Abstract
The molybdenum nitrogenase is responsible for most biological nitrogen fixation, a prokaryotic metabolic process that determines the global biogeochemical cycles of nitrogen and carbon. Here we describe the trafficking of molybdenum for nitrogen fixation in the model diazotrophic bacterium Azotobacter vinelandii. The genes and proteins involved in molybdenum uptake, homeostasis, storage, regulation, and nitrogenase cofactor biosynthesis are reviewed. Molybdenum biochemistry in A. vinelandii reveals unexpected mechanisms and a new role for iron-sulfur clusters in the sequestration and delivery of molybdenum.
The scarcity of molybdenum in the Earth's crust belies its importance for the metabolism of living organisms and for the global biogeochemical cycles of major elements such as nitrogen, sulfur and carbon (1, 2). In contrast, many elements that are present in considerably larger amounts have not apparent biological function (e. g. Al, Ti or Zr) (3). Despite its low abundance - Mo ranks 53rd in the Earth's crust - molybdenum is more available to biological processes than many other metals, which are found in chemical forms difficult to assimilate. Molybdate is the predominant source of MoVI at neutral and basic pH. Other common sources of molybdenum are the highly insoluble molybdenite (MoS2) and wulfenite (PbMo4) ores (4). In marine environments, molybdenum is present at around 110 nM, being the most abundant transition metal in the sea (1). In terrestrial environments, molybdenum distribution is irregular, usually lower than in the marine systems, and estimated to be 50 nM on average. The importance of molybdenum in soil ecosystems has recently been highlighted by a study showing that molybdenum scarcity severely limits biological nitrogen fixation in tropical forests (5). Limitation of nitrogen fixation by molybdenum might be common in highly weathered acidic soils, which would hinder the ability of some forests to balance carbon and nitrogen (6).
Comparative genomic studies reveal that molybdenum metabolism is widespread in nature (7). Molybdenum is utilized - to different extents - by most organisms belonging to the three domains of life: archea, bacteria, and eukaryotes. Phylogenetic analyses reveal that most prokaryotes (archea and bacteria) and higher eukaryotes utilize molybdenum, whereas many unicellular eukaryotes, including parasites and some yeast, have lost their ability to use this metal.
In addition to being the only second row transition metal essential for life, molybdenum provides an extremely versatile building tool for the coordination chemist. The element has a [Kr] 4d5 6s1 electron configuration, and its chemical properties thus center on its half-filled 4d shell. Molybdenum is able to adopt oxidation states from (-II) to (+VI) together with ligand coordination numbers ranging from 4 to 8 and a variety of coordination geometries. It forms compounds with most inorganic and organic ligands, and a wide range of metal and mixed-metal clusters have been synthesized. The chemistry of the higher molybdenum oxidation states (MoVI, MoV, MoIV, MoIII) is dominated by oxo species, such as molybdates and polymolybdates, and terminal oxygen containing species like MoVIO2, MoVO, MoIVO and MoIVO2 comprise the central cations in a range of complexes (8). Molybdenum also has a rich and diverse sulfur chemistry, which comprises ligand-based redox behavior, internal electron-transfer processes and ‘intermediate’ redox states (9), all largely a consequence of the small energy gap between the sulfur 3p and the Mo 4d orbitals (10). The importance of molybdosulfur complexes in biological and industrial catalysis has led to the study of an array of monomeric (11), dimeric (12) and cluster complexes (13, 14).
Biologically active molybdenum is found in the cofactors of molybdoenzymes. Much of our understanding of molybdenum sites in enzymes is derived from combining the results of EPR1 (15-17) and ENDOR (18, 19) spectroscopies on paramagnetic enzyme intermediates, with structural information from extended X-ray absorption fine structure (EXAFS) spectroscopy (16, 20-24) and X-ray crystallography (25-28). In addition, the chemical behavior of related inorganic compounds has been used to calibrate the spectroscopies and to suggest or confirm structural and mechanistic possibilities for the enzymes (11, 14). These studies have shown that all molybdenum cofactors, with the exception of the FeMo-co of nitrogenase, are based on a unique tricyclic pterin (molybdopterin) and are generically termed as Mo-co (16, 24, 29). Mo-co containing enzymes catalyze a range of oxidation/reduction reactions in carbon, sulfur and nitrogen metabolism, such as the oxidation of hypoxanthine to xanthine, the reduction of nitrate to nitrite and the oxidation of formate to carbon dioxide (16, 24).
Biological Nitrogen Fixation
Fixed nitrogen is an essential component of amino acids, proteins, and nucleic acids in all organisms. It is also present in other essential molecules that are abundant in the biosphere, such as chlorophyll and heme groups. Although nitrogen gas (N2) constitutes 78% by volume of Earth's atmosphere, it is unusable by most organisms, which can only assimilate fixed nitrogen molecules. The low reactivity of N2 limits its conversion into fixed nitrogen molecules. Nevertheless, a special group of prokaryotic organisms has developed the ability to fix N2 at moderate temperature and pressure conditions in a process known as biological nitrogen fixation. Nitrogen-fixing organisms (diazotrophs) reduce N2 into NH4+ that is subsequently assimilated by themselves and by other organisms, such as plants, fungi, and animals. Thus, the global balance of nitrogen on Earth's biosphere relies on the capacity of diazotrophic organisms to serve as primary input of fixed nitrogen into the ecosystems (30).
The enzyme responsible for all biological nitrogen fixation activity is termed nitrogenase. The catalytic site of nitrogenase contains a complex metallocluster where N2 binding and reduction into NH4+ takes place. There are four classes of nitrogenase enzymes characterized so far. Three of them are homologous enzymes with similar - not identical - protein subunit composition and metal cofactor structure (31, 32); these are the Mo-nitrogenase, V-nitrogenase, and Fe-only nitrogenase. The Mo-nitrogenase, which contains the iron-molybdenum cofactor or FeMo-co, is the most commonly distributed nitrogenase; it is also the most efficient in the conversion of N2 into NH4+. Although most diazotrophs only have the Mo-nitrogenase, some of them also synthesize alternative V-nitrogenase and/or Fe-only nitrogenase enzymes, which contain the FeMo-co-like FeV-co or FeFe-co metalloclusters at their active sites, respectively. There are no reported diazotrophs lacking a Mo-nitrogenase and carrying uniquely an alternative nitrogenase. Regulation of nitrogenase expression in bacteria carrying alternative nitrogenases is dependent on the availability of molybdenum, vanadium or iron in the medium (33). These three nitrogenases consist of two component proteins that, in the case of the Mo-nitrogenase, are denoted as NifDK2 and NifH. The NifDK component accommodates the active site cofactor whereas the NifH component serves as specific electron donor to NifDK.
The FeMo-co active site of the Mo-nitrogenase is a complex Mo-Fe-S metallocluster comprising an inorganic Fe6-S9 core that coordinates a central light atom X (C, N or O) and is capped by external Fe and Mo atoms. A molecule of R-homocitrate coordinates the molybdenum atom through its C-2 carboxyl and hydroxyl groups to complete the cofactor (Figure 1) (27, 28, 34, 35). The role of the molybdenum sub-site in FeMo-co is unclear (31, 36). For some time, molybdenum was considered a likely locus for substrate binding and catalysis as the Mo-nitrogenase is substantially more active in N2 reduction than its V or Fe analogs, and the R-homocitrate ligand is essential for catalysis (31, 32). In addition, molybdenum compounds can readily coordinate N2 and its reduced forms, and this chemistry has been systemized into a putative mechanism termed the “Chatt cycle” (37). However, recent combined spectroscopic and substrate/inhibitor binding studies strongly indicate that the [Fe6-S9-X] core is the initial substrate binding site (31, 32, 36) whereas current mechanistic models do not include a direct function for the molybdenum sub-site (31).
FIGURE 1.
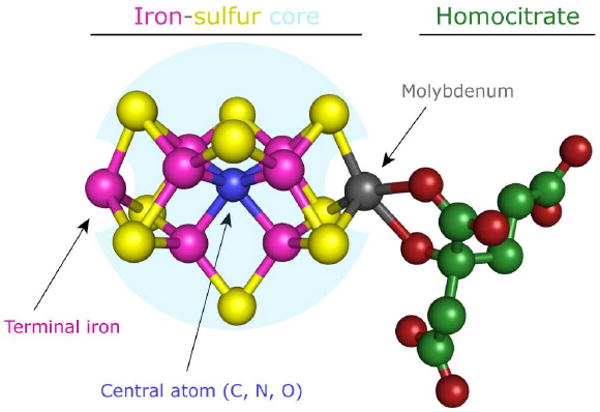
The iron-molybdenum cofactor (FeMo-co) of nitrogenase structured from a biosynthetic perspective. The location of the Fe6-S9-X cofactor core corresponding to the NifB-co precursor is highlighted by a cyan sphere. Terminal Fe and Mo atoms capping the open sites at the cofactor core are labeled. Atom colors: iron, magenta; molybdenum, grey; sulfur, yellow; oxygen, red; carbon, green; and central atom X, blue.
The fourth type of nitrogenase is a Mo-nitrogenase phylogenetically unrelated to the other three classes that has only been found in the bacterium Streptomyces thermoautotrophicus. This Mo-nitrogenase exhibits completely different biochemical features that consist of different protein composition, insensitivity to O2, low Mg·ATP requirement, and a Mo-co type of cofactor at the active site (Mo-molybdopterin cytosine dinucleotide or Mo-MCD) (38).
The bacterium Azotobacter vinelandii has traditionally been one of the preferred models to study molybdenum metabolism in prokaryots because it carries several Mo-co-containing enzymes and a Mo-nitrogenase. The genetic components involved in molybdenum capture, trafficking, storage, metabolism, and regulation that are encoded in this organism have been studied for decades (Figure 2). Although studies in other diazotrophs such as Klebsiella pneumoniae and Rhodobacter capsulatus have provided significant insights into the metabolism of molybdenum, A. vinelandii will be used as the reference organism for discussion in this review. The recent release of the A. vinelandii genome sequence (39) has revealed the presence of new genes related to molybdenum, for example those encoding an anaerobic Mo-co-dependent formate dehydrogenase. This review will focus on those proteins and processes related - albeit not necessarily in an exclusive way - to molybdenum-dependent nitrogen fixation.
FIGURE 2.
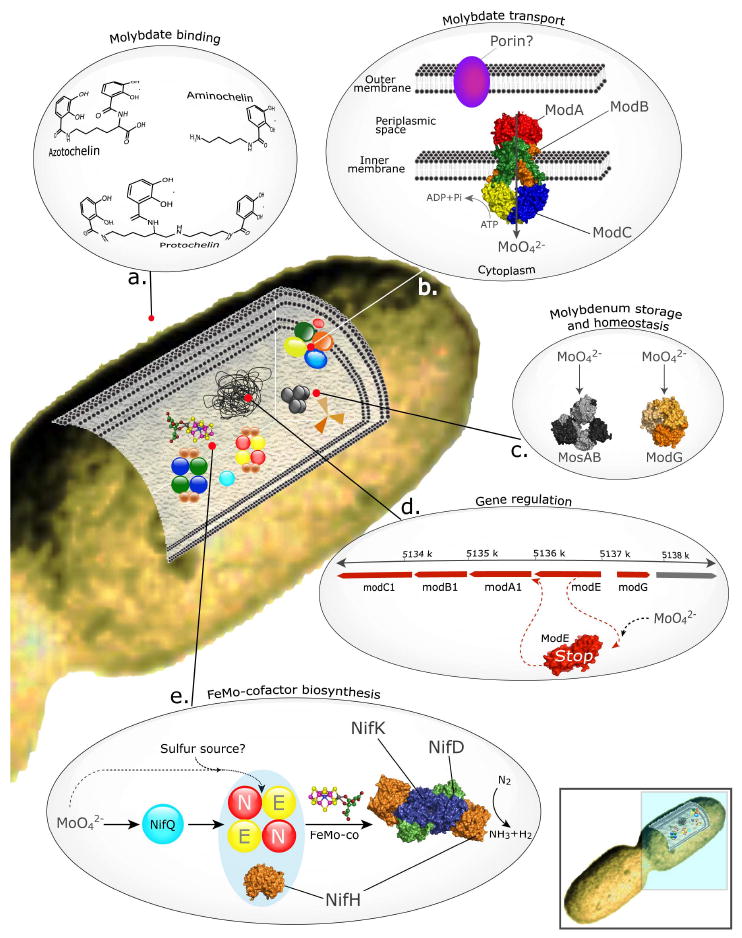
Molybdenum trafficking for nitrogen fixation in the bacterium Azotobacter vinelandii. The figure shows a pathway of molybdenum towards the molybdenum-nitrogenase enzyme and the protein components involved in this pathway. Some of these proteins are exclusively dedicated to the nitrogenase biogenesis (e.g. NifQ and NifEN); some other have general roles in the metabolism of molybdenum (e.g. the ModABC molybdate transport system and the molybdenum-dependent transcriptional regulator ModE). Excreted siderophores with capacity to bind molybdate anions are also depicted. The modABC structure corresponds to the molybdate transport system of Archaeoglobus fulgidus. The ModE, NifH, and NifHDK structures shown are from A. vinelandii. Panel e shows a simplified FeMo-co biosynthetic pathway illustrating the two putative pathways for molybdenum incorporation into the Mo-nitrogenase cofactor. A complete FeMo-co biosynthetic pathway is shown in Figure 3.
Molybdenum Trafficking for Nitrogen Fixation: The Path to the Nitrogenase Active Site
(i) Binding of extracellular molybdenum
The average molybdenum concentration in soils (ca. 15 μmol/kg or 1-2 ppm) is the lowest among all transition metals having a biological role (40). Molybdate (MoO42-, MoVI) is the main source of molybdenum because it is soluble at neutral and basic pH. Other forms of molybdenum described to efficiently serve as assimilable molybdenum sources are MoO3, Mo-cysteine dimer, Mo-glutathione dimer, MoCl6, MoCl4 dipyridine, and MoS2 (41-43). In contrast to iron, which is poorly available in oxic soils due to precipitation of iron oxides and iron hydroxides, the oxoanionic molybdate is highly soluble. Its negative electric charge prevents adsorption onto particle surfaces at neutral and basic pH resulting in the formation of weak complexes with most organic ligands with the exception of catechols (44). The catechol groups of organic matter are able to bind molybdate over a wide pH range retaining molybdenum in the top layer of the soil (45).
Many bacteria are known to produce and secrete siderophores. Siderophores are small, high-affinity FeIII-chelating compounds secreted to the extracellular medium by organisms subjected to low iron availability (46). Besides chelating FeIII, siderophores are able to chelate molybdate, vanadate, and tungstate. The genome of A. vinelandii encodes the genes for the production of five different siderophores: the mono-chatecols aminochelin and 2, 3-dihydroxybenzoic acid (DHBA), the bis-chatecol azotochelin, the tris-chatecol protochelin, and the pyoverdin-like azotobactin (47). Genes involved in the production and excretion of catechol siderophores, including csbC (Avin21220) (48), and csbX (Avin21230) (49) had been previously reported. The siderophore DHBA is the one most abundantly excreted by A. vinelandii, but has low affinity for metals and its participation in sequestering molybdenum is limited (50). Protochelin, also abundantly excreted, dominates the speciation of iron, molybdate, vanadate and tungstate present in the medium, whereas aminochelin and azotobactin become dominantly excreted at extremely low iron concentrations (48, 51). Catechol siderophores play a crucial role in trace metal nutrition by binding oxoanions of molybdenum, vanadium or tungsten present either in solution, or attached to naturally occurring ligands, or bound to organic matter (47, 52). In addition, catechol siderophores have been proposed to be involved in detoxification of those metals that are deleterious to cell metabolism (53).
(ii) Molybdate transport inside the cell
Bacteria scavenge molybdate from the environment by using high affinity ABC-type transport systems (54-58). These specialized Mo-uptake systems are required by bacteria to efficiently discriminate between molybdate and tungstate (53). Average soil tungstate concentrations are around 1 ppm, in the range of molybdate concentrations. Tungstate has similar structural properties to molybdate, but its uptake and incorporation into active-site cofactors yield inactive W-nitrogenase (59) and W-nitrate reductase (60) enzymes.
In addition to the high-affinity transporter specific for molybdate, the sulfate transporter has been suggested to serve as low affinity molybdate transport system, although this role has not been extensively characterized (33).
The high affinity ABC transporter is encoded by the modA, modB and modC genes present in the mod operon. The modA gene encodes a high-affinity molybdate-binding protein located in the periplasm; molybdate reaches the periplasm, presumably through porins, and then binds to ModA. The modB gene encodes an integral cytoplasmic membrane protein that provides the transport channel. The modC gene encodes the so-called conserved component, a membrane associated protein that binds and hydrolyzes Mg·ATP to provide energy for the active transport of molybdate. Unlike most microorganisms, A. vinelandii contains three copies of the modABC operon, revealing the importance and complexity of molybdenum metabolism in this microorganism. Even the closely related nitrogen-fixing strain Pseudomonas stutzeri contains a single copy of the modABC operon (61). The products of the A. vinelandii modA1B1C1 operon (Avin50670 to Avin50650) have been shown to be required for growth under conditions of molybdate limitation (58). The A. vinelandii genome sequence showed that the modA1B1C1 operon is located near the so-called minor nif cluster, which contains the nifB and nifQ genes among others. The second known mod operon, modA2B2C2 (Avin01300 to Avin01280), is located near the major nif operon. The genome sequence also revealed the existence of a third mod copy, modA3B3aB3bC3 (Avin50730 to Avin50700) located next to the modA1B1C1 operon (39).
In addition to the use of soluble molybdate ions, A. vinelandii cells are able to use up molybdate-siderophore complexes as molybdenum source (44). The mechanism of molybdate-siderophore utilization is not clear. Yet, on the basis of the known mechanism of iron transport systems, it has been suggested that siderophores transfer molybdate to the ModA protein located in the periplasm (47). Similarly, A. vinelandii cells are able to take up vanadate-siderophore complexes. On the other hand, the tungstate-siderophore complexes, which are deleterious to bacterial growth, are either poorly taken up (53) or not taken up at all (47).
(iii) Molybdenum cellular homeostasis
The A. vinelandii ModG protein has been proposed to be responsible for the homeostasis of molybdenum in the cytoplasm (58). The modG mutants exhibit pleiotropic effects in nitrate reductase and nitrogenase activities that suggest a role for ModG in balancing molybdenum availability for the biosyntheses of Mo-co and FeMo-co (58). The A. vinelandii ModG protein is encoded by the Avin50690 gene, which is clustered with and divergently transcribed from modEA1B1C1. Each ModG monomer consists of a tandem repeat of two 65-amino-acid-long Mop domains, a structural fold that specifically binds molybdate and discriminates against other oxoanions. In its native conformation, ModG is a trimer that binds up to eight molecules of molybdate (62).
Mop domains can be found as stand-alone structures or in combination with other types of domains as part of larger proteins, such as in ModA and ModE (see below). The Mop domains are widespread in bacteria and have been thoroughly studied in A. vinelandii, Clostridium pasteurianum, Escherichia coli and R. capsulatus (63, 64).
(iv) Molybdenum storage
The capacity of A. vinelandii to scavenge molybdate from the medium is remarkable. This bacterium can accumulate 25 times more molybdenum than it requires for maximum nitrogenase activity (42). The molybdenum storage protein (MoSto) is responsible for these high levels of molybdenum accumulation (65) and, to our knowledge, it has only been described in A. vinelandii.
The MoSto protein is an α3β3 hexamer of the mosA (Avin43200) and mosB (Avin43210) gene products that can store up to 100 molybdenum atoms per hexamer (66). MoSto can also incorporate tungsten. Metal storage within MoSto occurs in the form of a variety of compact polynuclear oxoanions (67). The incorporation of molybdate into MoSto is a nucleotide-dependent process (68). The release of molybdate from MoSto is, however, ATP-independent and appears to be pH-dependent and to occur stepwise, suggesting the involvement of several amino acid groups in the release mechanism (66, 69). No additional proteins seem to be required to load or unload molybdate from MoSto. Although MoSto expression is not controlled by nif regulatory factors (65), the level of molybdenum incorporation into MoSto increases in nitrogen fixing cells, consistent with a proposed role as a molybdenum reservoir destined to FeMo-co synthesis. Nitrogen fixation is a process requiring considerable amounts of molybdenum given that NifDK may represent up to 10% of the total protein content during diazotrophic growth.
(v) Molybdate-dependent gene regulation
The function and structure of the molybdate-responsive transcriptional regulator ModE has been well characterized in E. coli. ModE binds molybdate and represses the expression of modABC genes under molybdenum-replete growth conditions (70-72). In addition, ModE has been shown to coordinate expression of a number of molybdenum-dependent genes by repressing or activating their expression. The molybdate transporter genes, Mo-co biosynthesis genes (73), and several molybdoenzyme structural genes, such us the periplasmic and respiratory nitrate reductases (74, 75) or the dimethylsulfoxide reductase (76), are known to be regulated by ModE.
Each subunit of a ModE dimer consists of two Mop domains and a helix-turn-helix (HTH) DNA-binding domain (77, 78). Efficient DNA binding requires protein dimerization and molybdate binding; both functions are mediated by the Mop domains of ModE (78). Binding of molybdate to ModE drives extensive conformational changes in both the Mop domain and the DNA-binding domains that largely increase its affinity for specific operator sequences (72, 77, 79, 80).
ModE has been identified in a number of bacteria, including A. vinelandii (54). Moreover, ModE binding sequences have been found widespread in bacteria and archea (81). The A. vinelandii ModE is encoded by the Avin50680 gene located within the modEA1B1C1 operon. The modE mutant exhibits normal diazotrophic growth rates. However, A. vinelandii strains carrying mutations in both modE and modG genes are severely impaired in diazotrophic growth (58), indicating their involvement in nitrogenase biogenesis or regulation. The A. vinelandii genome carries an additional modE copy (Avin33430) adjacent to a V-nitrogenase transcriptional regulator vnfA2 (Avin33440). The role of this ModE homolog has not yet been characterized. The presence of putative ModE-binding sites upstream the vnfA2 and anfA genes, encoding regulatory proteins for the V-nitrogenase and the Fe-only-nitrogenase respectively, suggests that ModE coordinates the expression of all three nitrogenases in A. vinelandii in response to molybdenum availability (33).
(vi) FeMo-co biosynthesis
The biosynthesis of the complex FeMo-cofactor involves the activities of a large battery of nif and non-nif gene products, including proteins that exhibit catalytic activity, proteins that act as molecular scaffolds, and proteins which role is to carry and protect FeMo-co intermediates between the sequential assembly sites (82). The genes encoding proteins involved in FeMo-co biosynthesis in A. vinelandii are located within two chromosomal regions equidistant from the origin of replication denoted as the major nif region (Avin01360 to Avin01710) and the minor nif region (Avin50990 to Avin51060) (39, 83-85).
The model for the FeMo-co biosynthetic pathway depicted in Figure 3 illustrates the convergence of the FeMo-co building blocks towards a central catalytic node consisting of NifEN and NifH. The NifU/NifS/NifB/NifX branch of the pathway provides NifB-co, a FeMo-co biosynthetic intermediate proposed to comprise the [Fe6-S9-X] core of the cofactor (86). At this stage, molybdenum has not yet been incorporated into the precursor (87). The specific role of the cysteine desulfurase NifS and the scaffold protein NifU is to provide [Fe2-S2] and/or [Fe4-S4] clusters that would serve as metabolic substrates for NifB to synthesize NifB-co (88). The synthesis of NifB-co by NifB is a redox and S-adenosyl methionine-dependent process that involves radical chemistry (89). It has been proposed that very low-potential radical chemistry carried out by NifB is responsible for the incorporation of the X atom into NifB-co (89). Although not essential, the carrier protein NifX has been shown to optimize the transfer of NifB-co from NifB to NifEN (90). NifEN transforms NifB-co into the VK-cluster, which is proposed to be the next intermediate in the biosynthetic pathway (90). Similar to NifB-co, the VK-cluster does not contain molybdenum or homocitrate. However, these two intermediates are electronically and structurally different: while NifB-co is EPR silent (91), the VK-cluster exhibits EPR signals in the dithionite-reduced and the thionine-oxidized states (90). EXAFS and NRVS analyses indicate that NifB-co is an [Fe6-S9-X] cluster (86) whereas VK-cluster structures containing this core plus one or two additional terminal Fe atoms are favored (92).
FIGURE 3.
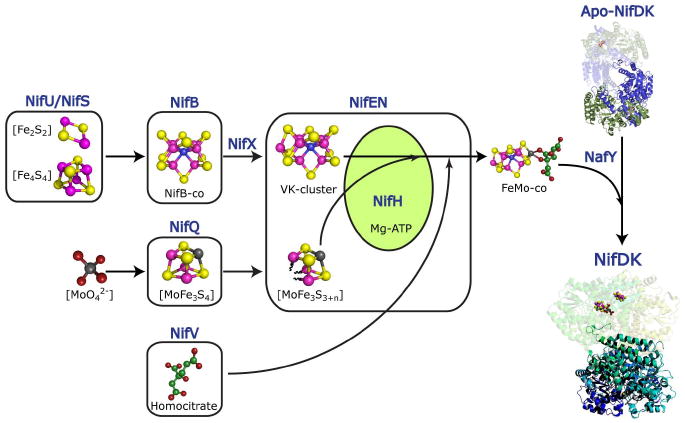
Schematic model of the FeMo-co biosynthetic pathway showing the convergence of FeMo-co biosynthetic precursors ([Fe6-S9-X], [MoFe3S4] and homocitrate) into a central node composed of the NifEN and NifH proteins. The [Fe-S] cluster biosynthetic branch involves the activities of NifS, NifU, NifB and NifX; the molybdenum branch involves the activity of NifQ; and the homocitrate is synthesized by NifV. The NifEN protein catalyzes the conversion of NifB-co into the VK-cluster, whereas the incorporation of molybdenum into FeMo-co – and probably that of homocitrate– requires the concerted activities of NifEN and NifH. Completed FeMo-co is then transferred by NafY to the cofactor-deficient apo-NifDK protein to generate active Mo-nitrogenase.
In the second branch, NifQ specifically donates molybdenum to the NifEN/NifH proteins (93). The molybdenum of NifQ is present in an [Fe-S] cluster environment (see below). It is currently unknown whether NifQ transfers its entire [MoFe3S4] cluster, a portion of the cluster or simply the Mo atom. The third branch of the pathway provides homocitrate synthesized by the condensation of acetyl coenzyme A and α-ketoglutarate, a reaction catalyzed by NifV (94).
The putative NifEN/NifH complex would integrate the building components provided by the three biosynthetic branches to complete the synthesis of FeMo-co in a redox-dependent reaction or series of reactions that also require Mg·ATP. The newly synthesized FeMo-cofactor is transferred to the cofactor deficient apo-NifDK protein to generate catalytically active NifDK. The transfer process is mediated by the nitrogenase accessory factor NafY, a non-nif metallochaperone that is thought to protect FeMo-co from degradation and to stabilize the FeMo-co deficient apo-NifDK (95, 96).
(vii) Molybdenum in NifQ
The nifQ gene product is an iron-sulfur protein known by genetic evidence to be involved in the incorporation of molybdenum into nitrogenase. NifQ is present in A. vinelandii cells grown in the presence of molybdenum and the absence of ammonium. At concentrations of molybdate in the medium around the nM range, nifQ mutants of A. vinelandii are impaired in molybdenum-dependent nitrogen fixation (97). It is significant that nifQ mutants are not defective in the activities of other molybdoenzymes (98) or the alternative V- or Fe-only-nitrogenases (85). Although nifQ mutant strains are not defective in molybdate uptake, it has been observed that they accumulate lower levels of molybdenum than the wild-type strain. The nifQ phenotype is suppressed by increasing the molybdate concentrations to μM levels or by adding excess cysteine to the medium, suggesting that the reaction catalyzed by NifQ might also occur non-enzymatically when the levels of the reactants are high. Similar nifQ phenotype and suppression profiles have been observed in K. pneumoniae (99, 100).
All nitrogen-fixing Proteobacteria –except some species of Rhizobia- contain NifQ homologues. Amino acid sequence alignments of NifQ proteins show a conserved Cx4Cx2Cx5C motif at the C-terminus of the protein that could be capable of coordinating the [Fe-S] cluster of NifQ. The native molecular weight of A. vinelandii NifQ (25.7 kDa) is similar to the weight deduced from the nifQ sequence (19.7 kDa), indicating a monomeric structure and precluding the possibility of having one [Fe-S] cluster coordinated by two NifQ subunits (93).
Recent biochemical and EPR spectroscopic evidence has shown that NifQ carries a metal cluster comprising a [MoFe3S4] core, and that the presence of this metal cluster in NifQ is correlated with its ability to support in vitro FeMo-co synthesis (93). As-isolated NifQ exhibits EPR properties similar to those of [Fe3-S4]+-containing proteins, whereas the EPR signals of reduced NifQ resemble those of [MoFe3S4] metal clusters prepared synthetically. Metal analysis, and the observation that [Fe3-S4] to [MoFe3S4] cluster conversion could be achieved in vitro by incubating NifQ with molybdate and sulfide under reducing conditions, confirmed that each NifQ molecule contains a single [Fe-S] cluster with the capability to carry a Mo atom.
Molybdate must undergo at least three chemical transformations before being incorporated into FeMo-co: replacement of O-ligands by S ligands, reduction of molybdenum from the MoVI oxidation state to the MoIV state found in the cofactor (101, 102), and insertion of molybdenum into an [Fe-S] cluster environment. The [Fe3S4] cluster observed in as-isolated NifQ could be the site for molybdenum binding and reduction. The reduced [Fe3-S4]0 clusters are known to be able to coordinate heterometals to complete [MFe3-S4] clusters in reversible equilibria (103). An attractive hypothesis is that NifQ would have its role in molybdenum trafficking through a cyclic process of molybdenum binding to the [Fe3S4] cluster by reductive coupling to molybdate and molybdenum releasing from the [MoFe3S4] cluster in response to an oxidation event (Figure 4). Consistently, EXAFS analysis strongly suggests that the molybdenum within NifQ is in the (IV) oxidation state (George, Hernandez and Rubio, unpublished results).
FIGURE 4.
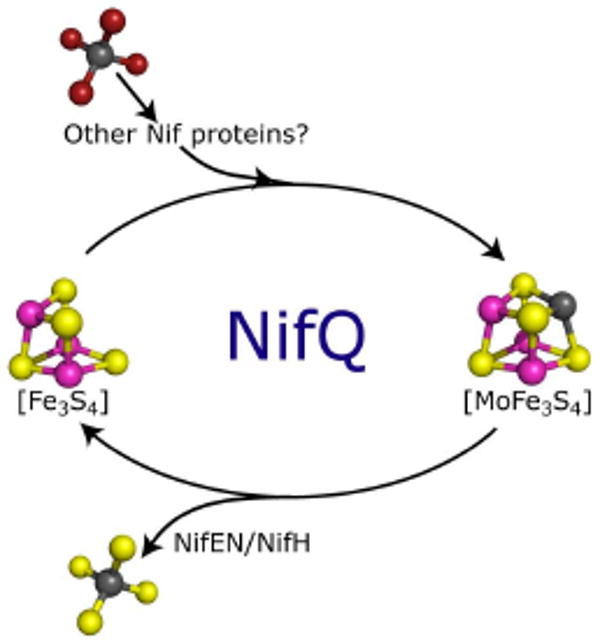
Hypothetical model for a redox-dependent cycle of cluster interconversion in NifQ. A [MoFe3S4] structure based on recent spectroscopy data was used for this model (93). The [Fe3-S4] cluster of NifQ serves as molybdenum binding site. Substitution of Mo-S for Mo-O bonds and reduction of molybdate would be required; other Nif proteins might assist NifQ during this process in vivo. The presence of NifEN/NifH promotes the release of molybdenum from NifQ. A sulfur donor might be required to release Mo from the [MoFe3S4] cluster. Atom colors: iron, magenta; sulfur, yellow; molybdenum, grey; oxygen, red.
The [MoFe3S4]-loaded form of NifQ serves as specific molybdenum donor for the NifEN/NifH proteins during FeMo-co biosynthesis. A puzzling fact is that molybdate too can serve as a Mo source for FeMo-co biosynthesis in vitro. However, genetic evidence indicates that NifQ has an essential role when the levels of available molybdenum are in a physiological range. Mobilization of molybdenum from NifQ requires the simultaneous participation of NifH and NifEN, suggesting that NifQ would be the physiological molybdenum donor to a putative NifEN/NifH complex.
(viii) Molybdenum in NifEN and NifH
The NifEN protein, as isolated from a strain lacking nifH, contains substoichiometric amounts of molybdenum (104). The lack of NifH impedes the final steps of FeMo-co biosynthesis causing accumulation of biosynthetic intermediates within the NifEN scaffold. The presence of molybdenum in NifEN has been a rather contentious finding because it appears to depend on the purification method used to isolate this protein (104, 105). However, two independent observations support that the molybdenum within NifEN is indeed relevant to FeMo-co synthesis: (i) the purified preparations of NifEN served as a sole molybdenum source for the in vitro FeMo-co biosynthesis assay in a defined reaction mixture containing only purified components (104). (ii) The EXAFS analysis showed that molybdenum in NifEN is part of a [MoFe3S3+n] cluster but not adventitiously bound molybdate (106).
Although the quality of EXAFS data was not high enough as to discard the possibility of the [MoFe3S3+n] being part of a larger [Mo-Fe-S] cluster, the lack of apparent long-range (5Å) metal interactions supports the hypothesis that this cluster is in fact a [MoFe3S4] cubane. In such a case, this cluster would be very similar to the one carried by NifQ, and it is possible to envision at least three biosynthetic origins. First, NifEN contains a “receiving” [Fe3S4] cluster that accepts a reduced Mo atom or a Mo-S molecule extracted from the [MoFe3S4] cluster of NifQ, which would act as a donor. This would be a redox dependent metal transfer similar to the ones reported to occur in reduced [Fe3-S4]0 clusters (103). The observation of EPR signals that show similarity to signals from [Fe3S4]+-containing proteins in preparations of NifEN lacking molybdenum is consistent with this scenario (105, 107). Second, the complete [MoFe3S4] cluster is transferred from NifQ to NifEN. Transfer of complete [Fe4S4] clusters from scaffold proteins to target apo-proteins is widespread in nature (108). Third, the NifEN [MoFe3S4] cluster could have its origin in the reductive coupling of molybdate into a “receiving” [Fe3S4] cluster in a NifQ-independent pathway. It is important to note that the NifEN was isolated from a strain that contains NifQ but lacks NifH, and that molybdenum transfer from NifQ to NifEN is not efficient in the absence of NifH (93). This pathway could be responsible for the suppression of the nifQ mutant phenotype observed by increasing the concentration of molybdate or cysteine in the medium (99, 100). In any case, the [MoFe3S3+n] cluster of NifEN has been shown to be a molybdenum donor for the VK-cluster, which is the other FeMo-co biosynthetic intermediate carried by NifEN (104).
NifH too has been proposed to serve as the entry point for molybdenum incorporation into the FeMo-co biosynthetic pathway. 99Mo radiolabeling experiments showed the incorporation of 99Mo into NifH in FeMo-co biosynthesis reaction mixtures containing purified NifEN, NifH, NifB-co and Mg·ATP (109). However, it should be noted that incorporation of 99Mo occurred also in NifEN, and that the presence of all components (NifEN, NifH, NifB-co and Mg·ATP) were required for 99Mo incorporation into both NifEN and NifH. Similar results were observed by EXAFS analysis of FeMo-co precursors associated with NifH after in vitro FeMo-co biosynthesis reactions (110). Since the presence of the [MoFe3S3+n] cluster of NifEN is not absolutely dependent on NifH (104), a possible explanation for the role of NifH would be that its activity is required to mobilize molybdenum from the [MoFe3S3+n] cluster into the NifB-co-derived VK-cluster within the NifEN protein thus generating a molybdenum-containing FeMo-co biosynthetic intermediate.
Perspectives
The finding that the [Fe-S] cluster of NifQ was involved in the sequestration and delivery of molybdenum during FeMo-co synthesis has triggered many questions (111). Perhaps, the most intriguing one is how would NifQ exert all the biochemical transformations upon molybdenum that are required for such trafficking. A molybdate-reducing system that replaces Mo-O bonds by Mo-S bonds, reduces molybdenum, and inserts the heterometal into the [Fe3-S4] cluster of NifQ must exist in order to generate a [MoFe3S4] cluster. We hypothesize that NifQ is the central protein of an enzymatic system with molybdate reductase activity. Adjacent to nifQ in the minor nif region are genes encoding a ferredoxin (Avin51020), NifO (Avin51030), a putative rhodanase (Avin51050), and a monothiol glutaredoxin (Avin51060).
Some glutaredoxins have been described to be involved in [Fe-S] cluster assembly (112). Thus, it is possible that the nif glutaredoxin has a role in the formation of the [Fe3-S4] cluster of NifQ, which serves as scaffold for molybdenum binding. Rhodanases have sulfur transferase activity and have been shown to direct the reconstitution of [Fe-S] clusters in ferredoxins and NifH (113, 114). Thus, it is possible that the nif rhodanase could be involved in the synthesis of the [Fe3-S4] cluster of NifQ. The generation of the [MoFe3S4] cluster could be achieved by reductive coupling of molybdate to the [Fe3-S4] cluster. The nif-specific ferredoxin could be involved in electron donation to reduce the [Fe3-S4] cluster of NifQ. Finally, the NifO protein is similar to the arsenate reductase ArsC, which uses reduced glutathione to convert arsenate to arsenite. Whether or not NifO could be involved in molybdate reduction remains open.
Table 1.
A. vinelandii genes, described in this review, related to molybdenum metabolism and nitrogen fixation.
| Gene number | Annotation | Function |
|---|---|---|
| Binding of extracellular molybdenum | ||
| Avin21220 | csbC | Isochorismate synthase |
| Avin21230 | csbX | Efflux pump |
| Molybdate transport inside the cell | ||
| Avin50670 | modA1 | Periplasmic molybdate-binding protein |
| Avin50660 | modB1 | Membrane channel protein |
| Avin50650 | modC1 | Cytoplasmic ATPase |
| Avin01300 | modA2 | Periplasmic molybdate-binding protein |
| Avin01290 | modB2 | Membrane channel protein |
| Avin01280 | modC2 | Cytoplasmic ATPase |
| Avin50730 | modA3 | Periplasmic molybdate-binding protein |
| Avin50720 | modB3a | Membrane channel protein |
| Avin50710 | modB3b | Membrane channel protein |
| Avin50700 | modC3 | Cytoplasmic ATPase |
| Molybdenum cellular homeostasis | ||
| Avin50690 | modG | Molybdenum homeostasis |
| Molybdenum storage | ||
| Avin43200 | mosA | Subunit of the Mo-storage protein |
| Avin43210 | mosB | Subunit of the Mo-storage protein |
| Molybdate-dependent gene regulation | ||
| Avin50680 | modE | Mo-responsive transcriptional regulator |
| Avin33430 | modE copy | Mo-responsive transcriptional regulator |
| Avin33440 | vnfA2 | Transcriptional activator of the vnf genes |
| FeMo-co biosynthesis and nitrogen fixation | ||
| Avin01360 to Avin01710 | Major nif | |
| Avin01380, Avin01390, Avin01400 | nifHDK | Mo-nitrogenase structural genes |
| Avin01450, Avin01470, Avin01480, Avin01640 | nifENXV | FeMo-co biosynthesis |
| Avin01620, Avin01630, | nifUS | Fe-S cluster biosynthesis |
| Avin50990 to Avin51060 | Minor nif | |
| Avin50910 | nafY | FeMo-co insertion and apo-NifDK stability |
| Avin51000, Avin50990 | nifAL | Transcriptional regulation of nif genes |
| Avin51010 | nifB | FeMo-co biosynthesis |
| Avin51020 | fdxN | Ferredoxin |
| Avin51030 | nifO | Unknown function |
| Avin51040 | nifQ | Incorporation of Mo into FeMo-co |
| Avin51050 | rhdN | Putative rhodanase |
| Avin51060 | grx5nif | Monothiol glutaredoxin |
Acknowledgments
We thank Carlos Echavarri-Erasun for helping in figure preparation.
Footnotes
This work was supported by ERC Starting Grant 205442 (L.M.R.), by Midwestern University Intramural Funds (J.A.H.), and by NIH GM-65440 grant (S.J.G.).
FeMo-co, iron-molybdenum cofactor; FeV-co, iron-vanadium cofactor; FeFe-co, iron-only cofactor; Mo-co, molybdenum cofactor; NifB-co, NifB-cofactor; VK-cluster, Vinod K. Shah cluster; nif, genes encoding proteins involved in nitrogen fixation; EPR, electron paramagnetic resonance; EXAFS, extended X-ray absorption fine spectroscopy; ENDOR, electron nuclear double resonance.
The NifDK component of nitrogenase is also referred to as dinitrogenase, component I, or MoFe protein. The NifH component is also referred to as dinitrogenase reductase, component II, or Fe protein.
References
- 1.Pau RN, Lawson DM. Molydenum and tungsten: their roles in biological processes. In: Sigel A, Sigel H, editors. Metal ions in biological systems. Marcel Dekker Inc.; New York: 2002. pp. 31–74. [Google Scholar]
- 2.Stiefel EI. Molybdenum and tungsten: their roles in biological processes. In: Sigel A, Sigel H, editors. Metal ions in biological systems. Marcel Dekker Inc.; New York: 2002. pp. 1–30. [Google Scholar]
- 3.Wedepohl KH. The composition of the continental crust. Geochim Cosmochim Acta. 1995;59:1217–1232. [Google Scholar]
- 4.Pope MT, Steele ER, Williams RJP. The comparison between the chemistry and biochemistry of molybdenum and related elements. In: Coughlan MP, editor. Molybdenum and Molybdenum-containing enzymes. Pergamon Press; Oxford: 1980. pp. 3–40. [Google Scholar]
- 5.Barron AR, Wurzburger N, Bellenger JP, Wright SJ, Kraepiel AML, Hedin LO. Molybdenum limitation of asymbiotic nitrogen fixation in tropical forest soils. Nat Geosci. 2009;2:42–45. [Google Scholar]
- 6.Hungate BA, Stiling PD, Dijkstra P, Johnson DW, Ketterer ME, Hymus GJ, Hinkle CR, Drake BG. CO2 elicits long-term decline in nitrogen fixation. Science. 2004;304:1291. doi: 10.1126/science.1095549. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Gladyshev VN. Molybdoproteomes and evolution of molybdenum utilization. J Mol Biol. 2008;379:881–899. doi: 10.1016/j.jmb.2008.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotton FA, Wilkinson G, Murillo CA, Bochmann M. Advanced Inorganic Chemistry. 6th. Wiley-Interscience; 1999. pp. 920–973. [Google Scholar]
- 9.Stiefel EI. Transition metal sulfur chemistry and its relevance to molybdenum and tungsten enzymes. Pure Appl Chem. 1998;70:889–896. [Google Scholar]
- 10.Stiefel EI, Halbert TR, Coyle CL, Wei L, Pan WH, Ho TC, Chianelli RR, Daage M. Molecules, clusters, solids and catalysts in early transition-metal sulfide systems. Polyhedron. 1989;8:1625–1629. [Google Scholar]
- 11.Enemark JH, Cooney JJA. Synthetic analogues and reaction systems relevant to the molybdenum and tungsten oxotransferases. Chem Rev. 2004;104:1175–1200. doi: 10.1021/cr020609d. [DOI] [PubMed] [Google Scholar]
- 12.Lee CC, Halbert TR, Pan WH, Harmer MA, Wei L, Leonowicz ME, Dim COB, Miller KF, Bruce AE, McKenna S, Corbin JL, Wherland S, Stiefel EI. Synthesis, reactivity and redox properties of dinuclear molybdenum-sulfur complexes. Inorganica Chim Acta. 1996;243:147–160. [Google Scholar]
- 13.Hernandez-Molina R, Sykes AG. Chalcogenide-bridged cuboidal clusters with M(4)Q(4) (M = Mo, W; Q = S, Se, Te) cores. Dalton Trans. 1999:3137–3148. [Google Scholar]
- 14.Lee SC, Holm RH. The clusters of nitrogenase: Synthetic methodology in the construction of weak-field clusters. Chem Rev. 2004;104:1135–1157. doi: 10.1021/cr0206216. [DOI] [PubMed] [Google Scholar]
- 15.George GN, Bray RC. Studies by electron-paramagnetic resonance spectroscopy of xanthine-oxidase enriched with Mo-95 and with Mo-97. Biochemistry. 1988;27:3603–3609. doi: 10.1021/bi00410a011. [DOI] [PubMed] [Google Scholar]
- 16.Hille R. The mononuclear molybdenum enzymes. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 17.Lowe DJ, Eady RR, Thorneley RNF. Electron-paramagnetic-resonance studies on nitrogenase of Klebsiella pneumoniae: evidence for acetylene-nitrogenase and ethylene-nitrogenase transient complexes. Biochem J. 1978;173:277–290. doi: 10.1042/bj1730277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howes BD, Bray RC, Richards RL, Turner NA, Bennett B, Lowe DJ. Evidence favoring molybdenum-carbon bond formation in xanthine oxidase action: O-17 and C-13-ENDOR and kinetic studies. Biochemistry. 1996;35:1432–1443. doi: 10.1021/bi9520500. [DOI] [PubMed] [Google Scholar]
- 19.Lukoyanov D, Pelmenschikov V, Maeser N, Laryukhin M, Yang TC, Noodleman L, Dean DR, Case DA, Seefeldt LC, Hoffman BM. Testing if the interstitial atom, X, of the nitrogenase molybdenum-iron cofactor is N or C: ENDOR, ESEEM, and DFT studies of the S=3/2 resting state in multiple environments. Inorg Chem. 2007;46:11437–11449. doi: 10.1021/ic7018814. [DOI] [PubMed] [Google Scholar]
- 20.Christiansen J, Tittsworth RC, Hales BJ, Cramer SP. Fe and Mo EXAFS of Azotobacter vinelandii nitrogenase in partially oxidized and singly reduced forms. J Am Chem Soc. 1995;117:10017–10024. [Google Scholar]
- 21.Cramer SP, Hodgson KO, Gillum WO, Mortenson LE. Molybdenum site of nitrogenase: preliminary structural evidence from X-ray absorption spectroscopy. J Am Chem Soc. 1978;100:3398–3407. [Google Scholar]
- 22.George GN, Kipke CA, Prince RC, Sunde RA, Enemark JH, Cramer SP. Structure of the active-site of sulfite oxidase: X-ray absorption spectroscopy of the Mo(IV), Mo(V), and Mo(VI) oxidation states. Biochemistry. 1989;28:5075–5080. doi: 10.1021/bi00438a026. [DOI] [PubMed] [Google Scholar]
- 23.Harris HH, George GN, Rajagopalan KV. High-resolution EXAFS of the active site of human sulfite oxidase: Comparison with density functional theory and X-ray crystallographic results. Inorg Chem. 2006;45:493–495. doi: 10.1021/ic0512274. [DOI] [PubMed] [Google Scholar]
- 24.Hille R. Structure and function of mononuclear molybdenum enzymes. J Biol Inorg Chem. 1996;1:397–404. [Google Scholar]
- 25.Romao MJ. Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans. 2009:4053–4068. doi: 10.1039/b821108f. [DOI] [PubMed] [Google Scholar]
- 26.Romao MJ, Archer M, Moura I, Moura JJG, Legall J, Engh R, Schneider M, Hof P, Huber R. Crystal structure of the xanthine oxidase-related aldehyde oxidoreductase from D. gigas. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- 27.Chan MK, Kim J, Rees DC. The nitrogenase FeMo-cofactor and P-cluster pair: 2.2 A resolution structures. Science. 1993;260:792–794. doi: 10.1126/science.8484118. [DOI] [PubMed] [Google Scholar]
- 28.Hoover TR, Robertson AD, Cerny RL, Hayes RN, Imperial J, Shah VK, Ludden PW. Identification of the V factor needed for synthesis of the iron-molybdenum cofactor of nitrogenase as homocitrate. Nature. 1987;329:855–857. doi: 10.1038/329855a0. [DOI] [PubMed] [Google Scholar]
- 29.Mendel RR, Bittner F. Cell biology of molybdenum. Biochim Biophys Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Falkowski PG. Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature. 1997;387:272–275. [Google Scholar]
- 31.Seefeldt LC, Hoffman BM, Dean DR. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eady RR. Structure-function relationships of alternative nitrogenases. Chem Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 33.Pau RN. Molybdenum uptake and homeostasis. In: Klipp W, Masepohl B, Gallon JR, Newton WE, editors. Genetics and regulation of nitrogen fixation in free-living bacteria. Kluwer Academic Publishers; Netherlands: 2004. pp. 225–256. [Google Scholar]
- 34.Shah VK, Brill WJ. Isolation of an iron-molybdenum cofactor from nitrogenase. Proc Natl Acad Sci USA. 1977;74:3249–3253. doi: 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Einsle O, Tezcan FA, Andrade SL, Schmid B, Yoshida M, Howard JB, Rees DC. Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 36.Seefeldt LC, Dance IG, Dean DR. Substrate interactions with nitrogenase: Fe versus Mo. Biochemistry. 2004;43:1401–1409. doi: 10.1021/bi036038g. [DOI] [PubMed] [Google Scholar]
- 37.Pickett CJ. The Chatt cycle and the mechanism of enzymic reduction of molecular nitrogen. J Biol Inorg Chem. 1996;1:601–606. [Google Scholar]
- 38.Ribbe M, Gadkari D, Meyer O. N2 fixation by Streptomyces thermoautotrophicus involves a molybdenum-dinitrogenase and a manganese-superoxide oxidoreductase that couple N2 reduction to the oxidation of superoxide produced from O2 by a molybdenum-CO dehydrogenase. J Biol Chem. 1997;272:26627–26633. doi: 10.1074/jbc.272.42.26627. [DOI] [PubMed] [Google Scholar]
- 39.Setubal JC, dos Santos P, Goldman BS, Ertesvag H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du Z, Godsy E, Goodner B, Hellner-Burris K, Hernandez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O'Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang B, Wheeler C, Zhu H, Dean DR, Dixon R, Wood D. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol. 2009;191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lepp NW, Edwards R, Jones KC. Other less abundant elements of potential environment significance. In: Alloway BJ, editor. Heavy metals in soils. 2nd. Blackie & Professional; London: 1995. pp. 306–352. [Google Scholar]
- 41.Schrauzer GN, Schlesinger G. Chemical evolution of a nitrogenase model. I. Reduction of acetylene and other substrates by a molybdenum-thiol catalyst system. J Am Chem Soc. 1970;92:1808–1809. [Google Scholar]
- 42.Shah VK, Ugalde RA, Imperial J, Brill WJ. Molybdenum in nitrogenase. Annu Rev Biochem. 1984;53:231–257. doi: 10.1146/annurev.bi.53.070184.001311. [DOI] [PubMed] [Google Scholar]
- 43.Werner D, Russell SA, Evans HJ. Reduction of acetylene and hydrazine with a molybdenum-glutathione complex. Proc Natl Acad Sci USA. 1973;70:339–342. doi: 10.1073/pnas.70.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellenger JP, Wichard T, Kustka AB, Kraepiel AM. Nitrogen fixing soil bacterium uses catechol siderophores for molybdenum and vanadium acquisition. Nat Geosci. 2008;1:243–246. [Google Scholar]
- 45.Wichard T, Mishra B, Kraepiel AML, Myneni SCB. Molybdenum speciation and bioavailability in soils. Geochim Cosmochim Acta. 2008;72:A1019. [Google Scholar]
- 46.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 47.Kraepiel AM, Bellenger JP, Wichard T, Morel FM. Multiple roles of siderophores in free-living nitrogen-fixing bacteria. Biometals. 2009;22:573–581. doi: 10.1007/s10534-009-9222-7. [DOI] [PubMed] [Google Scholar]
- 48.Tindale AE, Mehrotra M, Ottem D, Page WJ. Dual regulation of catecholate siderophore biosynthesis in Azotobacter vinelandii by iron and oxidative stress. Microbiology. 2000;146:1617–1626. doi: 10.1099/00221287-146-7-1617. [DOI] [PubMed] [Google Scholar]
- 49.Page WJ, Kwon E, Cornish AS, Tindale AE. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol Lett. 2003;228:211–216. doi: 10.1016/S0378-1097(03)00753-5. [DOI] [PubMed] [Google Scholar]
- 50.Neilands JB. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 51.Page WJ, Von Tigerstom T. Aminochelin, a catecholamine siderophore produced by Azotobacter vinelandii. J Gen Microbiol. 1988;134:453–460. [Google Scholar]
- 52.Liermann LJ, Guynn RL, Anbar A, Brantley SL. Production of a molybdophore during metal-targeted dissolution of silicates by soil bacteria. Chem Geol. 2005;220:285–302. [Google Scholar]
- 53.Wichard T, Bellenger JP, Loison A, Kraepiel AM. Catechol siderophores control tungsten uptake and toxicity in the nitrogen-fixing bacterium Azotobacter vinelandii. Environ Sci Technol. 2008;42:2408–2413. doi: 10.1021/es702651f. [DOI] [PubMed] [Google Scholar]
- 54.Grunden AM, Shanmugam KT. Molybdate transport and regulation in bacteria. Arch Microbiol. 1997;168:345–354. doi: 10.1007/s002030050508. [DOI] [PubMed] [Google Scholar]
- 55.Self WT, Grunden AM, Hasona A, Shanmugam KT. Molybdate transport. Res Microbiol. 2001;152:311–321. doi: 10.1016/s0923-2508(01)01202-5. [DOI] [PubMed] [Google Scholar]
- 56.Hu Y, Rech S, Gunsalus RP, Rees DC. Crystal structure of the molybdate binding protein ModA. Nat Struct Biol. 1997;4:703–707. doi: 10.1038/nsb0997-703. [DOI] [PubMed] [Google Scholar]
- 57.Maupin-Furlow JA, Rosentel JK, Lee JH, Deppenmeier U, Gunsalus RP, Shanmugam KT. Genetic analysis of the modABCD (molybdate transport) operon of Escherichia coli. J Bacteriol. 1995;177:4851–4856. doi: 10.1128/jb.177.17.4851-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mouncey NJ, Mitchenall LA, Pau RN. Mutational analysis of genes of the mod locus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J Bacteriol. 1995;177:5294–5302. doi: 10.1128/jb.177.18.5294-5302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siemann S, Schneider K, Oley M, Muller A. Characterization of a tungsten-substituted nitrogenase isolated from Rhodobacter capsulatus. Biochemistry. 2003;42:3846–3857. doi: 10.1021/bi0270790. [DOI] [PubMed] [Google Scholar]
- 60.Paneque A, Vega JM, Cárdenas J, Herrera J, Aparicio PJ, Losada M. 185W-labelled nitrate reductase from Chlorella. Plant Cell Physiol. 1972;13:175–178. [Google Scholar]
- 61.Yan Y, Yang J, Dou Y, Chen M, Ping S, Peng J, Lu W, Zhang W, Yao Z, Li H, Liu W, He S, Geng L, Zhang X, Yang F, Yu H, Zhan Y, Li D, Lin Z, Wang Y, Elmerich C, Lin M, Jin Q. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA. 2008;105:7564–7569. doi: 10.1073/pnas.0801093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delarbre L, Stevenson CE, White DJ, Mitchenall LA, Pau RN, Lawson DM. Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. J Mol Biol. 2001;308:1063–1079. doi: 10.1006/jmbi.2001.4636. [DOI] [PubMed] [Google Scholar]
- 63.Duhme AK, Meyer-Klaucke W, White DJ, Delarbre L, Mitchenall LA, Pau RN. Extended X-ray absorption fine structure studies on periplasmic and intracellular molybdenum-binding proteins. J Biol Inorg Chem. 1999;4:588–592. doi: 10.1007/s007750050381. [DOI] [PubMed] [Google Scholar]
- 64.Wang G, Angermuller S, Klipp W. Characterization of Rhodobacter capsulatus genes encoding a molybdenum transport system and putative molybdenum-pterin-binding proteins. J Bacteriol. 1993;175:3031–3042. doi: 10.1128/jb.175.10.3031-3042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pienkos PT, Brill WJ. Molybdenum accumulation and storage in Klebsiella pneumoniae and Azotobacter vinelandii. J Bacteriol. 1981;145:743–751. doi: 10.1128/jb.145.2.743-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fenske D, Gnida M, Schneider K, Meyer-Klaucke W, Schemberg J, Henschel V, Meyer AK, Knochel A, Muller A. A new type of metalloprotein: The Mo storage protein from Azotobacter vinelandii contains a polynuclear molybdenum-oxide cluster. Chembiochem. 2005;6:405–413. doi: 10.1002/cbic.200400263. [DOI] [PubMed] [Google Scholar]
- 67.Schemberg J, Schneider K, Demmer U, Warkentin E, Muller A, Ermler U. Towards biological supramolecular chemistry: a variety of pocket-templated, individual metal oxide cluster nucleations in the cavity of a Mo/W-storage protein. Angew Chem Int Ed Engl. 2007;46:2408–2413. doi: 10.1002/anie.200604858. [DOI] [PubMed] [Google Scholar]
- 68.Allen RM, Roll JT, Rangaraj P, Shah VK, Roberts GP, Ludden PW. Incorporation of molybdenum into the iron-molybdenum cofactor of nitrogenase. J Biol Chem. 1999;274:15869–15874. doi: 10.1074/jbc.274.22.15869. [DOI] [PubMed] [Google Scholar]
- 69.Schemberg J, Schneider K, Fenske D, Muller A. Azotobacter vinelandii metal storage protein: “classical” inorganic chemistry involved in Mo/W uptake and release processes. Chembiochem. 2008;9:595–602. doi: 10.1002/cbic.200700446. [DOI] [PubMed] [Google Scholar]
- 70.Grunden AM, Ray RM, Rosentel JK, Healy FG, Shanmugam KT. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol. 1996;178:735–744. doi: 10.1128/jb.178.3.735-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rech S, Deppenmeier U, Gunsalus RP. Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J Bacteriol. 1995;177:1023–1029. doi: 10.1128/jb.177.4.1023-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grunden AM, Self WT, Villain M, Blalock JE, Shanmugam KT. An analysis of the binding of repressor protein ModE to modABCD (molybdate transport) operator/promoter DNA of Escherichia coli. J Biol Chem. 1999;274:24308–24315. doi: 10.1074/jbc.274.34.24308. [DOI] [PubMed] [Google Scholar]
- 73.McNicholas PM, Rech SA, Gunsalus RP. Characterization of the ModE DNA-binding sites in the control regions of modABCD and moaABCDE of Escherichia coli. Mol Microbiol. 1997;23:515–524. doi: 10.1046/j.1365-2958.1997.d01-1864.x. [DOI] [PubMed] [Google Scholar]
- 74.Hasona A, Self WT, Ray RM, Shanmugam KT. Molybdate-dependent transcription of hyc and nar operons of Escherichia coli requires MoeA protein and ModE-molybdate. FEMS Microbiol Lett. 1998;169:111–116. doi: 10.1111/j.1574-6968.1998.tb13306.x. [DOI] [PubMed] [Google Scholar]
- 75.McNicholas PM, Gunsalus RP. The molybdate-responsive Escherichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability. J Bacteriol. 2002;184:3253–3259. doi: 10.1128/JB.184.12.3253-3259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNicholas PM, Chiang RC, Gunsalus RP. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol Microbiol. 1998;27:197–208. doi: 10.1046/j.1365-2958.1998.00675.x. [DOI] [PubMed] [Google Scholar]
- 77.Hall DR, Gourley DG, Leonard GA, Duke EM, Anderson LA, Boxer DH, Hunter WN. The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds. EMBO J. 1999;18:1435–1446. doi: 10.1093/emboj/18.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McNicholas PM, Mazzotta MM, Rech SA, Gunsalus RP. Functional dissection of the molybdate-responsive transcription regulator, ModE, from Escherichia coli. J Bacteriol. 1998;180:4638–4643. doi: 10.1128/jb.180.17.4638-4643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gourley DG, Schuttelkopf AW, Anderson LA, Price NC, Boxer DH, Hunter WN. Oxyanion binding alters conformation and quaternary structure of the C-terminal domain of the transcriptional regulator mode. Implications for molybdate-dependent regulation, signaling, storage, and transport. J Biol Chem. 2001;276:20641–20647. doi: 10.1074/jbc.M100919200. [DOI] [PubMed] [Google Scholar]
- 80.Schuttelkopf AW, Boxer DH, Hunter WN. Crystal structure of activated ModE reveals conformational changes involving both oxyanion and DNA-binding domains. J Mol Biol. 2003;326:761–767. doi: 10.1016/s0022-2836(02)01358-x. [DOI] [PubMed] [Google Scholar]
- 81.Studholme DJ, Pau RN. A DNA element recognised by the molybdenum-responsive transcription factor ModE is conserved in Proteobacteria, green sulphur bacteria and Archaea. BMC Microbiol. 2003;3:24. doi: 10.1186/1471-2180-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rubio LM, Ludden PW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 83.Jacobson MR, Brigle KE, Bennett LT, Setterquist RA, Wilson MS, Cash VL, Beynon J, Newton WE, Dean DR. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J Bacteriol. 1989;171:1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 85.Joerger RD, Bishop PE. Nucleotide sequence and genetic analysis of the nifB-nifQ region from Azotobacter vinelandii. J Bacteriol. 1988;170:1475–1487. doi: 10.1128/jb.170.4.1475-1487.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.George SJ, Igarashi RY, Xiao Y, Hernandez JA, Demuez M, Zhao D, Yoda Y, Ludden PW, Rubio LM, Cramer SP. Extended X-ray absorption fine structure and nuclear resonance vibrational spectroscopy reveal that NifB-co, a FeMo-co precursor, comprises a 6Fe core with an interstitial light atom. J Am Chem Soc. 2008;130:5673–5680. doi: 10.1021/ja0755358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allen RM, Chatterjee R, Ludden PW, Shah VK. Incorporation of iron and sulfur from NifB cofactor into the iron-molybdenum cofactor of dinitrogenase. J Biol Chem. 1995;270:26890–26896. doi: 10.1074/jbc.270.45.26890. [DOI] [PubMed] [Google Scholar]
- 88.Zhao D, Curatti L, Rubio LM. Evidence for nifU and nifS participation in the biosynthesis of the iron-molybdenum cofactor of nitrogenase. J Biol Chem. 2007;282:37016–37025. doi: 10.1074/jbc.M708097200. [DOI] [PubMed] [Google Scholar]
- 89.Curatti L, Ludden PW, Rubio LM. NifB-dependent in vitro synthesis of the iron-molybdenum cofactor of nitrogenase. Proc Natl Acad Sci USA. 2006;103:5297–5301. doi: 10.1073/pnas.0601115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hernandez JA, Igarashi RY, Soboh B, Curatti L, Dean DR, Ludden PW, Rubio LM. NifX and NifEN exchange NifB cofactor and the VK-cluster, a newly isolated intermediate of the iron-molybdenum cofactor biosynthetic pathway. Mol Microbiol. 2007;63:177–192. doi: 10.1111/j.1365-2958.2006.05514.x. [DOI] [PubMed] [Google Scholar]
- 91.Shah VK, Allen JR, Spangler NJ, Ludden PW. In vitro synthesis of the iron-molybdenum cofactor of nitrogenase. Purification and characterization of NifB cofactor, the product of NIFB protein. J Biol Chem. 1994;269:1154–1158. [PubMed] [Google Scholar]
- 92.Corbett MC, Hu Y, Fay AW, Ribbe MW, Hedman B, Hodgson KO. Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc Natl Acad Sci USA. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hernandez JA, Curatti L, Aznar CP, Perova Z, Britt RD, Rubio LM. Metal trafficking for nitrogen fixation: NifQ donates molybdenum to NifEN/NifH for the biosynthesis of the nitrogenase FeMo-cofactor. Proc Natl Acad Sci USA. 2008;105:11679–11684. doi: 10.1073/pnas.0803576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng L, White RH, Dean DR. Purification of the Azotobacter vinelandii nifV-encoded homocitrate synthase. J Bacteriol. 1997;179:5963–5966. doi: 10.1128/jb.179.18.5963-5966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rubio LM, Rangaraj P, Homer MJ, Roberts GP, Ludden PW. Cloning and mutational analysis of the γ gene from Azotobacter vinelandii defines a new family of proteins capable of metallocluster-binding and protein stabilization. J Biol Chem. 2002;277:14299–14305. doi: 10.1074/jbc.M107289200. [DOI] [PubMed] [Google Scholar]
- 96.Rubio LM, Singer SW, Ludden PW. Purification and characterization of NafY (apodinitrogenase γ subunit) from Azotobacter vinelandii. J Biol Chem. 2004;279:19739–19746. doi: 10.1074/jbc.M400965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodríguez-Quiñones F, Bosch R, Imperial J. Expression of the nifBfdxNnifOQ region of Azotobacter vinelandii and its role in nitrogenase activity. J Bacteriol. 1993;175:2926–2935. doi: 10.1128/jb.175.10.2926-2935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pienkos PT, Shah VK, Brill WJ. Molybdenum cofactors from molybdoenzymes and in vitro reconstitution of nitrogenase and nitrate reductase. Proc Natl Acad Sci USA. 1977;74:5468–5471. doi: 10.1073/pnas.74.12.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Imperial J, Ugalde RA, Shah VK, Brill WJ. Role of the nifQ gene product in the incorporation of molybdenum into nitrogenase in Klebsiella pneumoniae. J Bacteriol. 1984;158:187–194. doi: 10.1128/jb.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ugalde RA, Imperial J, Shah VK, Brill WJ. Biosynthesis of the iron-molybdenum cofactor and the molybdenum cofactor in Klebsiella pneumoniae: effect of sulfur source. J Bacteriol. 1985;164:1081–1087. doi: 10.1128/jb.164.3.1081-1087.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee HI, Hales BJ, Hoffman BM. Metal-ion valencies of the FeMo cofactor in CO-inhibited and resting state nitrogenase by 57Fe Q-band ENDOR. J Am Chem Soc. 1997;119:11395–11400. [Google Scholar]
- 102.Venters RA, Nelson MJ, McLean PA, True AE, Levy MA, Hoffman BM, Orme-Johnson WH. ENDOR of the resting state of nitrogenase molybdenum-iron proteins from Azotobacter vinelandii, Klebsiella pneumoniae, and Clostridium pasteurianum: 1H, 57Fe, 95Mo, and 33S studies. J Am Chem Soc. 1986;108:3487–3498. [Google Scholar]
- 103.Butt JN, Armstrong FA, Breton J, George SJ, Thomson AJ, Hatchikian EC. Investigation of metal-ion uptake reactivities of [3Fe-4S] clusters in proteins. Voltammetry of coadsorbed ferredoxin aminocyclitol films at graphite-electrodes and spectroscopic identification of transformed clusters. J Am Chem Soc. 1991;113:6663–6670. [Google Scholar]
- 104.Soboh B, Igarashi RY, Hernandez JA, Rubio LM. Purification of a NifEN protein complex that contains bound Mo and a FeMo-co precursor from an Azotobacter vinelandii ΔnifHDK strain. J Biol Chem. 2006;281:36701–36709. doi: 10.1074/jbc.M606820200. [DOI] [PubMed] [Google Scholar]
- 105.Hu Y, Fay AW, Ribbe MW. Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc Natl Acad Sci USA. 2005;102:3236–3241. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.George SJ, Igarashi RY, Piamonteze C, Soboh B, Cramer SP, Rubio LM. Identification of a Mo-Fe-S cluster on NifEN by Mo K-edge extended X-ray absorption fine structure. J Am Chem Soc. 2007;129:3060–3061. doi: 10.1021/ja0663428. [DOI] [PubMed] [Google Scholar]
- 107.Goodwin PJ. Biochemistry Department. Virginia Tech; Blacksburg: 1999. Biosynthesis of the nitrogenase FeMo-cofactor from Azotobacter vinelandii: involvement of the NifNE complex, NifX and the Fe protein; p. 210. [Google Scholar]
- 108.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 109.Rangaraj P, Ludden PW. Accumulation of 99Mo-containing iron-molybdenum cofactor precursors of nitrogenase on NifNE, NifH, and NifX of Azotobacter vinelandii. J Biol Chem. 2002;277:40106–40111. doi: 10.1074/jbc.M204581200. [DOI] [PubMed] [Google Scholar]
- 110.Hu YL, Corbettt MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Nitrogenase Fe protein: A molybdate/homocitrate insertase. Proc Natl Acad Sci USA. 2006;103:17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dos Santos PC, Dean DR. A newly discovered role for iron-sulfur clusters. Proc Natl Acad Sci USA. 2008;105:11589–11590. doi: 10.1073/pnas.0805713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pagani S, Bonomi F, Cerletti P. Enzymic synthesis of the iron-sulfur cluster of spinach ferredoxin. Eur J Biochem. 1984;142:361–366. doi: 10.1111/j.1432-1033.1984.tb08295.x. [DOI] [PubMed] [Google Scholar]
- 114.Pagani S, Eldridge M, Eady RR. Nitrogenase of Klebsiella pneumoniae Rhodanese-catalysed restoration of activity of the inactive 2Fe species of the Fe protein. Biochem J. 1987;244:485–488. doi: 10.1042/bj2440485. [DOI] [PMC free article] [PubMed] [Google Scholar]


