotency: a less than 2-fold increase causes differentiation into a mixture of primitive endoderm and mesoderm lineages, and a decrease in Oct4 levels causes dedifferentiation to trophectoderm (2). Our experiments show that Dax1 overexpression or knockdown results in subtle changes in Oct4 mRNA and protein levels. Specifically, in luciferase reporter and gene expression studies, Oct4-Luc and Oct4 mRNA levels never change more than 2-fold. A confounding variable in these overexpression experiments is that endogenous Dax1 levels in mES cells are high, with Dax1 being one of the top 20 mRNAs enriched in mES cells (21). Thus, overexpression of Dax1 may have only minor effects if Dax1 is not limiting. However, we suggest that these subtle changes in Oct4 mRNA are anticipated due to the tight requirements for Oct4 expression. Additionally, with the numerous levels of transcriptional regulation that control Oct4 levels, as experimental manipulation elevates Oct4 levels, other compensatory mechanisms may attempt to down-regulate its expression to maintain pluripotency.
Although we have achieved overexpression of Dax1, we have not observed differentiation (as mentioned above). Sun et al. (11), however, found that stable Dax1-overexpressing mES cells dedifferentiated into trophectodermal lineage, expressing the markers Cdx2 and Rhox6. This is consistent with a down-regulation of Oct4, which they also observed. Their study showed that Dax1 inhibits Oct4-mediated transcription; because Oct4 has been shown to up-regulate its own transcription, it was suggested that Dax1 may down-regulate Oct4 through interaction with Oct4 on its own promoter. Additionally, previous studies have shown an interaction of Dax1 with Nanog, which has also been shown to activate Oct4 expression (22,23). Indeed, the ChIP studies for Dax1 indicate that Dax1 is localized to two sites on the Oct4 promoter. However, it is worth noting that our data indicate that the overall net effect of Dax1 on Oct4 expression appears to be activation.
In a separate report, Oct4 was also shown to participate in the regulation of Dax1 expression in mES cells through a site within the Dax1 intron (24). Together these data allow speculation that there may be a positive feedback loop in which higher levels of Dax1 induced by Oct4 may serve to up-regulate Oct4 expression; however, because Dax1 can also inhibit the transcriptional activity of Oct4, this could serve two functions. First, this could provoke negative feedback to keep levels of Oct4 in check via its autoinduction, and second, Dax1 would inhibit Oct4 activity, thereby preventing high Oct4 levels from causing differentiation. These dual mechanisms would serve to rapidly regulate the actions of Oct4 in the cell.
Previous work characterizing the effects of Dax1 knockdown in mES cells showed that after 24 h of knockdown, 90% of altered genes in a microarray were up-regulated, indicating that Dax1 likely repressed these genes (7). Thus, 10% of these genes that changed were down-regulated, suggesting a role for Dax1 as a coactivator on some promoters. Additionally, the report demonstrated that, in a luciferase reporter assay using a construct containing domains of Dax1 fused to the yeast Gal4 DNA-binding domain, this construct repressed some artificial promoters but failed to repress others, showing the context specificity of the actions of Dax1. However, it was stated that Oct4 levels did not change in their experiment. Interestingly, supplemental data from an additional publication are consistent with these data showing that knockdown of Dax1 results in a down-regulation of endogenous Oct4 levels (22).
Whereas many studies have examined Dax1 in the context of genomic experiments, characterizing the overall importance of Dax1 in mES cells, this report is only the second study to show a specific mechanism by which Dax1 regulates gene expression. Although it is important to appreciate the critical role of Dax1 in these cells as reflected by the number of genomic sites to which it is localized and the numerous protein interactions in which it participates, understanding the mechanisms of Dax1 action is critical for full understanding of the role of Dax1 in mES cell biology. Thus, this study, showing Dax1 coactivation of Oct4 through interaction with LRH-1 and SRA and localization of LRH-1 and Dax1 to many common target genes in mES cells, is an additional contribution to the complex network of transcriptional control of mES cell pluripotency.
Materials and Methods
Cell culture and transfection
The D3 line of mouse embryonic stem (ES) cells (a kind gift from K. Sue O’Shea) was cultured on 0.1% gelatin-coated substrates in ES medium consisting of DMEM (Life Technologies, Gaithersburg, MD) supplemented with 10% ES-tested fetal bovine serum (Hyclone Laboratories, Logan, UT), 10−4 m β-mercaptoethanol (Sigma Chemical Co., St. Louis, MO), 0.224 μg/ml l-glutamine (Life Technologies), 1.33 μg/ml HEPES (Life Technologies), 100 U penicillin, 100 μg/ml streptomycin, and 1000 U/ml LIF (Chemicon, Temecula, CA). F9 embryonal carcinoma cells were maintained on 0.1% gelatin-coated substrates in DMEM with 10% bovine serum (Life Technologies) and penicillin-streptomycin. All cells were grown at 37 C in a humidified atmosphere of 5% CO2. Transient transfection was performed with Lipofectamine 2000 at a ratio of 3 μl:1 μg DNA (D3 cells) or 7 μl:1 μg DNA (F9 cells). Differentiation was performed with either 1 μm retinoic acid or removal of LIF from the media for 48 h.
Plasmids
pDax-1-Myc and pCMV-3tag-4A have been previously described (13). pGL3-Oct4 (hereafter called Oct4-Luc) was cloned by insertion of a 1141-bp PCR-amplified Oct4 promoter fragment into the KpnI/XhoI sites of pGL3 Basic. Primers used are as follows: 5′-CCGGGTACCCCCATGGCTGGACACCTGGCTTCA-3′ and 5′-CCGCTCGAGACCCCAAAACTTCAGGTTCTCTTGTCT-3′. A mutated Oct4-Luc construct (Oct4-Luc mPP) was generated using QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: 5′-ggggccagaggtcaaacctagagggtgggatt-3′ and 5′-aatcccaccctctaggtttgacctctggcccc-3′. pEGFP Dax1 was constructed by subcloning Dax1 cDNA with HindIII/XbaI into pEGFP C1 (CLONTECH Laboratories, Inc., Palo Alto, CA). shRNA expression constructs (pGIPZ shDax1, pGIPZ scramble; Open Biosystems, Huntsville, AL) were obtained from the University of Michigan shRNA Core Facility (http://fgc.lsi.umich.edu/). pcDNA LRH-1 was a generous gift from Dr. William Rainey (Medical College of Georgia). pSuperior shSRA and pSuperior scramble were used as described previously (13).
AP staining
For AP staining, cells were washed with PBS and then fixed for 2 min with 4% formalin in PBS. Cells were then equilibrated in AP buffer for 5 min (100 mm Tris-HCl, pH 9.5; 100 mm NaCl; and 10 mm MgCl2). After equilibration, AP staining was performed using 90 μl Nitro-Blue Tetrazolium Chloride/5-Bromo-4-Chloro-3′-Indolyphosphate p-Toluidine Salt (Roche, Indianapolis, IN) in 10 ml AP buffer for 10 min. Images were captured using stereo or inverted microscopes (Leica, Inc., Deerfield, IL) in the Microscopy and Image-analysis Laboratory (MIL) at the University of Michigan Department of Cell & Developmental Biology. Images were subjected to cell counting using Photoshop CS3.
Luciferase assays
D3 or F9 cells were plated at a density of 10 × 104 cells per well in 24-well plates. Cells were transiently transfected, 24 h after plating, with luciferase reporter constructs as noted in each figure, and harvested 48 h after transfection (except where noted). Cell lysates were assayed for luciferase activity using the Dual Luciferase Assay (Promega Corp., Madison, WI) with an injector luminometer. Luciferase activity was normalized by transfection of pRL-TK Renilla luciferase (Promega).
Gene overexpression and knockdown assays
Overexpression experiments were carried out as follows: 250 × 103 cells were plated in six-well plates, and 24 h later transfected with 2–3 μg DNA. After 48 h, cells were harvested, RNA was isolated, cDNA was synthesized, and QPCR was carried out as described previously (25). To analyze for up-regulation of protein, 3 × 106 cells were plated in 10-cm plates and 24 h later transfected with 10 μg pcDNA3 Dax1 or empty vector. Cells were harvested 48 h later, as described under Western Blotting procedures.
The method of stable knockdown of endogenous SRA in mouse ES cells was modified from previously described methods (13). The shRNA construct targeting mouse SRA and the scramble-sequence shRNA control were described previously (13). The SRA and control shRNAs were expressed from the retroviral vector pSuperior.retro.puro (OligoEngine). The retroviruses were grown in and harvested from Phoenix cells (kindly provided by G. Bommer, University of Michigan,) and then used to infect mES cells three times with 8- to 12-h intervals. The infected cells were selected with 1 μg /ml of puromycin. The SRA-silencing effects of shRNAs were confirmed by QPCR using mouse SRA-specific primers (13).
For transient knockdown studies, experiments were carried out as follows: cells were plated in 10-cm plates, and 24 h later transfected with 10 μg pGIPZ shDax1 or scramble control. Cells were harvested 24 h after transfection and sorted by flow cytometry (using a FACSDiva) for expression from the GFP cassette in the shRNA plasmids. Cells were immediately harvested for RNA isolation, and QPCR was carried out using primer pairs listed in Table 2.
Table 2.
QPCR primers
| Genes | Forward | Reverse |
|---|---|---|
| Dax1 | accgtgctctttaacccaga | ccggatgtgctcagtaagg |
| Oct4 | tgaggctacaggacacctttc | Gtgccaaagtccggacct |
| SRA | ggctggagggaagttgtcaatac | Ccactggtgatctaaaagttcttg |
| Tdgf1 | ttttacgagccgtcgaagat | Aattcaaacgcactggaaatg |
| Phc1 | tcattgaaggctttgttatcca | Tctttcaggaactgagaacatcc |
| Trp53 | atgcccatgctacagaggag | Agactggcccttcttggtct |
| Ezh1 | catgacccagaacttttgtgaa | Gacaaccaggaaagcgattc |
| Mcl1 | ggtatttaagctagggtcatttgaa | Tgcagccctgactaaaggtc |
| Msh6 | cagctggcagtgtgtgatg | Gataaataagcctcatgcacctc |
| Mmp14 | aacttcgtgttgcctgatga | Tttgtgggtgaccctgactt |
| Spp1 | ggaggaaaccagccaagg | Tgccagaatcagtcactttcac |
| Nodal | ccaaccatgcctacatcca | Cacagcacgtggaaggaac |
| Aqp3 | ctggggaccctcatcctt | Tggtgaggaagccaccat |
| Pdk1 | gttgaaacgtcccgtgct | Gcgtgatatgggcaatcc |
| Fscn1 | gccaacgagaggaacgtg | Ggtgcgaaaggcacactt |
| Socs3 | atttcgcttcgggactagc | Aacttgctgtgggtgaccat |
| Zfp36l1 | ttcacgacacaccagatcct | Tgagcatcttgttacccttgc |
| Nanog | ttcttgcttacaagggtctgc | Agaggaagggcgaggaga |
| GAPDH | aatgtgtccgtcgtggatct | Cccagctctccccatacata |
Co-IP and Western blotting
D3 cells in 10-cm plates were harvested and cellular protein was collected by lysis in a buffer containing 40 mm HEPES, 120 mm sodium chloride, 10 mm sodium pyrophosphate, 10 mm sodium glycerophosphate, 1 mm EDTA, 50 mm sodium fluoride, 0.5 mm sodium orthovanadate, 1% Triton X-100 buffer, and protease inhibitor cocktail (Sigma), followed by rotation for 1 h at 4 C. Soluble protein was collected from centrifuged total lysates and quantified by Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA). Protein lysates were resolved on a 10% SDS-PAGE and transferred to nitrocellulose membrane by standard procedures. Proteins were detected using anti-Dax1 antibody (1:1000, R&D Systems, Minneapolis, MN), anti-Oct4 antibody (1:200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or anti-β-actin antibody (1:5000, Sigma), followed by blotting with goat antimouse horseradish peroxidase (Pierce) or rabbit antigoat horseradish peroxidase (Thermo Scientific, Rockford, IL), and detection was performed using Super Signal West Dura Extended Duration Substrate (Pierce).
For co-IP, lysates were cleared by incubation with Protein A agarose beads (Invitrogen, Carlsbad, CA) and subsequent washing. Immunoprecipitations were performed on 100 μg protein using 3 μg anti-LRH-1 (Santa Cruz) or normal serum and 40 μl Protein A agarose followed by stringent washing with lysis buffer. Bound proteins were resolved by SDS-PAGE and immunoblot performed as described above.
ChIP
ChIP assays were performed on mES cells as previously described (26). Anti-Dax antibody (2 μg) (Santa Cruz) was used for immunoprecipitation. Results shown are representative and from independent experiments, quantitated by QPCR or visualized by PCR and agarose gel electrophoresis. Primer pairs used for ChIP assays are listed in Table 3.
Table 3.
ChIP primers
| Forward | Reverse | |
|---|---|---|
| Oct4 promoter LRH PP (14) | cctccgtctggaagacacaggcagatagcg | cgaagtctgaagccaggtgtccagccatgg |
| Oct4 promoter LRH PE (13) | gctggggaagtcttgtgtga | gcttccagcctagttcctgg |
| Control | agagggtcaaggatggaatgatt | cagtgtgctccctcccacc |
PE, Proximal enhancer.
RNA-immunoprecipitation
For the immunoprecipitation of Dax1 to evaluate enrichment of SRA, mouse ES cells were transiently transfected with pDax1-Myc or empty pCMV-3tag-4A vector. The subsequent immunoprecipitation procedures were modified from previously described methods (13). Briefly, cells were washed with PBS and cross-linked with 0.1% formaldehyde for 10 min at room temperature. After the addition of 0.25 m glycine for 5 min, cells were harvested and lysed in radioimmune precipitation assay buffer containing protease inhibitor cocktail tablet (Roche), and 40 U/μl RNasin (Promega) and sonicated (13,27). After immunoprecipitation with anti-Myc antibody (Immunology Consultants Laboratory), reversal of cross-linking and deoxyribonuclease I treatment were performed as described previously (13). RNA was then isolated by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation containing glycogen (Ambion, Inc., Austin, TX), and the total amount was used for cDNA synthesis. QPCR was carried out using 2 μl of the resulting cDNA and primers that amplify mouse SRA. Data were normalized to housekeeping gene expression.
Intersection of LRH-1 and Dax1 ChIP data sets
We obtained the locations of 3346 overrepresented genomic regions, typically 250–700 bases long, from Heng et al. (28) chromatin immunoprecipitation using an antihemagglutinin to enrich for chromatin bound by hemagglutinin-tagged LRH-1, followed by sequencing. The data were obtained from GEO series GSE19019. Using the National Center for Biotechnology Information build 38 of the mouse genome, we associated these segments with 722 transcripts by asking if the segment midpoints were within −10,000 to 3000 bases from transcript start sites. These transcripts represented 668 distinct Entrez genes. For Dax1, we obtained 1982 locations in terms of gene symbols and distance from transcript start sites from a chromatin immunoprecipitation study using biotin-tagged Dax-1 followed by hybridization to an Affymetrix mouse promoter (1.0R) array (9). We collapsed these locations to 1658 distinct Entrez genes. The two gene lists contained 288 genes in common, which is 5.6 times as many as expected by chance considering 21,577 total Entrez genes, indicating a significant overlap in promoter occupancy for LRH-1 and Dax-1 (P = 1 × 10−6, two-sided Fisher’s Exact test).
Statistics
Experiments were each performed at least in triplicate, and error bars represent standard deviation. Statistical analyses were performed by ANOVA and/or Student’s t test. P values are defined in the figure legends.
Supplementary Material
Acknowledgments
We thank K. Sue O’Shea, Michelle Wood, and Joanne Heaton for critical reading of this manuscript and K. Sue O’Shea for critical intellectual input. We also thank the Biomedical Research Core Facilities (Flow cytometry and Sequencing) at the University of Michigan.
Footnotes
This work was supported by National Institututes of Health (NIH) Grant DK062027 from National Institute of Diabetes and Digestive and Kidney Diseases (to G.D.H.); V.R.K. was supported, in part, by NIH Grant T32 HD07048 to the Training Program in Reproductive Sciences at the University of Michigan. In addition, this research used the shRNA Core of the Michigan Diabetes Research and Training Center supported by NIH Grant DK20572.
Disclosure Summary: The authors have nothing to disclose.
First Published Online October 13, 2010
Abbreviations: AP, Alkaline phosphatase; ChIP, chromatin immunoprecipitation; co-IP, coimmunoprecipitation; ES, embryonic stem; GFP, green fluorescent protein; LIF, leukemia-inhibitory factor; LRH-1, liver receptor homolog 1; mES, mouse embryonic stem; PP, proximal promoter; QPCR, quantitative PCR; Sf1, steroidogenic factor 1; shRNA, short hairpin RNA; SRA, steroid receptor RNA activator.
References
- Niwa H 2007 How is pluripotency determined and maintained? Development 134:635–646 [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG 2000 Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24:372–376 [DOI] [PubMed] [Google Scholar]
- Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, McCabe ER, Meitinger T, Monaco AP, Sassone-Corsi P, Camerino G 1994 An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372:635–641 [DOI] [PubMed] [Google Scholar]
- McCabe ER 2007 DAX1: increasing complexity in the roles of this novel nuclear receptor. Mol Cell Endocrinol 265–266:179–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipsham R, Niakan K, McCabe ER 2004 Nr0b1 and its network partners are expressed early in murine embryos prior to steroidogenic axis organogenesis. Gene Expr Patterns 4:3–14 [DOI] [PubMed] [Google Scholar]
- Niakan KK, Davis EC, Clipsham RC, Jiang M, Dehart DB, Sulik KK, McCabe ER 2006 Novel role for the orphan nuclear receptor Dax1 in embryogenesis, different from steroidogenesis. Mol Genet Metab 88:261–271 [DOI] [PubMed] [Google Scholar]
- Khalfallah O, Rouleau M, Barbry P, Bardoni B, Lalli E 2009 Dax-1 knockdown in mouse embryonic stem cells induces loss of pluripotency and multilineage differentiation. Stem Cells 27:1529–1537 [DOI] [PubMed] [Google Scholar]
- Yu RN, Ito M, Saunders TL, Camper SA, Jameson JL 1998 Role of Ahch in gonadal development and gametogenesis. Nat Genet 20:353–357 [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008 An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132:1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P 1997 DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature 390:311–315 [DOI] [PubMed] [Google Scholar]
- Sun C, Nakatake Y, Akagi T, Ura H, Matsuda T, Nishiyama A, Koide H, Ko MS, Niwa H, Yokota T 2009 Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol Cell Biol 29:4574–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yu R, Jameson JL 1997 DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol 17:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang WH, Gerin I, Hu CD, Hammer GD, Koenig RJ 2009 Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol 29:1719–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ 2005 Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol 25:3492–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Dorn C, Sadovsky Y, Milbrandt J 1998 Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol Cell Biol 18:2949–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altincicek B, Tenbaum SP, Dressel U, Thormeyer D, Renkawitz R, Baniahmad A 2000 Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem 275:7662–7667 [DOI] [PubMed] [Google Scholar]
- Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA 1998 Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93:445–454 [DOI] [PubMed] [Google Scholar]
- Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ 2008 The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci USA 105:18390–18395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW 1999 A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97:17–27 [DOI] [PubMed] [Google Scholar]
- Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, Tonini G, Ferrante E, Chiumello G, McCabe ER, Fraccaro M, Zuffardi O, Camerino G 1994 A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet 7:497–501 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S 2003 The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631–642 [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH 2006 A protein interaction network for pluripotency of embryonic stem cells. Nature 444:364–368 [DOI] [PubMed] [Google Scholar]
- Pan G, Li J, Zhou Y, Zheng H, Pei D 2006 A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J 20:1730–1732 [DOI] [PubMed] [Google Scholar]
- Sun C, Nakatake Y, Ura H, Akagi T, Niwa H, Koide H, Yokota T 2008 Stem cell-specific expression of Dax1 is conferred by STAT3 and Oct3/4 in embryonic stem cells. Biochem Biophys Res Commun 372:91–96 [DOI] [PubMed] [Google Scholar]
- Gummow BM, Scheys JO, Cancelli VR, Hammer GD 2006 Reciprocal regulation of a glucocorticoid receptor-steroidogenic factor-1 transcription complex on the Dax-1 promoter by glucocorticoids and adrenocorticotropic hormone in the adrenal cortex. Mol Endocrinol 20:2711–2723 [DOI] [PubMed] [Google Scholar]
- Winnay JN, Hammer GD 2006 Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol Endocrinol 20:147–166 [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA 2002 Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26:182–190 [DOI] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH 2010 The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6:167–174 [DOI] [PubMed] [Google Scholar]
- Duval D, Reinhardt B, Kedinger C, Boeuf H 2000 Role of suppressors of cytokine signaling (Socs) in leukemia inhibitory factor (LIF) -dependent embryonic stem cell survival. FASEB J 14:1577–1584 [DOI] [PubMed] [Google Scholar]
- Costello I, Biondi CA, Taylor JM, Bikoff EK, Robertson EJ 2009 Smad4-dependent pathways control basement membrane deposition and endodermal cell migration at early stages of mouse development. BMC Dev Biol 9:54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamgüney T, Zhang C, Fiedler D, Shokat K, Stokoe D 2008 Analysis of 3-phosphoinositide-dependent kinase-1 signaling and function in ES cells. Exp Cell Res 314:2299–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba R, Niwa H, Masui S, Ohtsuka S, Carter MG, Sharov AA, Ko MS 2006 Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One 1:e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liguori G, Adamson ED, Persico MG 1998 Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev Biol 196:237–247 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li H, Liu Y, Mattson MP, Rao MS, Zhan M 2008 Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS One 3:e3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH 2008 EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost S, Bellamy CO, Clarke AR, Wyllie AH, Harrison DJ 1998 p53-independent DNA repair and cell cycle arrest in embryonic stem cells. FEBS Lett 425:499–504 [DOI] [PubMed] [Google Scholar]
- Roos WP, Christmann M, Fraser ST, Kaina B 2007 Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O(6)-methylguanine due to high E2F1 regulated mismatch repair. Cell Death Differ 14:1422–1432 [DOI] [PubMed] [Google Scholar]
- Carter MG, Stagg CA, Falco G, Yoshikawa T, Bassey UC, Aiba K, Sharova LV, Shaik N, Ko MS 2008 An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns 8:181–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmüller D, Raineri I, Gross B, Oakeley EJ, Moroni C 2007 A cassette system to study embryonic stem cell differentiation by inducible RNA interference. Stem Cells 25:1178–1185 [DOI] [PubMed] [Google Scholar]
- Jincho Y, Sotomaru Y, Kawahara M, Ono Y, Ogawa H, Obata Y, Kono T 2008 Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol Reprod 78:568–576 [DOI] [PubMed] [Google Scholar]
- Grandela C, Pera MF, Grimmond SM, Kolle G, Wolvetang EJ 2007 p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res 1:116–128 [DOI] [PubMed] [Google Scholar]
- Tiedemann H, Asashima M, Grunz H, Knöchel W 2001 Pluripotent cells (stem cells) and their determination and differentiation in early vertebrate embryogenesis. Dev Growth Differ 43:469–502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.