Abstract
Recent studies have shown that activated aryl hydrocarbon receptor (AHR) induced the recruitment of estrogen receptor-α (ERα) to AHR-regulated genes and that AHR is recruited to ERα-regulated genes. However, these findings were limited to a small number of well-characterized AHR- or ERα-responsive genes with little knowledge of what was occurring at other genomic regions. In this study, we showed using chromatin immunoprecipitation followed by hybridization to promoter focused microarrays (ChIP-chip) that 2,3,7,8-tetrachlorodibenzo-p-dioxin treatment significantly increased the overlap of genomic regions bound by both AHR and ERα. Conventional and sequential ChIPs confirmed the recruitment of AHR and ERα to many of the identified regions. Transcription factor binding site analysis revealed an overrepresentation of aryl hydrocarbon receptor response elements in regions bound by both AHR and ERα, suggesting that AHR was the important factor determining the recruitment of ERα to these regions. RNA interference-mediated knockdown of AHR confirmed its requirement for the recruitment of ERα to some, but not all, of the shared regions. Our findings demonstrate not only that dioxin induces the recruitment of ERα to AHR target genes but also that AHR is recruited to estrogen-responsive regions in a gene-specific manner, suggesting that AHR utilizes both of these mechanisms to modulate estrogen-dependent signaling.
Keywords: aryl hydrocarbon receptor, estrogen receptor-α, ChIP-chip, receptor crosstalk, dioxin
Estrogen receptor-α (ERα) and estrogen receptor-β (ERβ) (gene symbols ESR1 and ESR2) play important roles in a number of biological functions including the reproductive tract, the skeletal-immune system, and the central nervous system (Heldring et al., 2007). Disruption of estrogen receptor (ER) signaling pathways can have devastating physiological, developmental, and pathological outcomes (McLachlan et al., 2006). A number of environmental contaminants are known or suspected to adversely affect ER function either through direct binding to the receptor or by the activation of other receptor pathways that modulate ER activity, in what is widely referred to as receptor crosstalk (Safe and Wormke, 2003). Dioxins are persistent environmental contaminants that adversely affect estrogen action. The prototypical dioxin, 2,3,7,8-tetrachlorodi-benzo-p-dioxin (TCDD or dioxin) inhibits ER activity through its binding and activation of the aryl hydrocarbon receptor (AHR) (Wormke et al., 2003).
The AHR and ERs are ligand-activated transcription factors and members of the basic helix-loop-helix/PAS (Per-ARNT-Sim where Per=Period; ARNT=aryl hydrocarbon nuclear translocator; Sim=Single-minded)-family and nuclear receptor super family, respectively. AHR binds a wide range of endogenous and xenobiotic compounds (Denison and Nagy, 2003; Safe, 1993). In the absence of ligand, the AHR is located in the cytoplasm bound to a multichaperone protein complex (Petrulis and Perdew, 2002). Upon ligand binding, the AHR translocates into the nucleus, heterodimerizes with aryl hydrocarbon nuclear translocator (ARNT) and binds to specific DNA sequences termed aryl hydrocarbon receptor response elements (AHREs). The activated AHR/ARNT heterodimer recruits coregulators leading to changes in target gene expression, including cytochrome P4501A1 (CYP1A1) and CYP1B1 (Hankinson, 1995; Nebert et al., 2004).
Estrogen action is mediated by binding to ERα and ERβ (Heldring et al., 2007; Nilsson and Gustafsson, 2000; Nilsson et al., 2001). Ligand binding results in homodimerization followed by binding to specific DNA sequences termed estrogen-responsive elements (EREs) located in the proximal and distal regulatory regions of their target genes (Nilsson and Gustafsson, 2000). ERs can also interact with other transcription factors through protein-protein interactions to modulate gene expression utilizing a protein tethering pathway (Paech et al., 1997). Once bound to DNA, either directly via EREs or through tethering to other transcription factors, ERs recruit additional accessory proteins known as coregulators resulting in changes in gene expression (Nilsson and Gustafsson, 2000).
Crosstalk has been observed between AHR and a number of different signaling pathways (Puga et al., 2002; Tan et al., 2004; Tian et al., 2002), but its antiestrogenic effects are perhaps the most well documented (Matthews and Gustafsson, 2006; Safe and Wormke, 2003). The precise molecular mechanism of AHR/ERα crosstalk remains elusive and is thought to be through a combination of several different pathways (Matthews and Gustafsson, 2006; Ohtake et al., 2007; Safe and Wormke, 2003). Moreover, many well-characterized TCDD-responsive and AHR-regulated genes such as CYP1A1, CYP1B1, and TCDD-inducible poly-ADP-ribose polymerase (TiPARP) have also been reported to be estrogen responsive in immortalized human breast cancer cells (Frasor et al., 2004; Tsuchiya et al., 2004), suggesting that AHR and ERs regulate the expression of many of the same genes.
AHR-mediated transcription has only been intensively studied on a small number of endogenous target promoters, with the majority of the data coming from AHR-mediated regulation of CYP1A1 (Hankinson, 1995; Whitlock, 1999). Expression-based microarrays have identified hundreds of genes potentially regulated by AHR (Puga et al., 2000). However, these studies do not identify direct genomic targets of AHR but rather messenger RNA (mRNA) transcripts that can be directly or indirectly regulated by AHR.
Recent genome-wide analyses using chromatin immunoprecipitation (ChIP) in combination with DNA microarrays (chip; ChIP-chip) or DNA sequencing (ChIP-Seq) have provided new insight into the whole-genome binding patterns of ERs in cell lines and tissues (Carroll et al., 2005, 2006; Laganiere et al., 2005; Lin et al., 2007; Liu et al., 2008). These studies identified new genomic targets for ER and caused a reevaluation of the mechanisms of ER-mediated transcription that considers the role of distal enhancer elements in the regulation of ER target genes.
Previous studies from our laboratory showed that activation of AHR induced the recruitment of ERα to well-characterized AHR-regulated genes, CYP1A1 and CYP1B1, in an AHR ligand–dependent manner (Matthews et al., 2005, 2007). However, it is unclear whether this recruitment is limited to CYP1A1 and CYP1B1 or whether AHR ligands induce ERα recruitment to other AHR-regulated genes. To address this, we investigated TCDD-induced occupancy of ERα and AHR to a comprehensive set of human promoters, using ChIP-chip. Our data show that the number of regions bound by both AHR and ERα were significantly increased after TCDD treatment and that RNA interference (RNAi)-mediated knockdown of AHR reduced the TCDD-dependent occupancy of ERα to some but not all of the co-occupied regions.
MATERIALS AND METHODS
Chemicals
Antibodies used for ChIP experiments include; ERα, HC-20 and AHR, H-211 (Santa Cruz Biotechnology, Santa Cruz, CA). Primary antibodies used for Western blots were ERα, HC-20, and AHR, SA-210 (Biomol International Inc., Plymouth Meeting, PA). TCDD was purchased from Accustandard (New Haven, CT) and dimethyl sulfoxide (DMSO) was purchased from Sigma (St Louis, MO). Cell culture media, fetal bovine serum (FBS), and trypsin were all purchased from Invitrogen (Carlsbad, CA). All other chemicals and biochemicals were of the highest quality available from commercial vendors.
ChIP-chip experiments
T-47D human breast carcinoma cells were cultured in 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and F12 Ham’s nutrient mixture medium supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin. Cells were maintained at 37°C and 5% CO2. T-47D cells were subcultured every 2–3 days or when they reached 80–90% confluency. For the ChIP-chip studies, T-47D cells were seeded in 10-cm dishes in 1:1 mixture of DMEM:F12 supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin. After 48 h, cells were treated with either 10 nM TCDD or DMSO (final concentration 0.1%) for 1 h and the ChIP assay was performed as described previously (Matthews et al., 2005). For re-chip experiments, the first antibody-bound protein-chromatin complexes were released by incubation with 50 mM of dithiothreitol/1% SDS for 1 h at 37°C. The supernatant was collected and diluted 40×. Two micrograms of the second antibody was added to the supernatant and rotated for 2 h at room temperature. For chip experiments done in dextran-coated charcoal (DCC) (Sigma)-stripped serum, T-47D cells were seeded in 10-cm dishes using 1:1 mixture of DMEM:F12 phenol red free supplemented with 5% (vol/vol) of stripped serum and 1% penicillin/streptomycin. After 72 h, cells were treated with either DMSO (final concentration 0.1%) or 10 nM TCDD and ChIPs were performed as previously described (Matthews et al., 2005). Immunoprecipitated DNA from 10-cm dish per antibody was linearly amplified using a random hexamer linear amplification protocol with primer A: GTTTCCCAGTCACGGTC(N)9 and primer B: GTTTCCCAGTCACGGTC according the manufacturer’s instructions (Affymetrix, Santa Clara, CA). Linearly amplified DNA (7.5 μg) was fragmented by limited DNAseI digestion and hybridized to Affymetrix human promoter tiling arrays 1.0R (Affymetrix). Hybridization and washing steps were performed according to the manufacturer’s protocol by The Centre for Applied Genomics at the Hospital for Sick Children (Toronto, Canada). Data were normalized and analyzed using CisGenome (Ji et al., 2008). Enriched peaks at a false detection rate of 0.2 were determined by comparing triplicate samples of AHRTCDD and ERαTCDD to triplicate IgGTCDD using TileMap v2 by a moving average approach using default settings (Ji and Wong, 2005). Regions were merged if the gap between them was < 300 bp, and the number of probes failing to reach the cutoff was < 5. Regions were discarded if they were < 120 bp or did not contain at least five continuous probes above the cutoff. Quantitative real-time PCR (Q-PCR) primers used to amplify the regions of interest and mRNA target gene sequences are provided in Supplementary Tables 1 and 2. Bed files containing the genomic coordinates of AHRTCDD regions (Supplementary Table 3) and ERαTCDD regions (Supplementary Table 4) are provided in Supplementary data.
Overlap of regions and closest gene definition
We first clustered the ChIP regions that overlapped between experiments and merged regions between experiments that overlapped with > 50% of the width of the smallest region. We then labeled each region with the gene symbol of the closest transcript, regardless of strand, using the all_mRNA and unigene annotation from the UCSC Genome Browser (Karolchik et al., 2008). Note that this can potentially give multiple regions per gene. In most cases, this assignment was trivial since the tiling array was focused on promoter regions, but there are cases of miss-annotation. Therefore, we excluded gene-region pairs that were further away than 10 kb.
Transcription factor binding site searching and heat map visualization
Transcription factor binding site (TFBS) prediction has been reviewed previously (Wasserman and Sandelin, 2004). We searched for potential TFBSs using the motif models from the JASPAR database (Bryne et al., 2008) and the ASAP framework (Marstrand et al., 2008) as previously described (Liu et al., 2008). We counted all hits that scored above a threshold t, set to 85% of the total scoring range for a given model as described previously (Wasserman and Sandelin, 2004). We then performed two types of over-representation tests, one using a background and one comparing differences among the three data sets (inter-set test). To deal with the different lengths of the sets, we used two different strategies. When comparing to background, we constructed a specific background similar in length and position to the set it was compared to. Specifically, these were constructed using the transcription start sites (TSSs) of 10,000 transcripts sampled randomly from Ensembl (Flicek et al., 2008). In order to avoid bias caused by proximity to genes (for instance higher CG content), we sampled the background sequence relative to the TSS of this transcript based on empirically observed distributions of distances (to the closest TSS) and region lengths for each set. We then counted the number of sequences that had at least one subsequence that scored above the threshold (t). For the inter-set test, we used simulations to achieve Z scores as described in (Liu et al., 2008). By using this matrix of Z values, we clustered the transcription factor models (rows) and the data sets (columns) with a hierarchical clustering method. The Z value was transformed into a color and presented in a heat map. A trace was added to make it easier to distinguish similar colors. The heatmap.2 function in the R gplots() package (Ihaka and Gentleman, 1996) was used to draw the heat map. To focus only on the most strongly overrepresented and underrepresented patterns, we filtered out any pattern that did not have an overrepresentation or underrepresentation of at least 3 SD.
RNA isolation and real-time PCR
T-47D cells were seeded in six-well plates and grown in a 1:1 mixture of DMEM and F12 Ham’s nutrient mixture. Cells were treated with either 10 nM TCDD or DMSO for 1.5, 3, 6, or 24 h. RNA was isolated using RNeasy spin columns (Qiagen, Valencia, CA) and reverse transcribed as previously described (Matthews et al., 2006). Briefly, 0.5 μg of RNA was reverse transcribed using Superscript II, and Q-PCR was performed using 1 μl complementary DNA and SYBR Green (Bio-Rad, Hercules, CA). All target gene transcripts were normalized to ribosomal 18s RNA content, and fold inductions were calculated using time-matched DMSO samples.
Western blot
Proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane in 25 mM Tris base (pH 8.3) containing 19.2 nM glycine and 20% (vol/vol) methanol. The membrane was blocked in 2% (wt/vol) ECL-Advanced blocking agent for 1 h at room temperature with constant rocking and then incubated with 1:5000 anti-ERα (HC-20) or 1:5000 anti-AHR (SA-210) overnight at 4°C with constant rocking. The membrane was then washed three times in PBS/0.1% Tween 20 and incubated with 1:200,000 horseradish peroxidase (HRP)–conjugated anti-rabbit secondary antibody for 1 h at room temperature with constant rocking. For detection of β-actin, a 1:500,000 dilution of primary mouse anti-β-actin antibody (Sigma) was incubated for 2 h at room temperature, followed by a 1-h washing with PBS/0.1% Tween 20 prior to incubation with 1:200,000 HRP-conjugated anti-mouse secondary antibody for 1 h at room temperature. After washing, the bands were visualized using ECL-Advanced chemiluminescent substrate according to the manufacturer’s instructions (GE Healthcare, Piscataway, NJ).
Transient transfection and small interfering RNA
AHR (L-004990-00-0020) and ERα (L-003401-00-0020) ON-TARGETplus SMART pool small interfering RNA (siRNA) and DharmaFECT1 transfection reagent were purchased from Dharmacon (Lafayette, CO). Briefly, T-47D cells were seeded 300,000 per well in a six-well plate containing 2 ml of medium. After 24 h, 2 μM of siRNA against AHR (L-004990-00-0020), ERα (L-003401-00-0020), or nontargeting pool (NTP) (D-0011810-10-20) (Dharmacon) were transfected using 4 μl of DharmaFECT and 400 μl Opti-MEM. ChIP assays, mRNA isolation, and whole-cell extracts were prepared 48 h after transfection.
RESULTS
Defining AHR- and ERα-Bound Regions Isolated by ChIP-chip
In order to determine if AHR-induced recruitment of ERα occurs at all or just a subset of AHR target genes, we performed ChIP-chip assays on T-47D human breast cancer cells grown for 48 h in 10% FBS-containing medium prior to 1-h treatment with 10 nM TCDD or solvent control DMSO. We used AHR- and ERα-specific antibodies for ChIP and isolated precipitated chromatin as previously described (Matthews et al., 2005). Isolated and linearly amplified DNA was hybridized to Affymetrix Human promoter tiling 1.0R micro-arrays. We performed three biological replicates, and data were normalized and analyzed as described in the “Materials and Methods” section. This analysis resulted in the identification of 412 regions bound by AHRTCDD (Supplementary Table 3) and 364 regions bound by ERαTCDD (Supplementary Table 4). The genomic coordinates for these data sets are provided in Supplementary data. AHR and ERα regions are referred to as AHR_number and ERα_number where the number indicates the relative rank of the region within each of the respective analyses. Since the enriched regions were determined by comparing AHR- or ERα-bound regions to IgG, these regions may or may not be dependent on TCDD treatment.
Overlap Between AHR- and ERα-Bound Regions and Putative Target Genes
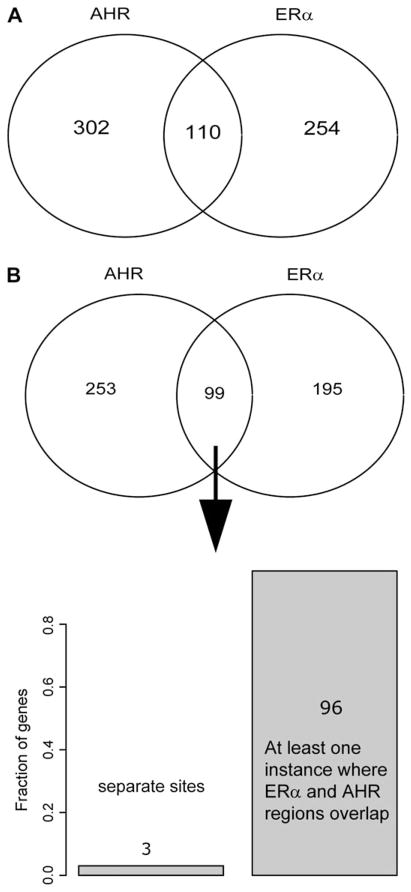
As described above, we merged regions that overlapped between the ERα and the AHR data sets (see “Materials and Methods” section) resulting in a total of 110 regions that were bound by both ERα and AHR. We will refer to these subsets of regions as the intersect set, AHR-only set and ERα-only set. A Venn diagram illustrating the relation among these sets is shown in Figure 1A.
FIG. 1.
Overlap between ChIP sets and target genes. (A) Venn diagram showing the number of ChIP regions from respective experiments that overlap with more than 50% of the length of the smallest region. (B) Overlap of the experiments in terms of the identity of the closest gene. In 96 of the 99 genes where the gene has both AHR and ERα chip regions, the ChIP regions overlap physically.
We then investigated putative target genes for the identified regions by determining the gene with the closest TSS on any strand (see “Materials and Methods” section). In this process, we noted that several known AHR target genes, CYP1A1 and CYP1B1, as well as known ERα target genes cyclin G2 (CCNG2), ERα (ESR1), gene regulated in breast cancer 1 (GREB1), and carbonic anhydrase XII (CA12) were identified in both the AHRTCDD and the ERαTCDD data sets. Since each of the isolated regions could be labeled with a target gene, we also assessed the overlap between the experiments in terms of target genes. This type of analysis is not the same as simply considering enriched regions since many ChIP regions may be located in the upstream regulatory region of a single gene (Fig. 1B). Interestingly, in ~97% of cases (96/99) where a gene was targeted by both ERα and AHR, the two regions in question also overlapped.
The annotated function of the target genes (Gene Ontology terms) in the respective sets was not significantly different when comparing the sets to one another, although compared to a general background (all genes) some differences were observed (see Supplementary data and Supplementary Table 5).
Validation of ChIP-chip Regions Using Conventional ChIP
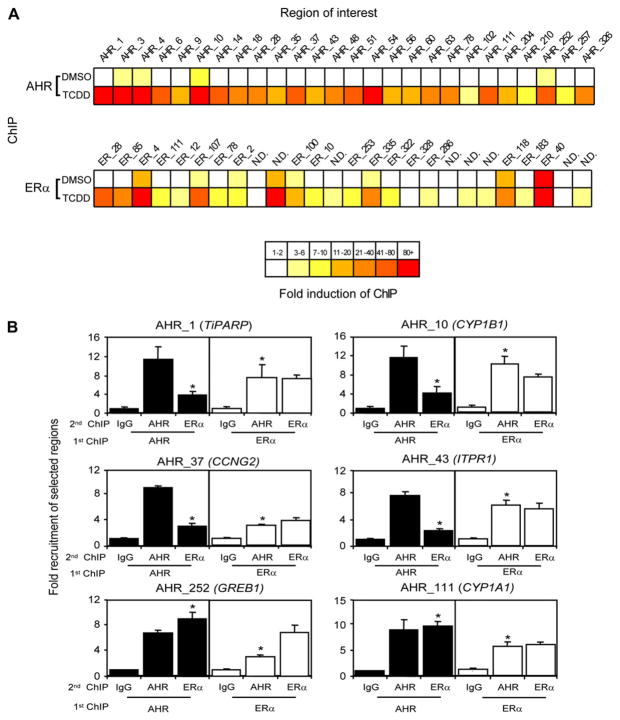
To validate the enriched regions identified by ChIP-chip, conventional ChIPs were performed on a subset of 26 identified regions using qPCR. The regions were chosen to cover a range of enrichment values but also included regions near a select number of known AHR- and ERα-regulated genes. All 26 regions verified the recruitment of AHR and ERα (or lack thereof) from the ChIP-chip study with the level of enrichment varying among the regions. The results shown in Figure 2A reveal a strong ligand-dependent recruitment of AHR to a subset of the total identified AHR-bound regions. For the most part, there was weak binding of AHR to the tested regions in the absence of TCDD. In agreement with other studies, AHR occupied the CYP1B1 (AHR_10) upstream regulatory region in the absence of TCDD when compared to IgG (Yang et al., 2008). Significant ligand-independent AHR occupancy was also observed at the upstream regulatory regions of synaptotagmin XII (SYT12, AHR_4), transmembrane protein 30A (TMEM30a, AHR_3), pregnancy-specific beta-1-glycoprotein 9 (PSG9, AHR_51), and gene amplified in breast cancer 1 (GREB1, AHR_252). AHR has been reported to exist simultaneously in the nucleus and cytoplasm in human breast cancer cells (Wang et al., 1998), which might explain the occupancy of AHR at these regions in the absence of TCDD. The highest ranked region bound by AHR was also bound by ERα and mapped to a sequence ~100 kb downstream of the TiPARP (AHR_1) transcriptional start site, suggesting that this gene might be regulated by a distal 3′ enhancer. A 3′ enhancer has also been reported to regulate the CYP1A2 (Okino et al., 2007). We also identified a number of novel AHR-bound sites upstream of prospero homeobox 1 (PROX1, AHR_326), Forkhead box N4 (FOXN4, AHR_35), homeobox 10 (HOXC10, AHR_63), sortilin-related receptor (SORL1, AHR_14), inositol 1,4,5-triphosphate receptor, type 1 (ITPR1, AHR_43) genes.
FIG. 2.
TCDD-induced recruitment of both AHR and ERα to ChIP-chip identified regions. (A) Quantification of AHR and ERα binding was determined as fold induction above IgG DMSO and is expressed as the mean of three independent replicates. Regions were chosen to cover a range of enrichment values and included a select number of sites near AHR and ERα target genes. N.D. refers to regions that were not detected in the ERα ChIP-chip experiment. T-47D cells were treated with 10 nM TCDD for 1 h. ChIP assays were performed with the indicated antibodies, and the immunoprecipitated DNA was measured by quantitative PCR using primers targeting regions isolated in the ChIP-chip study. (B) T-47D cells were treated with 10 nM TCDD for 1 h. Sequential ChIPs were performed with the indicated antibodies. Immunoprecipitated DNA was measured by quantitative PCR using primers targeting regions isolated in the ChIP-chip study. Quantification of binding was determined as fold induction above IgG DMSO. Each error bar represents the SE of the mean of three independent replicates. Asterisks indicate statistically significant differences (p < 0.05) compared with IgG DMSO control samples.
There was good agreement between the ChIP-chip regions and the confirmation of TCDD-induced ERα recruitment to shared regions in the intersect group. In contrast to the AHR confirmation data (Fig. 2A), promoter occupancy of ERα in DMSO samples was observed at a number of analyzed regions. This was due to the fact that cells were cultured in 10% FBS and not DCC-treated or steroid-reduced serum, which is required to observe robust estrogen-dependent responses in breast cancer cell lines. However, steroid deprivation is not necessary to observe robust TCDD-dependent activation of AHR transcription (Hankinson, 1995). The occupancy of ERα at the promoter regions of the well-characterized ER target genes GREB1 and ESR1 was consistent with previously published ChIP-chip studies (Carroll et al., 2006; Kwon et al., 2007; Laganiere et al., 2005).
TCDD-dependent recruitment of ERα was observed to a number of regions including those upstream of RAS-like, estrogen-regulated growth inhibitor (RERG, ERα_22), CCNG2 (ERα_100), CYP1B1 (ERα_107), and synaptotagmin XII (SYT12, ERα_4). TCDD also induced recruitment of AHR to known estrogen-responsive genes including GREB1 (AHR_252), RERG (AHR_18), CCNG2 (AHR_37), and ESR1 (AHR_204). These findings indicate not only that AHR influenced the recruitment of ERα to AHR-regulated genes but also that AHR is recruited to genomic regions occupied by ERα where the binding of ERα is independent of AHR activation. We observed three false negatives in that our ChIP-chip experiment failed to detect recruitment of ERα to CYP1A1 (AHR_111), transducin (beta)-like 1 X-linked receptor 1 (TBL1XR1; AHR_48), and Janus kinase 1 (JAK1; AHR_102), but ERα binding to these regions was detected by conventional ChIP. This may have been due to the thresholds applied in the ChIP-chip experiments. Sequential ChIPs were done on a subset of six regions in the intersect set (bound by both AHR and ERα) confirming the simultaneous binding of both AHR and ERα to the regions examined (Fig. 2B).
Chromatin Binding and Correlation With Gene Expression of TCDD-Responsive Genes
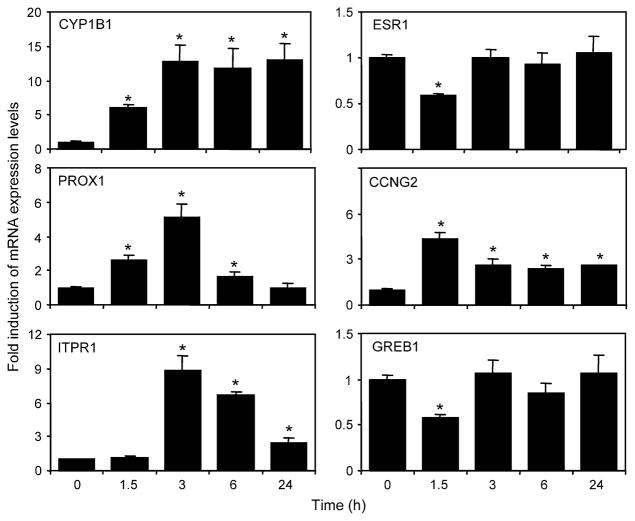
We were then interested to determine if the binding of AHR and/or ERα to genomic regions resulted in changes in mRNA levels of the closest genes that map to the isolated genomic fragments. We treated T-47D cells with 10 nM TCDD for 1.5, 3, 6, and 24 h, isolated RNA, and determined changes in mRNA levels using qPCR. A subset of the examined genes is shown in Figure 3. We observed that mRNA expression of the predicted target genes displayed TCDD-dependent increases, decreases, or no change at the time points examined. Temporal ChIP analysis of AHR binding following treatment with 3-methyl-cholanthrene or TCDD every 30 min from 0 to 4.5 h revealed an oscillatory recruitment pattern of both AHR and ERα that correlated well with the oscillatory mRNA expression pattern of the genes examined (Pansoy and Matthews, in preparation). A table summarizing the mRNA changes for the closest genes corresponding to the confirmed ChIP-chip regions is provided in Supplementary Table 6. As expected, TCDD increased the mRNA expression levels of CYP1A1 and CYP1B1. We also observed TCDD-dependent increases in CCNG2, PROX-1, and ITPR1 mRNA expression levels (Fig. 3). In support of the antiestrogenic action of TCDD, the estrogen-responsive genes GREB1 and ESR1 were both inhibited by TCDD treatment but quickly rebounded at the later time points.
FIG. 3.
Chromatin profiles correlate with expression status in TCDD-responsive genes. After TCDD treatment for the indicated time periods, RNA was isolated and reverse transcribed. mRNA expression was then determined using quantitative PCR. Data were normalized against time-matched DMSO and to ribosomal 18s levels. Each error bar represents the SE of the mean of three independent replicates. Asterisks indicate statistically significant differences (p < 0.05) compared to time-matched DMSO control samples.
TFBS Analysis of the ChIP Regions
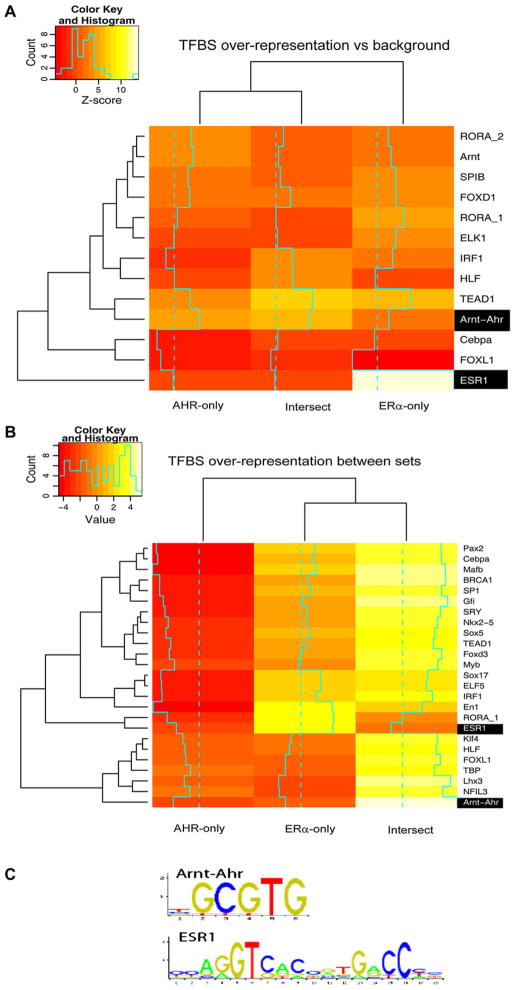
We then investigated the density of putative TFBSs in the AHR-only, intersect, and ERα-only regions and calculated overrepresentation or underrepresentation of TFBSs compared to either a sampled promoter background (Fig. 4A) or among sets (Fig. 4B). In the first type of analysis, we obtained an “absolute” measure of overrepresentation, where one factor can be overrepresented in all sites, while in the second type of analysis, we determined the different binding sites among the different data sets.
FIG. 4.
TFBS analysis. Heat maps showing the most overrepresented and underrepresented TFBS patterns in each set, either compared to large promoter background (A) or compared between sets (B). Heat map A can be viewed as an “absolute” measure of overrepresentation, while heat map B shows what patterns that are significantly different between at least two sets in terms of occurrence. Overrepresentation or underrepresentation is expressed as a Z score, where a negative value means underrepresentation (coded red) and high values indicate overrepresentation (coded white). Z scores were translated into a color range from red to white. Rows (transcription factor binding patterns from JASPAR) and columns (ChIP regions as in Fig. 1A) are ordered by similarity to each other. Patterns having no preference to any set are not shown (see “Materials and Methods” section). The Arnt-Ahr pattern (corresponding to an AHRE) and the ESR1 pattern (corresponding to an ERE) are highlighted. (C) Sequence logo for the Arnt-Ahr and ESR1 matrices from JASPAR.
We measured the overrepresentation as a Z score statistic and visualized which TFBSs were significantly overrepresented and underrepresented by hierarchically clustered heat maps as in (Liu et al., 2008), where the rows are the JASPAR database (Bryne et al., 2008) TFBSs and the columns are the ChIP sets. As expected, when compared to a generic promoter sequence background, the ERα-only set had a strong overrepresentation of the ERE pattern represented by the ESR1 JASPAR model, whereas the AHRE pattern, represented by the Arnt-Ahr JASPAR model, was overrepresented in all sets but most evident in the intersect set (Fig. 4A). This was also consistent when assessing overrepresentation between sets (Fig. 4B). In that analysis, the intersect set was strongly enriched in AHREs (Arnt-Ahr JASPAR model) when compared to the AHRTCDD or the ERαTCDD sets, while as expected, the ERE (ESR1 JASPAR model) was strongest in the ERαTCDD set. Of the 110 regions in the intersect group, 57 contained an AHRE, 24 contained an ERE, but surprisingly only 10 regions contained both response elements. Since the AHRE was particularly overrepresented in the intersect set, we hypothesized that it was likely that AHR was contributing at least in part to the recruitment of ERα to regions in the intersect set.
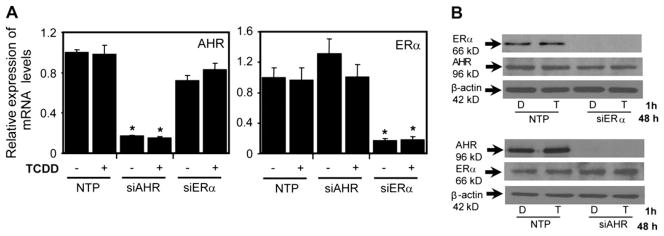
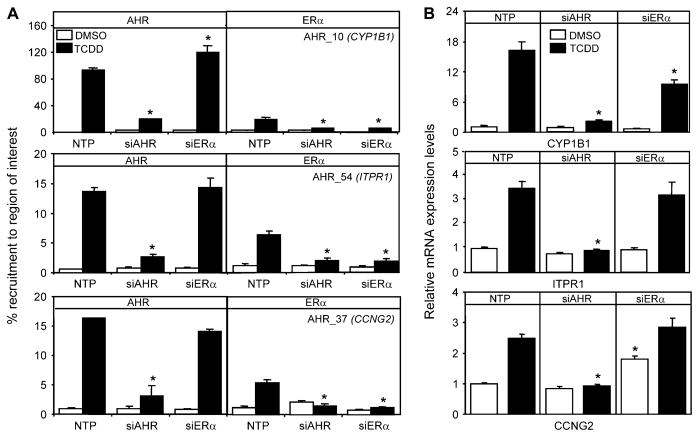
AHR Modulates Recruitment of ERα to the Shared Regions
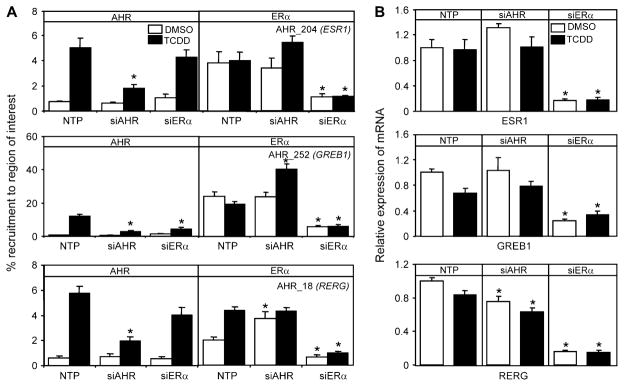
To test the hypothesis that AHR was influencing the recruitment of ERα to the regions in the intersect set, we used RNAi-mediated knockdown of AHR or ERα and determined the recruitment of each of these factors to a subset of regions in the intersect group as well as changes in mRNA expression levels. Following transfection of siRNA oligos into T-47D, we determined that 48-h post-transfection both AHR and ERα protein levels were undetectable and mRNA expression levels were reduced to 20% compared to controls (Figs. 5A and B). Western blots of ERα levels after 1-h TCDD treatment showed that any reduction in recruitment levels of ERα were not due to TCDD-dependent proteolysis of ERα (Wormke et al., 2003) (Fig. 5B). ChIP assays and RNA isolation were then done on siRNA-transfected T-47D cells exposed to 10 nM TCDD for 1 and 6 h, respectively. As expected, knockdown of AHR or ERα reduced their respective recruitment to the genomic regions examined. AHR knockdown reduced the TCDD-dependent recruitment of ERα to AHR_10 (CYP1B1), AHR_54 (ITPR1), and AHR_37 (CCNG2) compared to NTP controls (Fig. 6A). All three of these genes have been reported to also be responsive to estrogen treatment (Kirkwood et al., 1997; Stossi et al., 2006; Tsuchiya et al., 2004). Further studies completed in DCC-stripped serum confirmed the TCDD-dependent recruitment of ERα to these regions (Supplementary Fig. 7). Knockdown of ERα had no effect on TCDD-dependent induction of ITPR1 and CCNG2 (Fig. 6B) but caused a significant reduction of TCDD-induced CYP1B1 (Fig. 6B) mRNA levels. Knockdown of ERα also resulted in a significant reduction in TCDD-induced CYP1A1 mRNA (data not shown). Interestingly, knockdown of ERα resulted in increased basal mRNA levels of CCNG2 (Fig. 6B). CCNG2 is negatively regulated by ERα, which may explain the increase in basal expression following ERα knockdown (Stossi et al., 2006). As expected, the occupancy of ERα at all regions examined was significantly reduced in cells transfected with siERα compared to controls. AHR knockdown, however, had no effect on the recruitment of ERα to upstream regulatory regions for AHR_252 (GREB1), AHR_204 (ESR1), and AHR_18 (RERG), demonstrating that AHR exhibits region-specific modulation of ERα genomic binding profiles (Fig. 7A). These three genes have been reported to be estrogen target genes (Castles et al., 1997; DeNardo et al., 2005; Finlin et al., 2001; Lin et al., 2004), and ERα occupied these regions in the absence of TCDD, which may explain why AHR had no effect on the recruitment of ERα to these genes. TCDD increased the overlap of ERα and AHR to these genes through the recruitment of AHR to genomic sequences bound by ERα in the presence of DMSO. The recruitment of AHR was unaffected by knockdown of ERα for all regions examined with the following exceptions; TCDD-dependent recruitment of AHR to GREB1 (Fig. 7A) was decreased, while recruitment of AHR was increased at CYP1B1 (Fig. 6A). These results indicate that ERα influences the AHR transcription in a promoter- and context-specific manner. These data also show not only that TCDD-mediated activation of AHR modulates the recruitment of ERα to a number of genomic regions in a gene-specific manner but also that AHR is recruited to many genomic regions regulated by ERα.
FIG. 5.
Analysis of AHR and ERα knockdown in T-47D cells: protein and transcript levels. (A) T-47D cells were transfected with specific siRNA against AHR and ERα for 48 h. RNA was isolated and reverse transcribed. mRNA expression was then determined using quantitative PCR. Data were normalized against time-matched DMSO and to ribosomal 18s levels. Each error bar represents the SE of the mean of three independent replicates. Significance was determined by comparison to NTP TCDD treatment p < 0.05. (B) Western blot analysis of AHR and ERα knockdown in T-47D cells following 48-h transfection then 1-h treatment with either DMSO or 10 nM TCDD. Cell extracts were probed with rabbit antibody against AHR and ERα. β-actin was used as loading control.
FIG. 6.
AHR is required for TCDD-dependent recruitment of ERα to a subset of co-occupied AHR and ERα target genes. (A) T-47D cells were transfected for 48 h with siRNA and then treated for 1 h with TCDD. ChIP assays were performed with the indicated antibodies, and the immunoprecipitated DNA was measured by quantitative PCR using primers targeting regions isolated in the ChIP-chip study. Quantification of binding was determined as a percent of input DNA and is expressed as the mean of three independent replicates. (B) Gene expression profiles were completed on T-47D cells transfected for 48 h with siRNA and then treated for 6 h with TCDD. RNA was isolated and reverse transcribed. mRNA expression was then determined using quantitative PCR. Data were normalized against time-matched DMSO and to ribosomal 18s levels. Each error bar represents the SE of the mean of three independent replicates. Asterisks indicate statistically significant differences (p < 0.05) compared to NTP treatment–matched samples.
FIG. 7.
AHR is not necessary for ERα binding to a subset of co-occupied AHR and ERα target genes. (A) T-47D cells were transfected for 48 h with siRNA and then treated for 1 h with TCDD. ChIP assays were performed with the indicated antibodies, and the immunoprecipitated DNA was measured by quantitative PCR using primers targeting regions isolated in the ChIP-chip study. Quantification of binding was determined as a percent of input DNA and is expressed as the mean of three independent replicates. (B) Gene expression profiles were completed on T-47D cells transfected for 48 h with siRNA and then treated for 6 h with TCDD. RNA was isolated and reverse transcribed. mRNA expression was then determined using quantitative PCR. Data were normalized against time-matched DMSO and to ribosomal 18s levels. Each error bar represents the SE of the mean of three independent replicates. Asterisks indicate statistically significant differences (p < 0.05) compared to NTP treatment–matched samples.
DISCUSSION
In this study, we used genome-wide but promoter focused human tiling arrays to identify regions bound only by AHR, ERα, or both in T-47D human breast cancer cells treated with TCDD. This analysis identified a number of novel TCDD-responsive genes that were directly regulated by AHR and revealed that TCDD increased the co-occupancy of ERα and AHR to many target regions. Our findings provide new insight into AHR signal transduction and increase our understanding of how AHR ligands modulate ERα action at the genomic level.
Our ChIP-chip analysis of TCDD-treated human breast cancer cells resulted in a number of important findings. AHR binding was detected at many known AHR-responsive genes, including CYP1A1 and CYP1B1, but several new AHR target genes were also identified. TFBS analysis revealed that an AHRE was enriched in the AHRTCDD-isolated regions, although it was only present in ~30% of these regions using rather conservative thresholds in JASPAR. A recent study using ChIP combined with high-throughput southwestern chemistry-based enzyme-linked immunosorbent assay to identify TCDD-dependent AHR-binding regions in mouse hepatoma Hepa-1c1c7 cells (Kinehara et al., 2008) isolated 77 sites with approximately half of them containing an AHRE. Collectively, these findings demonstrate that TCDD-activated AHR binds to promoter regions that do not necessarily contain an AHRE. Therefore, the co-occupancy of these receptors at target promoters may also be mediated through tethering to other transcription factors and not necessarily through direct binding to DNA.
AHR was recruited to genes whose expression was increased or decreased in response to TCDD, which is consistent with AHR serving either as an activator or repressor of transcription in a context-specific manner (Okey, 2007). In a few cases, we observed AHR occupancy at genes that were not TCDD responsive (Supplementary Table 6). This implies that AHR binding was not the limiting factor for the regulation of these genes and suggests potential cell type–specific regulation. Similar findings have been reported for genome-wide glucocorticoid receptor binding in response to dexamethasone (So et al., 2007) and from a recent study examining TCDD-induced AHR binding in mouse Hepa1c1c7 cells (Kinehara et al., 2008).
Treatment with TCDD increased the overlapping genomic binding patterns of ERα and AHR, resulting in the identification of 110 regions or 27% of the AHRTCDD or 30% of the ERαTCDD regions. These large fractions are unlikely due to stringent cutoffs, but they also indicate that co-binding of AHR and ERα was not absolute. For example, there were ERα binding events where AHR played no major part in ERα recruitment. At these regions, co-occupancy was achieved by TCDD-induced recruitment of AHR to regions already bound by ERα.
AHR and ERα are known to regulate transcription through tethering to other transcription factors such as activating protein 1 (AP-1), stimulating protein 1 (Sp-1), and nuclear factor kappa B (Gillesby et al., 1997; Kobayashi et al., 1996; Paech et al., 1997; Saville et al., 2000; Tian et al., 1999; Zhou et al., 2005). The binding of AHR to ER-regulated genes is an important mechanism by which AHR inhibits estrogen-responsive gene expression (Wormke et al., 2003). It is thought to occur through direct competition for DNA binding with ERα at endogenous EREs, competition for DNA binding to GC-rich sites between AHR, AP-1 and/or Sp-1 transcription factors or AHR recruitment may interfere with the proper assembly of the preinitiation complex (Gillesby et al., 1997; Klinge et al., 1999; Wang et al., 2001). However, the AHRE core pentanucleotide (GCGTG) located in the upstream regulatory region of estrogen-responsive genes has been reported to be critical for the AHR-mediated antiestrogenicity (Safe et al., 1998). In agreement with these findings, we observed an overrepresentation of AHREs in the intersect set (shared genomic regions between AHR and ERα). Moreover, TCDD induced the recruitment of AHR to an AHRE located in the upstream regulatory region of ESR1 and caused a slight decrease in ERα mRNA levels, revealing ESR1 to be a direct AHR target gene. This result supports studies in rodents where TCDD treatment reduced ERα mRNA expression in the liver, ovary, and uterus of treated mice (Tian et al., 1998). AHR was also recruited to upstream regulatory regions of GREB1 and resulted in a slight reduction in GREB1 mRNA levels; however, an ERE but no AHREs were identified in this region. It is possible that AHR modulates ERα activity through distal regulatory regions of GREB1, which were not represented on our promoter focused array. Genome-wide analysis of ERα-binding sites revealed that ERα regulates GREB1 through distal enhancer elements 100 kb upstream of the start site (Carroll et al., 2006). Recruitment of AHR to GREB1 but not to ESR1 was dependent on ERα expression, suggesting that for some genomic sequences ERα influences the recruitment of AHR to those regions.
The ChIP-chip assays described in the present study were done at only a single time point in one cell type using promoter focused microarrays limiting our analysis to the regions represented on the arrays. Emerging data indicate that complete genomic binding profiles for sequence-specific DNA-binding proteins cannot be obtained from one ChIP-chip experiment in a single cell line or tissue (John et al., 2008; Krum et al., 2008). For example, ligand-dependent recruitment of ERα and AHR exhibit oscillatory recruitment to their target regions (Shang et al., 2000; Wihlen et al., 2009), which may not occur with the same kinetics for all ERα- and AHR-bound regions. Moreover, activation of AHR or ERα by different ligands that produce different receptor conformations of either receptor might produce a distinct set of receptor-bound regions from those identified in our study. We and others have shown that other AHR ligands induce recruitment of ERα to CYP1A1 and CYP1B1, but it is not clear whether these ligands influence the recruitment profiles of AHR or ERα at other AHR-responsive loci. The data presented herein and the recently reported ChIP-chip analysis of AHR binding in Hepa1c1c7 mouse hepatoma cells (Sartor et al., 2009) will help to build a map of the genomic binding profiles of AHR and add to the existing map for ERα (Carroll et al., 2005, 2006; Laganiere et al., 2005; Liu et al., 2008). A more comprehensive genomic binding profile for either of these factors will require genome-wide and temporal analysis in a variety of cell types.
The molecular mechanism and physiological significance of the co-occupancy of AHR and ERα are unknown. TCDD does not directly bind to ERα (Matthews et al., 2007) nor does ERα interact with AHREs (Klinge et al., 1999); thus, the precise mechanism by which TCDD induces recruitment of ERα to AHR target genes remains elusive. We have previously suggested that the recruitment of ERα to AHR-regulated genes is a mechanism by which AHR inhibits ERα activity by diverting it away from estrogen target genes through facilitated recruitment to AHR target genes (Matthews and Gustafsson, 2006; Matthews et al., 2005). Alternatively, the recruitment of ERα to activated AHR might target ERα for AHR-mediated proteolytic degradation by an E3 ligase ubiquitination pathway and thus contributing to the well-documented inhibitory action of AHR on ERα activity (Matthews and Gustafsson, 2006; Ohtake et al., 2007; Safe and Wormke, 2003). However, expression-based microarray analysis has shown that AHR-mediated inhibition of estrogen-regulated genes occurs at some but not all estrogen-responsive genes (Boverhof et al., 2008). These findings coupled with the evidence presented here would argue against proteolytic degradation of ERα as the sole antiestrogenic activity of AHR since it would be expected to completely reduce estrogen activity but rather our results support a gene-specific inhibition of ER activity.
We observed that knockdown of ERα reduced the TCDD-dependent induction of CYP1B1 and CYP1A1, whereas the TCDD-dependent regulation of other AHR target genes were unaffected. E2-dependent induction of CYP1A1 and CYP1B1 has also been observed (Frasor et al., 2004), and ERα has recently been shown to be an important factor in the elongation of RNA polymerase II at the CYP1B1 promoter (Kininis et al., 2007). In support of these findings, we observed reduced TCDD-mediated induction of CYP1B1 and CYP1A1 after siRNA-mediated knockdown of ERα. However, knockdown of AHR also significantly reduced recruitment of ERα to CYP1B1 in response to TCDD and AHR agonists strongly induce CYP1B1 expression in ERα-negative cell lines (Angus et al., 1999). Thus, the role of ERα in CYP1B1 expression is influenced by cell type, culture conditions, and AHR expression levels. However, knockdown of ERα did not affect the TCDD-dependent induction of target gene expression at the other loci examined. These findings suggest that interpretation of AHR-ERα crosstalk from the analysis of CYP1A1 and CYP1B1 regulation is not representative of all AHR-regulated genes. These data also show that recruitment of ERα to AHR-regulated target genes does not necessarily equate to changes in AHR-mediated transcription.
Collectively, our findings reveal not only that dioxin-type environmental contaminants modulate ERα-binding patterns at the genomic level but also show that ERα and AHR co-occupy several target genes in response to TCDD. It will be crucial to extend this analysis to nontumorogenic and other breast cancer cell lines, as well as appropriate transgenic mouse models and human breast cancer tissue samples to determine the physiological significance of this crosstalk at a genome-wide level.
Supplementary Material
Acknowledgments
S.A. is supported by an Ontario Graduate Student Scholarship and a Canadian Institute of Health Research Doctoral Research Award. J.M. is the recipient of a Canadian Institute of Health Research New Investigator Award.
FUNDING
Canadian Institute of Health Research operating grant (MOP-82715) to J.M., E.V, and A.S were supported by a grant from the Novo Nordisk Foundation to the Bioinformatics Center. The European Research Council has provided financial support to A.S. under the Community’s Seventh Framework Program (FP7/2007–2013) European Research Council (204135).
Footnotes
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
References
- Angus WG, Larsen MC, Jefcoate CR. Expression of CYP1A1 and CYP1B1 depends on cell-specific factors in human breast cancer cell lines: Role of estrogen receptor status. Carcinogenesis. 1999;20:947–955. doi: 10.1093/carcin/20.6.947. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Williams KJ, Zacharewski TR. Inhibition of estrogen-mediated uterine gene expression responses by dioxin. Mol Pharmacol. 2008;73:82–93. doi: 10.1124/mol.107.040451. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Castles CG, Oesterreich S, Hansen R, Fuqua SA. Auto-regulation of the estrogen receptor promoter. J Steroid Biochem Mol Biol. 1997;62:155–163. doi: 10.1016/s0960-0760(97)00023-x. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Kim HT, Hilsenbeck S, Cuba V, Tsimelzon A, Brown PH. Global gene expression analysis of estrogen receptor transcription factor cross talk in breast cancer: Identification of estrogen-induced/activator protein-1-dependent genes. Mol Endocrinol. 2005;19:362–378. doi: 10.1210/me.2004-0267. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Finlin BS, Gau CL, Murphy GA, Shao H, Kimel T, Seitz RS, Chiu YF, Botstein D, Brown PO, Der CJ, et al. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J Biol Chem. 2001;276:42259–42267. doi: 10.1074/jbc.M105888200. [DOI] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: Discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- Gillesby BE, Stanostefano M, Porter W, Safe S, Wu ZF, Zacharewski TR. Identification of a motif within the 5′ regulatory region of pS2 which is responsible for AP-1 binding and TCDD-mediated suppression. Biochemistry. 1997;36:6080–6089. doi: 10.1021/bi962131b. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, et al. Estrogen receptors: How do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wong WH. TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinehara M, Fukuda I, Yoshida K, Ashida H. High-throughput evaluation of aryl hydrocarbon receptor-binding sites selected via chromatin immunoprecipitation-based screening in Hepa-1c1c7 cells stimulated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Genes Genet Syst. 2008;83:455–468. doi: 10.1266/ggs.83.455. [DOI] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood KL, Homick K, Dragon MB, Bradford PG. Cloning and characterization of the type I inositol 1,4,5-trisphosphate receptor gene promoter. Regulation by 17beta-estradiol in osteoblasts. J Biol Chem. 1997;272:22425–22431. doi: 10.1074/jbc.272.36.22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM, Bowers JL, Kulakosky PC, Kamboj KK, Swanson HI. The aryl hydrocarbon receptor (AHR)/AHR nuclear translocator (ARNT) heterodimer interacts with naturally occurring estrogen response elements. Mol Cell Endocrinol. 1999;157:105–119. doi: 10.1016/s0303-7207(99)00165-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Sogawa K, Fujii-Kuriyama Y. Cooperative interaction between AhR. Arnt and Sp1 for the drug-inducible expression of CYP1A1 gene. J Biol Chem. 1996;271:12310–12316. doi: 10.1074/jbc.271.21.12310. [DOI] [PubMed] [Google Scholar]
- Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. Unique ERalpha cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22:2393–2406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Strom A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gao H, Marstrand TT, Strom A, Valen E, Sandelin A, Gustafsson JA, Dahlman-Wright K. The genome landscape of ERalpha- and ERbeta-binding DNA regions. Proc Natl Acad Sci U S A. 2008;105:2604–2609. doi: 10.1073/pnas.0712085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marstrand TT, Frellsen J, Moltke I, Thiim M, Valen E, Retelska D, Krogh A. Asap: A framework for over-representation statistics for transcription factor binding sites. PLoS ONE. 2008;3:e1623. doi: 10.1371/journal.pone.0001623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J, Gustafsson JA. Estrogen receptor and aryl hydrocarbon receptor signaling pathways. Nucl Recept Signal. 2006;4:e016. doi: 10.1621/nrs.04016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J, Wihlen B, Heldring N, MacPherson L, Helguero L, Treuter E, Haldosen LA, Gustafsson JA. Co-planar 3,3′,4,4′,5-pentachlorinated biphenyl and non-co-planar 2,2′,4,6,6′-penta-chlorinated biphenyl differentially induce recruitment of oestrogen receptor alpha to aryl hydrocarbon receptor target genes. Biochem J. 2007;406:343–353. doi: 10.1042/BJ20070585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl hydrocarbon receptor-mediated transcription: Ligand-dependent recruitment of estrogen receptor alpha to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Mol Cell Biol. 2005;25:5317–5328. doi: 10.1128/MCB.25.13.5317-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J, Wihlen B, Tujague M, Wan J, Strom A, Gustafsson JA. Estrogen receptor (ER) beta modulates ERalpha-mediated transcriptional activation by altering the recruitment of c-Fos and c-Jun to estrogen-responsive promoters. Mol Endocrinol. 2006;20:534–543. doi: 10.1210/me.2005-0140. [DOI] [PubMed] [Google Scholar]
- McLachlan JA, Simpson E, Martin M. Endocrine disrupters and female reproductive health. Best Pract Res Clin Endocrinol Metab. 2006;20:63–75. doi: 10.1016/j.beem.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Gustafsson JA. Estrogen receptor transcription and transactivation: Basic aspects of estrogen action. Breast Cancer Res. 2000;2:360–366. doi: 10.1186/bcr81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- Okey AB. An aryl hydrocarbon receptor odyssey to the shores of toxicology: The Deichmann Lecture, International Congress of Toxicology-XI. Toxicol Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- Okino ST, Quattrochi LC, Pookot D, Iwahashi M, Dahiya R. A dioxin-responsive enhancer 3′ of the human CYP1A2 gene. Mol Pharmacol. 2007;72:1457–1465. doi: 10.1124/mol.107.039826. [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- Petrulis JR, Perdew GH. The role of chaperone proteins in the aryl hydrocarbon receptor core complex. Chem Biol Interact. 2002;141:25–40. doi: 10.1016/s0009-2797(02)00064-9. [DOI] [PubMed] [Google Scholar]
- Puga A, Maier A, Medvedovic M. The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol. 2000;60:1129–1142. doi: 10.1016/s0006-2952(00)00403-2. [DOI] [PubMed] [Google Scholar]
- Puga A, Marlowe J, Barnes S, Chang CY, Maier A, Tan Z, Kerzee JK, Chang X, Strobeck M, Knudsen ES. Role of the aryl hydrocarbon receptor in cell cycle regulation. Toxicology. 2002;181–182:171–177. doi: 10.1016/s0300-483x(02)00276-7. [DOI] [PubMed] [Google Scholar]
- Safe S. Toxicology, structure-function relationship, and human and environmental health impacts of polychlorinated biphenyls: Progress and problems. Environ Health Perspect. 1993;100:259–268. doi: 10.1289/ehp.93100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Wang F, Porter W, Duan R, McDougal A. Ah receptor agonists as endocrine disruptors: Antiestrogenic activity and mechanisms. Toxicol Lett. 1998;102–103:343–347. doi: 10.1016/s0378-4274(98)00331-2. [DOI] [PubMed] [Google Scholar]
- Safe S, Wormke M. Inhibitory aryl hydrocarbon receptor-estrogen receptor alpha cross-talk and mechanisms of action. Chem Res Toxicol. 2003;16:807–816. doi: 10.1021/tx034036r. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, et al. Functional and physical interactions between the estrogen receptor Sp1 and nuclear aryl hydrocarbon receptor complexes. Environ Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville B, Wormke M, Wang F, Nguyen T, Enmark E, Kuiper G, Gustafsson JA, Safe S. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Sp1) promoter elements. J Biol Chem. 2000;275:5379–5387. doi: 10.1074/jbc.275.8.5379. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- Tan Z, Huang M, Puga A, Xia Y. A critical role for MAP kinases in the control of Ah receptor complex activity. Toxicol Sci. 2004;82:80–87. doi: 10.1093/toxsci/kfh228. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem. 1999;274:510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Thomas T, Meeker RJ, Gallo MA. Transcriptional suppression of estrogen receptor gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) J Steroid Biochem Mol Biol. 1998;67:17–24. doi: 10.1016/s0960-0760(98)00067-3. [DOI] [PubMed] [Google Scholar]
- Tian Y, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions: Mechanisms and physiological implications. Chem Biol Interact. 2002;141:97–115. doi: 10.1016/s0009-2797(02)00068-6. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Kyo S, Kanaya T, Inoue M, Yokoi T. Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 2004;64:3119–3125. doi: 10.1158/0008-5472.can-04-0166. [DOI] [PubMed] [Google Scholar]
- Wang F, Hoivik D, Pollenz R, Safe S. Functional and physical interactions between the estrogen receptor Sp1 and nuclear aryl hydrocarbon receptor complexes. Nucleic Acids Res. 1998;26:3044–3052. doi: 10.1093/nar/26.12.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Samudio I, Safe S. Transcriptional activation of cathepsin D gene expression by 17beta-estradiol: Mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Cell Endocrinol. 2001;172:91–103. doi: 10.1016/s0303-7207(00)00379-8. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Wihlen B, Ahmed S, Inzunza J, Matthews J. Estrogen receptor subtype- and promoter-specific modulation of aryl hydrocarbon receptor-dependent transcription. Mol Cancer Res. 2009;7:977–986. doi: 10.1158/1541-7786.MCR-08-0396. [DOI] [PubMed] [Google Scholar]
- Wormke M, Stoner M, Saville B, Walker K, Abdelrahim M, Burghardt R, Safe S. The aryl hydrocarbon receptor mediates degradation of estrogen receptor alpha through activation of proteasomes. Mol Cell Biol. 2003;23:1843–1855. doi: 10.1128/MCB.23.6.1843-1855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Solomon S, Fraser LR, Trombino AF, Liu D, Sonenshein GE, Hestermann EV, Sherr DH. Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue. J Cell Biochem. 2008;104:402–417. doi: 10.1002/jcb.21630. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Eppenberger-Castori S, Eppenberger U, Benz CC. The NFkappaB pathway and endocrine-resistant breast cancer. Endocr Relat Cancer. 2005;12(Suppl 1):S37–S46. doi: 10.1677/erc.1.00977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.