Abstract
Purpose
Esophageal cancer is one of the most aggressive and deadly forms of cancer; highlighting the need to identify biomarkers for early detection and prognostic classification. Our recent studies have identified inflammatory gene and microRNA signatures derived from tumor and nontumor tissues as prognostic biomarkers of hepatocellular, lung, and colorectal adenocarcinoma. Here, we examine the relationship between expression of these inflammatory genes and miRNA expression in esophageal adenocarcinoma and patient survival.
Experimental Design
We measured the expression of 23 inflammation-associated genes in tumors and adjacent normal tissues from 93 patients (58 Barrett's and 35 Sporadic adenocarcinomas) by quantitative reverse transcription-polymerase chain reaction. These data were used to build an inflammatory risk model, based on multivariate Cox regression, to predict survival in a training cohort (n=47). We then determined if this model could predict survival in a cohort of 46 patients. Expression data for miRNA-375 was available for these patients and was combined with inflammatory gene expression.
Results
IFNγ, IL-1α, IL-8, IL-21, IL-23, and PRG expression in tumor and nontumor samples were each associated with poor prognosis based on Cox regression ([Z-score]>1.5) and therefore, were used to generate an inflammatory risk score (IRS). Patients with a high IRS had poor prognosis compared to those with a low IRS in the training (P=0.002) and test (P=0.012) cohorts. This association was stronger in the group with Barrett's history. When combining with miRNA-375, the combined IRS/miR signature was an improved prognostic classifier than either one alone.
Conclusion
Transcriptional profiling of inflammation-associated genes and miRNA expression in resected esophageal Barrett's associated adenocarcinoma tissues may have clinical utility as predictors of prognosis.
Keywords: Inflammation, Cancer, Barrett's, Esophagus
INTRODUCTION
Esophageal cancer (EC) is one of the most lethal malignancies of the digestive tract, and at the time of diagnosis most patients are at an advanced stage (1). Despite advanced surgical techniques combined with various treatment modalities, such as radiotherapy and chemotherapy, the overall 5-year survival rate of EC remains at 15% to 24%, highlighting the need for identification of prognostic biomarkers and new therapeutic strategies (2).
There are two main histological subtypes of EC, squamous cell carcinoma (SCC) and adenocarcinoma (ADC). The incidence of ADC has increased remarkably over the past two decades and has supplanted SCC as the dominant phenotype in western countries (3). ADC frequently arises from Barrett's esophagus (BE), a chronic inflammatory condition characterized by a change in the normal esophagus epithelium into intestinal metaplasia as a result of gastroesophageal reflux (4–5) and genomic instability (6).
Both epidemiologic and functional studies have implicated chronic inflammation in the development of many human cancers, including liver, esophagus, stomach, colon, and bladder cancer (7). During this process, deregulated cytokine production and signaling between the immune and neoplastic cells can alter cell growth, differentiation, and apoptosis (8–9). We and others have reported that inflammation-associated gene signatures can be used as indicators of prognosis and tumor burden in liver (10), lung (11), colorectal cancer patients (12–14).
Several studies demonstrate that expression of inflammatory genes is associated with esophageal cancer progression and prognosis (15–20). In addition, experimental evidence indicates that tumor cells can produce cytokines to enhance their growth and to counteract the host immune response (21–22). Most of these studies, however, examined relatively few genes and focused on patients with SCC, with far less published information on ADC. Furthermore, there are also limited data regarding the association between inflammation and prognosis in patients who receive preoperative adjuvant therapy.
Although inflammation-related cytokines have shown as potential diagnostic and prognostic markers, emerging data suggest that miRNAs also provide independent prognostic information in cancer patients. MiRNAs are small, noncoding RNA molecules that its expression signatures have been associated with the etiology and prognosis of many cancer types (23). Detection of miRNAs in patients with cancer has been shown to be associated with cancer progression and shortened survival (24). For example, we recently reported that Barrett's associated adenocarcinoma patients with reduced expression of miR-375 in cancerous tissue had worse prognosis (25) and tumor expression of miR-16-2, miR-30e, and miR-200a were individually associated with shorter overall and disease-free survival in esophageal adenocarcinoma patients (26).
Combining a marker of miRNA-with cytokines in a multimarker strategy appears to be a useful approach for improving risk assessment in cancer patients. Colorectal cancer patients with increased miR-21 expression and a high inflammatory risk score (IRS) have been shown to have an increased mortality risk compared with those with both reduced miR-21 expression and lower IRS (12). Integrating this multimarker approach into the routine assessment of colorectal cancer patients may allow clinicians to more accurately identify high-risk patients who may derive increased benefit from intensive management strategies.
These findings support a model that the tumor inflammatory microenvironment combined with miRNAs can be a prognostic biomarker for esophageal cancer patients. In this study, we examined the expression of 23 inflammatory genes and use these data to develop an IRS that could be used as a prognostic classifier of ADC patients. We then used our previously reported data of miR-375 to determine if the combination between this miRNA and IRS could be a better prognostic classifier than either alone. The possible role of multimarker gene signatures as a prognostic classifier of response to neoadjuvant chemo-radiation therapy has also been examined.
PATIENTS AND METHODS
Clinical Samples
Our cohort comprised 93 patients who underwent surgical resection of primary esophageal ADC between 1993 and 2001 at the University of Maryland Medical Center, Baltimore Veterans Administration Medical Center (Baltimore, Maryland, USA) and the Queen Elizabeth II Health Science Centre (Halifax NS, Canada). The use of specimens from these centers was approved by Institutional Review Boards. All patients provided written consent, and samples were collected after surgical resection. The nontumor samples were taken from tumor-free margins of resected tumors and were histologically normal. Histopathological diagnoses of Barrett's associated adenocarcinoma (BAC) were verified on the basis of H&E-stained sections according to the Vienna Classification of gastrointestinal epithelial neoplasia (27). Diagnoses of Barrett's esophagus were made if intestinal metaplasia was found histologically. All BACs originated in the lower esophagus or gastro-esophageal junction, corresponding to ADC of the esophageal-gastric junction type. Tumor stage at the time of diagnosis and surgery was based on 2004 World Health Organization classification guidelines. Forty patients in the U.S.A. cohort received induction chemo-radiation therapy, comprising Cisplatin and 5-Fluorouracil concurrent with 45–50.4 Gy external beam radiation. Response to therapy was assessed according to Response Evaluation Criteria in Solid Tumors criteria (28). Cancer-specific mortality was used as the endpoint for all analyses.
RNA Isolation and Quantitative Reverse Transcription- Polymerase Chain Reaction Analysis (qRT-PCR)
Surgical resected specimens used for RNA extraction were transported immediately to the laboratory and a senior pathologist provided representative sections of tumor and adjacent nontumor tissues, which were immediately snap-frozen in liquid nitrogen. Paraffin-embedded slides were H&E stained to confirm histological diagnosis and determine tumor cellularity. Most samples have minimal 80% cancer cells with little or no necrosis. Total RNA was isolated using standard TRIZOL (Invitrogen, Carlsbad, CA) and then reverse-transcribed using AB High-Capacity cDNA Archive Kit (Applied Biosystems or AB, Foster City, CA). Expression levels for individual inflammatory genes were evaluated in duplicate using Taqman Gene Expression Assays (AB). Primers for 23 genes were purchased from AB, including interleukin 1α (IL-1α, assay ID Hs00174092_m1), IL-1β (ID Hs00174097_m1), IL-2 (ID Hs00174114_m1), IL-5 (D Hs00174200_m1), IL-6 (ID Hs00174131_m1), IL-8 (ID Hs00174103_m1), IL-10 (ID Hs00174086_m1), IL-12p35 (ID Hs00168405_m1), IL-12p40 (ID Hs00233688_m1), IL-15 (ID Hs00542571_m1), IL-17 (ID Hs00174383_m1), IL-18 (ID Hs00155517_m1), IL-21 (ID Hs00222327_m1), IL-22 (ID Hs00220924_m1), IL-23 (ID Hs00372324_m1), IL-24 (ID Hs01114274_m1), interferon gamma (IFN-γ, assay ID Hs00174143_m1) and tumor necrosis factor-α (TNF- α, assay ID Hs00174128_m1), the major histocompatibility complex class II antigens (HLA-DRA1, assay ID Hs00219575_m1; HLA-DPA1, assay ID Hs00410276_m1), annexin A1 (ANXA1, assay ID Hs00167549_m1), platelet proteoglycan (PRG1, assay ID Hs00160444_m1), colony stimulating factor 1 (CSF1, assay ID Hs00174164_m1), and 18S ribosomal RNA (18S rRNA, assay ID Hs99999901_s1). Double-stranded cDNA for each patient sample was amplified for 40 cycles using the TaqMan Universal PCR Master Mix using the manufacturer's protocol on the 7500HT Sequence Detection System (AB). For quality control, any samples with either an 18S rRNA cycle value >20 or a gene cycle value >36 were considered of poor quality and removed. In addition, a patient was removed from the analysis if either its tumor or its matched nontumor sample failed quality control. The tumor:nontumor ratios were then calculated using the comparative method (2−ΔΔCt), where Ct = threshold cycle and ΔΔCt = (Ct gene tumor – Ct 18S rRNA tumor) - (Ct gene nontumor – Ct 18S rRNA nontumor), as previously described (12).
MiRNA Measurement
MicroRNA-375 expression level was previously measured in all patient samples and was described in detail there (25). Briefly, miRNA-375 expression level was measured by quantitative reverse transcriptase-PCR (qRT-PCR) in triplicate using Taqman microRNA assays (Applied Biosystems) according to the manufacturer's instructions. High or low tumor miR-375 expression was defined based on the median expression separately done for each cohort.
Statistical Analysis
BRB ArrayTools version 3.6.0 (NCI, NIH, Bethesda, MD) was used to explore the differential expression between tumor samples and their matched nontumor samples. A differentially expressed gene was considered to be up- or down-regulated if P ≤0.05 and False Discovery Rate (FDR) ≤5%, using the Wilcoxon matched pairs signed rank test.
The association between IRS, miRNA and survival was analysed by Kaplan – Meier and assessed for significance by the log-rank test using WINSTAT 2007 (R Fitch Software, Bad Krozingen).
All samples were randomly divided into a training or test cohort to identify a gene expression model associated with cancer-specific mortality. There were similar numbers for each sub-groups presented in both the training and test cohorts. Univariate Cox regression analysis on the training cohort was used to select genes associated with cancer-specific mortality (|Z-score| >1.5; P < 0.13) to include in multivariate risk models using previously reported methods (12). All genes were included for these purposes, and expression values for all analyses are continuous variables. For multivariate Cox regression models, missing values for genes were replaced with the average values. In the training cohort, selected genes were used to build multivariate models using tumor and nontumor ratios. Coefficients from these models were multiplied with gene expression values and summed to build risk scores. Individuals were defined as high risk if they had highest tertile scores for combined tumor and noncancerous ratios, separately for each cohort to account for possible confounding factors in clinical management from different institutions. Cox regression was carried out in Stata 9.2 (StataCorp LP). For survival analysis, only the year of diagnosis was used as a continuous variable, and all of the other variables were used as categorical variables. The proportional hazard assumption tested using Schoenfeld residual was met for all models reported. Multivariate models were based on stepwise addition and removal of clinical covariates found to be associated with poor survival in univariate models (P<0.10). A Wald statistic of P<0.05 was used as the criterion for inclusion in final multivariate models. All P values reported are 2-sided. Correlation analysis between miR-375 expression and cytokine genes was done using Stata 9.2 (StataCorp LP).
RESULTS
Differential Gene Expression in Esophageal Cancer
Ninety three patients (66 U.S.A. and 27 Canadians) were recruited into this study and were randomly assigned to a training cohort (n=47) or a test cohort (n=46) (Table 1). The median follow-up times were 72 months for both cohorts. Both cohorts were similar in age, gender, race, TNM stage, Barrett's, neoadjuvant treatment status, smoking, drinking status, and cancer-specific mortality (Table 1).
Table 1.
Characteristics of the study population
| Training cohort (n = 47) | Test cohort (n = 46) | P value¶ | |
|---|---|---|---|
| Age at surgery (years) | |||
| Mean (SD) | 61.7 (10.6) | 62.7 (10.4) | |
| Range | 41–79 | 32–80 | |
| Nationality, no. (%) | |||
| U.S.A | 34 (72) | 32 (70) | 1.0 |
| Canada | 13 (28) | 14 (30) | |
| Gender, no. (%) | |||
| Male | 43 (91) | 40 (87) | 0.8 |
| Female | 4 (9) | 6 (13) | |
| Race, no. (%) | |||
| White | 46 (98) | 45 (98) | 1.0 |
| Non-white | 1 (2) | 1 (2) | |
| TNM staging,* no. (%) | |||
| 0 | 10 (21) | 10 (21) | 0.48 |
| I | 10 (21) | 9 (20) | |
| II | 11 (23) | 12 (26) | |
| III | 13 (28) | 12 (26) | |
| IV | 3 (7) | 3 (7) | |
| History of Barret's esophagus,§no. (%) | |||
| Yes | 27 (57) | 31 (67) | 0.68 |
| No | 20 (43) | 15 (33) | |
| Neoadjuvant treatment (CRT),¥no. (%) | |||
| Yes | 22 (47) | 18 (39) | 0.94 |
| No | 25 (53) | 28 (61) | |
| Drinking history, no. (%) | |||
| Yes | 32 (68) | 33 (72) | 0.64 |
| No | 8 (17) | 9 (19) | |
| Unknown | 7 (15) | 4 (9) | |
| Smoking history, no. (%) | |||
| Yes | 30 (64) | 31 (67) | 0.94 |
| No | 9 (19) | 8 (17) | |
| Unknown | 8 (17) | 7 (16) |
TNM staging at the time of surgery was based on the 2004 World Health Oganization classification
Diagnosis of Barrett's esophagus was verified on the basic of H&E stained section according to the Vienna Classification of gastrointestinal epithelial neoplasia
CRT composed of Cisplatin and 5-Fluorouracil concurrent with 45-50.4 Gy external beam radiation
Fisher's exact test
We measured the expression of 23 inflammation related genes in all 93 tumor and paired non-tumor tissues using qRT-PCR (Supplemental Table S1). Nineteen of these genes were selected from the previous studies of hepatocellular (10) lung (11) and colon carcinoma (12). We added 4 more genes (IL-18, IL-21 IL-22, and IL-24) based on literature supporting their roles in esophageal carcinogenesis (29–33).
Inflammatory genes were differentially expressed between tumor and the adjacent nontumor tissues. Three genes — IL-8, IL-21, and IL-24 — were significantly increased in tumors, whereas expression of six genes — ANXA1, IL-1α, IL-2, IL-12p35, IL-18, and TNFα — were significantly decreased within the ADC tumor (P≤0.05; paired t-test; FDR ≤5%; Supplemental Table S2). Of note, the top differentially expressed genes between tumor and nontumor were — IL-24 (9-fold increase) and ANXA1 (12-fold decrease). These findings reflect the possible diverse effects that these inflammatory genes can have in the tumor microenviroment of esophageal cancer cells.
Inflammatory Risk Score and Cancer-Specific Mortality in the Training Cohort
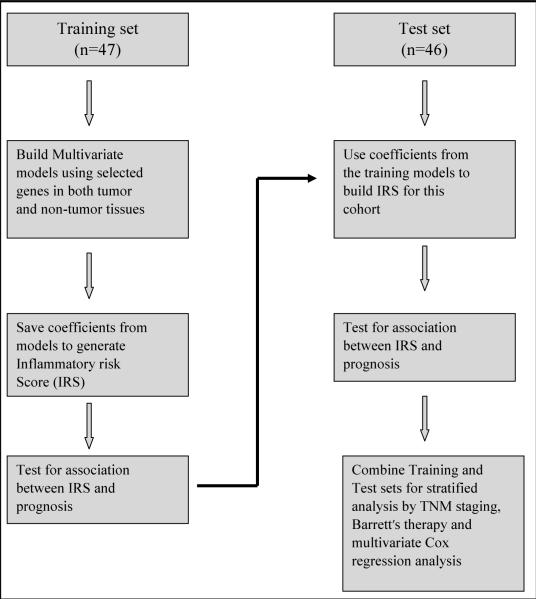
Similar to our previous analysis in colon cancer (12), we constructed a multi-gene signature using several genes with moderate association with prognosis in an attempt to develop an inflammatory risk score (IRS) in the training cohort which was then validated using the test cohort. A flow chart of this algorithm is shown in Figure 1. First, the 93 specimens (66 U.S.A. and 27 Canadians) were randomly assigned to a training set (n=47) or a test set (n=46; Table 1). We then applied the Cox proportional hazard regression to each of the 23 inflammatory genes in the training cohort to identify essential genes that were correlated with cancer-related death. We identified six genes — IFN-γ, IL-1α, IL-21, IL-23 (strong association), IL-8, and PRG (moderate association) — that were associated with patient survival {|Z-score| >1.5; Supplemental Fig. S1}. We then used these six genes to construct a signature by the risk score method. A risk-score formula (IRS) was calculated as [(0.079 × IFN-γ) + (0.085 × IL-1α) + (0.299 × IL-21) + (0.072 × IL-23) + (0.049 × IL-8) − (0.01 × PRG)]. This risk-score method was used to calculate the IRS for all patients in the training cohort. We then ranked patients in the training cohort according to their IRS and divided them into a high-risk group (high IRS) and a low-risk group (low IRS) using the upper tertile risk-score as the cut off point. Patients with higher than the upper tertile IRS were considered as having a high IRS, while all others were considered low IRS. In the context of survival, we found patients with elevated IRS had a significantly shorter survival than patients with a low IRS (P=0.00026, Kaplan-Meier log rank; Training cohort; Fig. 2A).
Fig. 1.
Model for building inflammatory risk scores. Genes chosen for IRS in the training set was selected based on univariate Cox regression. Multivariate Cox regression was used to build the IRS model. This model was then tested on an independent cohort (test cohort).
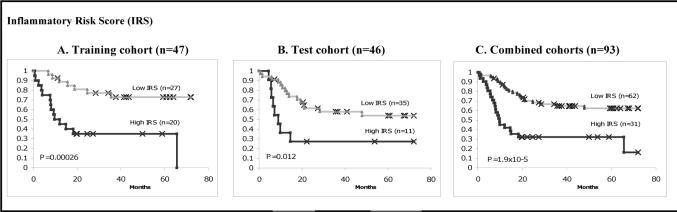
Fig. 2.
High IRS is associated with poor prognosis in the training, test and a combined cohort.
High IRS expression was classified based on the highest tertile using tumor and nontumor ratio. Am The association of high IRS with poor prognosis in the training cohort. B, High IRS associated with poor survival was validated in a test cohort. C, The association of high IRS and poor survival in a combined cohort. Log-rank P values are from Kaplan-Meier analysis.
Validation of IRS as a Prognostic Classifier in the Test Cohort
To validate these findings in our test cohort, we used the same risk-score formula obtained from the training cohort to calculate IRS for each of the 46 patients in the test cohort. We then dichotomized them into the high-risk group (high IRS) and the low risk-group (low IRS) using the same cut off point as in the training set. The test cohort was similar in all clinical characteristics to the training cohort (Table 1). Similar to what was observed in the training cohort, patients with elevated IRS had poor prognosis than patients with low IRS (P=0.01, Kaplan-Meier log rank; Fig. 2B). Given the similarity seen in both the training and test cohorts, we decided to combine these two cohorts for any subgroup analysis. Likewise, patients with high IRS associated with poor survival than the ones with low IRS (P=1.9 × 10−5; Kaplan-Meier log rank; Fig. 2C).
IRS Is Associated with Patient Survival within Histological Subgroups
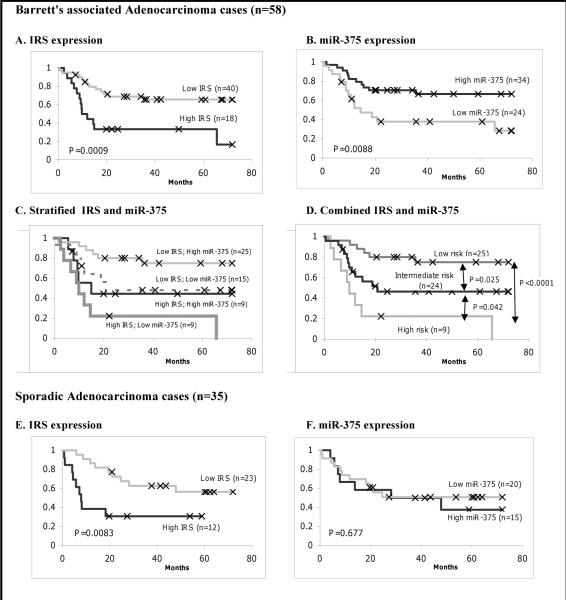
Since ADC can arise in very different biological settings, such as in the presence or absence of Barrett's esophagus (BE), we next examined the relationship between BAC (58 of 93 patients = 62%) and these inflammatory genes. Of all the genes tested, none was differentially expressed at a statistically significant level between BAC and SAC (Supplemental Table S3). In the context of survival, we found BAC patients with elevated IRS had significantly poor prognosis than those with low IRS (P=0.0009, Kaplan-Meier log rank; Fig. 3A). We observed a similar trend with SAC patients (P=0.0083, Kaplan-Meier log rank; Fig. 3E)
Fig. 3.
Combined IRS and miR-375 expression predicts esophageal cancer-specific mortality better than either alone. Graphs are Kaplan-Meier analysis between expression and survival.
For Barrett's associated adenocarcinoma: A, Kaplan-Meier (KM) analysis for IRS. B, KM for miR-375 C, stratified IRS and miR-375 together. D, combined IRS and miR-375. For sporadic adenocarcinoma: E, KM for IRS. F. KM for miR-375. High IRS expression was classified based on the highest tertile using tumor and nontumor ratio. High miR-375 expression was defied according to the higher than median expression in tumor samples.
Next, we examined the expression of these genes in a subset of 53 ADC patients in the combined cohort (n=93) that did not receive neoadjuvant treatment (NAT). This allows an assessment of the prognostic impact of these inflammatory genes, independent of any effects of therapy. In comparison with patients who had low IRS, i.e., below the upper tertile, those with high IRS experienced a statistically significant reduction in cancer-specific survival (P=3.3×10−6, Kaplan-Meier log rank; Supplemental Figure. 2A).
In our combined cohort, 40 of 93 patients (43%) received preoperative chemo-radiation therapy, of which 15 (32.5%), 10 (25%), and 15 (32.5%) patients showed pathological complete, partial, and non-respondent to treatment, respectively. In the case of complete responders, there were still microscopic tumors seen on the slides. Kaplan-Meier showed that elevated IRS was associated with shorter survival, although it was not statistically significant (Supplemental Figure. 2B). In addition, minimal TNR difference among IFN-γ, IL-1α, IL-21, IL-23, IL-8, and PRG was observed between NAT and NAT-naïve patients (Supplemental Table S4). Furthermore, NAT patients did not receive any survival benefit compared to NAT-naïve patients [(hazard ratio (HR), 1.83; 95% confidence interval (95% CI), 0.80–4.16; p = 0.15]. In summary, high IRS was associated with poor prognosis and a similar trend was observed for both NAT-naïve and BAC patients.
Combined IRS, and miR-375 is a Better Prognostic Classifier of Cancer-Specific Mortality in BAC Patients
Quantitative real-time polymerase chain reaction data for miR-375 expression was available for all 93 ADC tumor and nontumor samples from our previous study (25). We previously reported that reduced tumor expression of miR-375 was associated with poor prognosis in BAC, but not SAC patients (25). In this study, we showed that elevated IRS was associated with poor survival in ADC patients. Since both of these studies shared the same patient population, we asked whether or not combining these two signatures (IRS and miRNA) could improve prognostic utility of either method alone.
IRS and miRNA-375 were not associated with one another (P=0.402 for all cases or P=0.661 for all Barrett's cases, Fisher's exact test). Therefore, combination of these two markers might provide a good survival prediction power over either method alone. Furthermore, we used linear regression to examine the association of miR-375 expression with imflammatory genes in tumor and nontumor tissues. None showed to be either positively or negatively correlated with miR-375 expression in a statistical manner.
Next, we analyzed miR-375 in tumor samples of all patients using the median expression cut off where a high value was defined as having higher than the median value for all samples in the same cohort, consistent with our previous method (25). As we previously reported, low miR-375 expression was associated with poor survival in BAC patients (P=0.0088, Kaplan-Meier log rank; Fig. 3B). Given that no significant association between miR-375 and survival observed in SAC patients (P=0.677, Kaplan-Meier log rank; Fig. 3F), we decided to focus on BAC for further analysis from here on.
We then performed an analysis of IRS and miR-375 in the BAC group. Patients with high IRS/low miR-375 had the worst prognosis. Patients with either high IRS/high miR-375 or low IRS/low miR-375 had an intermediate prognosis. Patients with low IRS/high miR-375 had the best prognosis (Fig. 3C). From the survival curve shown in Figure 3C, we found that the survival curve for the low IRS/low miR-375 superimposed the curve for the high IRS/high miR-375. Therefore, we combined these two groups into a new group called “Intermediate risk”. The high IRS/low miR-375 was defined as “High risk” and the low IRS/high miR-375 was defined as “Low risk”. Patients classified as either high risk or low risk had significantly worst survival than the intermediate risk group (P=0.042 and P= 0.025, Kaplan-Meier log rank; respectively Fig. 3D). The “High risk” group was expected to have a shorter survival than the “Low risk” group (P=1.6×10−5, Kaplan-Meier log rank; Fig. 3D).
To further determine the additive predictive power of the combined method after accounting for all clinical variables in BAC patients, Cox proportional hazards analysis was performed using tumor stage, age, and therapy status in the 58 BAC patients cohort, which demonstrated that high IRS [hazard ratio (HR), 4.04; 95% confidence interval (95% CI), 1.80–9.09], low miR-375 (HR, 2.78; 95% CI, 1.27–6.10), and TNM stage (HR, 4.27; 95% CI, 1.78–10.21) were each associated with poor prognosis (Table 2; Univariate). Multivariate regression analysis indicated that both IRS (HR, 3.74; 95% CI, 1.62–8.63) and miR-375 (HR, 2.73; 95% CI, 1.17–6.39) were independent of one another and TNM staging. We found that the multivariate model including IRS, miR-375 and TNM staging was a significantly better prognostic classifier than a model of without IRS or a model without miR-375 (P = 0.003 and P = 0.026, likelihood ratio test; respectively). These results indicate that IRS and miR-375 have potential to be used in combination as a prognostic classifier of BAC, although this result requires validation in another cohort.
Table 2.
Cox regression analysis of IRS and miR-375 expression with cancer-specific mortality of Barrett's associated adenocarcinoma samples (n=58)
| Variables | Univariate | Multivariate* | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| IRS (high/low) | 4.04 (1.80–9.09) | 0.001 | 3.74 (1.62–8.63) | 0.002 |
| miR-375 (low/high) | 2.78 (1.27–6.10) | 0.011 | 2.73 (1.17–6.39) | 0.021 |
| Tumor stage (III–IV/I–II) | 4.27 (1.78–10.21) | <0.001 | 6.84 (2.40–19.51) | <0.001 |
| Age (<65/>65) | 0.86 (0.47–1.57) | 0.62 | ||
| Therapy (yes/no) | 1.83 (0.80–4.16) | 0.15 | ||
All analyses are adjusted for nationality (American vs. Canadian).
Multivariate analysis include IRS, miR-375 and TNM staging. Multivariate analysis used stepwise addition and removal of clinical covariates found to be associated with survival in univariate models (P<0.10) and final models include only those variables significantly associated with survival (Wald statics, P<0.05).
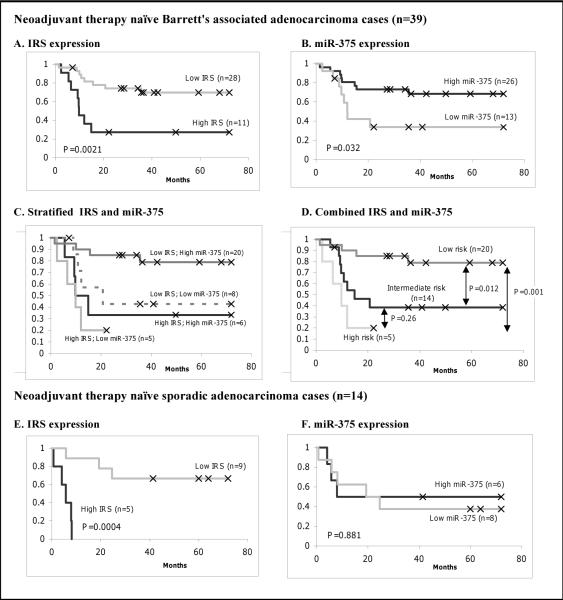
In this study, both IRS and miR-375 were significantly associated with cancer-specific mortality only in patients who did not receive any neoadjuvant therapy (NAT -naïve) (Figure 4). Therefore, we restricted our analysis to this group of NAT-naïve patients in order to investigate the association of IRS, miR-375 and survival independent of any therapeutic effects in BAC. In our combined cohort, 39 of 58 BAC patients did not receive any preoperative chemo-radiation therapy. Similar to what was seen before, either high IRS or low miR-375 was associated with poor survival (P=0.0021 and P=0.032, Kaplan-Meier log rank; Figure 4A–B; respectively). We then performed an analysis of IRS and miR-375 in the BAC NAT-naive group. Risk groups were defined as they were for the BAC analysis. Patients defined as low risk had the best prognosis (Figure 4C). Patients classified as high risk had the worst prognosis than low risk group (P=0.001; Kaplan-Meier log rank; Figure 4D).
Fig. 4.
Combined IRS and miR-375 expression predicts esophageal cancer-specific mortality in patients without any neoadjuvant treatment. Graphs are Kaplan-Meier analysis between expression and survival. For Barrett's associated adenocarcinoma: A, Kaplan-Meier (KM) analysis for IRS. B, KM for miR-375 C, stratified IRS and miR-375 together. D, combined IRS and miR-375. For sporadic adenocarcinoma: E, KM for IRS. F. KM for miR-375. High IRS expression was classified based on the highest tertile using tumor and nontumor ratio. High miR-375 expression was defined according to the higher than median expression in tumor samples.
Significant association in SAC NAT-naïve patients was observed between IRS and survival (P=0.0004, Kaplan-Meier log rank; Figure 4E), but not miR-375 and survival (P=0.881, Kaplan-Meier log rank; Figure 4F). In summary, we found significant association between IRS and miR-375 with survival in the BAC-NAT naïve patients. Therefore, combining IRS and miRNA expression might be used together as a prognostic classifier of Barrett's associated esophageal adenocarcinoma patients.
DISCUSSION
In this study, we analyzed 23 inflammation-associated genes in ADC patients with respect to survival. We identified gene expression signatures containing PRG, IL-1α, IL-8, IL-21, IL-23, IFN-γ, and miR-375 that was associated with survival in BAC patients. We confirmed these findings in a test cohort of ADC patients. Patients with a high IRS in their tumor and non-tumor samples and a low miR-375 in its tumors had shortened survival independent of other clinical and pathological parameters. These findings suggest that inflammatory genes and microRNA may play an important role in the molecular pathogenesis, clinical cancer progression, and prognosis of BAC patients.
Our findings here are consistent with other reports evaluating the connection between inflammation and cancer. Our previous studies support a role for inflammatory genes, in both tumor and nontumor tissues play important roles in tumor progression and metastasis. For example, a 17-cytokine gene signature (primarily composed of Th1 and Th2 cytokines) obtained from noncancerous hepatocellular carcinoma tissues predicted metastasis and recurrence (10). Similar gene signatures obtained from both tumor and non-tumor tissues were used to predict prognosis in lung and colon adenocarcinoma (11–12). In the present study, six of these inflammatory genes are associated with survival outcome in EC. This suggests that the patterns of inflammatory gene expression among lung, liver, colon, and esophagus are similar and support the mechanistic relationships between inflammation and carcinogenesis.
Our findings are also consistent with published studies on inflammation and EC. For example, elevated IFNγ expression was correlated with advanced SCC stage (20).
Cytokines such as IL-8 and IL-1β were markedly increased in biopsies from adenocarcinoma patients (15, 18). Metaplastic Barrett's epithelia cells were shown to produce IL-8, IL-10, and IL-1β (34). Other groups also found an association between Th1 inflammation and the pathogenesis of Barrett's dysplasia (35–36). For example, IL-1β enhances Barrett's metaplasia by increasing CDX1 (a regulator of intestinal epithelial development) (37).
ANXA1 and IL-24 identified in this study showed the highest fold-difference in tumors compared to nontumor tissues, suggesting their clinical utility as potential diagnostic markers. Although this finding does not address the causal relationship between inflammation and cancer, these changes are consistent with data in the current literature. For example, we showed in this study that ANXA1 had the largest reduction in esophageal tumors. Its expression has been shown to be reduced in a number of cancers, including those from the esophagus origin (38–40). ANXA1 is a mediator of apoptosis and inhibitor of cell proliferation in variety of cancer settings (41). Conversely, IL-24 had the largest fold-increase of all genes tested in our ADC samples. A newly discovered member of the IL-10 family, IL-24 was originally described in the setting of melanoma, where its function as a tumor suppressor (42). Its antitumor activities have since been expanded to include many different cancer models (43–44). To our knowledge, there are no published studies of IL-24 and esophageal cancer. Although our finding might appear counterintuitive, the anti-tumor effect of IL-24 is almost always tested in the setting of forced overexpression by the cancer cells themselves and the physiologic roles of IL-24 is not well understood. Whether IL-24 has any influence to the biology of ADC is beyond the scope of this study and would, therefore, warrant further investigation to access the etiologic role of IL-24 in esophageal carcinogenesis.
The current clinical-pathological staging method has limited success in predicting patient survival. For patients with identical clinical-pathological characteristics or different histological subtypes, great uncertainties remain regarding how some patients will be cured while other patients will have cancer recurrence, metastasis, or death after multi modalities treatment. Applying our IRS and miR-375 signature to the combined samples of the training and the test cohorts, we found a clear separation between the low- and the high risk curves. We showed that the combined signature can distinguish high-risk versus low-risk Barrett's patients only. Given the fact that the difference in survival between sporadic vs. Barrett's associated adenocarcinoma has rarely been examined before and thus is of great interest. This finding may potentially enable doctors to identify and select high-risk patients for effective therapy in addition to standard surgery in order to improve the treatment outcome of BAC patients.
Neoadjuvant therapy is now common practice in the treatment of EC given that more than 50% of patients present with bulky tumors and radiographic metastasis at the time of diagnosis (45). Although esophageal cancer patients may respond to NAT, overall survival remains generally poor with the exception of a subset of patients who achieve a complete pathologic response. However, there is currently no clinically useful indicator to select patients who should receive additional interventions. Unfortunately, because of the lack of pretreatment tissues in this study, we were unable to investigate therapeutic responses in NAT patients. In patients with resected tumors, particularly in the absence of any NAT, the major determinant of long-term cancer-specific survival is expected to be micrometastatic disease. Therefore, our observation that IRS and miR-375 were good prognostic classifiers in the BAC-NAT naïve patients was an important finding because it might help identify individuals that are likely to have micrometastasis and would therefore, require more aggressive adjuvant therapy. Although this specific observation requires an independent validation, the biomarkers defined here might aid in the identification of patients who did not received any neoadjuvant therapy and therefore, should be treated more aggressively post surgical resection.
A strength of this study lies in the fact that a small number of genes and miRNA was measured using real time qRT-PCR, a technique that is considered the most sensitive, specific, fast and economical RNA quantification method. This gene/miRNA set might be clinically applicable procedure. This methodology could be easily translated to the clinic by quantifying RNA from surgical specimens (46). A constraint of this study, on the other hand, is that there are still possibilities of false positives in the selection of these genes for the correlation of clinical outcome in ADC patients. Therefore, the potentially clinical application of this study should be examined in a larger independent cohort, although the validation of this signature in our test cohort provides confidence in the validity of this gene signature as a predictor of clinical outcome.
In conclusion, our finding provides evidence that elevated expression of imflammatory genes in association with miRNA could predict clinical outcome in an independent cohort. Our results suggest that this criteria defined here may have prognostic and therapeutic implications for the management of esophageal cancer patients.
TRANSLATIONAL RELEVANCE.
In this report, we provide evidence that elevated expression of imflammatory genes in tumor and paired non-tumor tissues was an independent prognostic marker for Barrett's associated esophageal adenocarcinoma patients. We also report that combining independent markers can be an improved prognostic classifier than either one alone. By combining the inflammatory gene signature and miRNA-375 expression, we could better predict clinical outcome of Barrett's associated adenocarcinoma patients. Our results suggest that this criteria defined here may have prognostic and therapeutic implications for the management of esophageal cancer patients.
Supplementary Material
Abbreviations
- EC
Esophageal cancer
- AC
adenocarcinoma
- BAC
Barrett's associated adenocarcinoma
- SAC
Sporadic adenocarcinoma
- NAT
neoadjuvant treatment
- TNR
tumor:nontumor ratios
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2007;57:43–6. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Rice TW, Adelstein DJ, Zuccaro G, et al. Advances in the treatment of esophageal carcinoma. Gastroenterologist. 1997;5:278–294. [PubMed] [Google Scholar]
- 3.Blot WJ, Devesa SS, Kneller RW, et al. Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA. 1991;265:1287–89. [PubMed] [Google Scholar]
- 4.DeMeester SR, DeMeester TR. Columnar mucosa and intestinal metaplasia of the esophagus: fifty years of controversy. Ann Surg. 2000;231:303–21. doi: 10.1097/00000658-200003000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell. 2004;6:11–6. doi: 10.1016/j.ccr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Izzo JG, Luthra R, Wu TT, et al. Molecular mechanisms in Barrett's metaplasia and its progression. Semin Oncol. 2007;34:S2–S6. doi: 10.1053/j.seminoncol.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Balkwill F, Coussens LM. Cancer: an inflammatory link. Nature. 2004;431:405–6. doi: 10.1038/431405a. [DOI] [PubMed] [Google Scholar]
- 8.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- 9.Dunn GP, Bruce AT, Ikeda H, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 10.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Seike M, Yanaihara N, Bowman ED, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst. 2007;99:1257–69. doi: 10.1093/jnci/djm083. [DOI] [PubMed] [Google Scholar]
- 12.Schetter A, J Nguyen GH, Bowman ED, et al. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–8. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagès F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;355:2654–66. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 14.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 15.Tsuboi K, Miyazaki T, Nakajima M, et al. Serum interleukin-12 and interleukin-18 levels as a tumor marker in patients with esophageal carcinoma. Cancer Lett. 2004;205:207–14. doi: 10.1016/j.canlet.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Wang KL, Wu TT, Resetkova E, et al. Expression of annexin A1 in esophageal and esophagogastric junction adenocarcinomas: association with poor outcome. Clin Cancer Res. 2006;12:4598–604. doi: 10.1158/1078-0432.CCR-06-0483. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher K, Haensch W, Röefzaad C, et al. Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer Res. 2001;61:3932–6. [PubMed] [Google Scholar]
- 18.Deans DA, Wigmore SJ, Gilmour H, et al. Elevated tumour interleukin-1beta is associated with systemic inflammation: A marker of reduced survival in gastro-oesophageal cancer. Br J Cancer. 2006;95:1568–75. doi: 10.1038/sj.bjc.6603446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosch SB, Izbicki JR, Pichlmeier U, et al. Expression and prognostic significance of immunoregulatory molecules in esophageal cancer. Int J Cancer. 1997;74:582–7. doi: 10.1002/(sici)1097-0215(19971219)74:6<582::aid-ijc4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Liu D, Chen P, et al. Negative feedback regulation of IFN-gamma pathway by IFN regulatory factor 2 in esophageal cancers. Cancer Res. 2008;68:1136–43. doi: 10.1158/0008-5472.CAN-07-5021. [DOI] [PubMed] [Google Scholar]
- 21.Oka M, Hirose K, Iizuka N, et al. Cytokine mRNA expression patterns in human esophageal cancer cell lines. J Interferon Cytokine Res. 1995;15:1005–9. doi: 10.1089/jir.1995.15.1005. [DOI] [PubMed] [Google Scholar]
- 22.Partridge M, Chantry D, Turner M, et al. Production of interleukin-1 and interleukin-6 by human keratinocytes and squamous cell carcinoma cell lines. J Invest Dermatol. 1991;96:771–6. doi: 10.1111/1523-1747.ep12471723. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 25.Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Correa AM, Hoque A, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2010 doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251–255. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors (RECIST Guidelines) J Natl Cancer Inst. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 29.Vidal-Vanaclocha F, Mendoza L, Telleria N, et al. Clinical and experimental approaches to the pathophysiology of interleukin-18 in cancer progression. Cancer Metastasis Rev. 2006;25:417–34. doi: 10.1007/s10555-006-9013-3. [DOI] [PubMed] [Google Scholar]
- 30.Sivakumar PV, Foster DC, Clegg CH, et al. Interleukin-21 is a T-helper cytokine that regulates humoral immunity and cell-mediated anti-tumour responses. Immunology. 2004;112:177–82. doi: 10.1111/j.1365-2567.2004.01886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber GF, Gaertner FC, Erl W, et al. IL-22-mediated tumor growth reduction correlates with inhibition of ERK1/2 and AKT phosphorylation and induction of cell cycle arrest in the G2-M phase. J Immunol. 2006;177:8266–72. doi: 10.4049/jimmunol.177.11.8266. [DOI] [PubMed] [Google Scholar]
- 32.Shan B, Yu L, Shimozato O, et al. Expression of interleukin-21 and -23 in human esophageal tumors produced antitumor effects in nude mice. Anticancer Res. 2004;24:79–82. [PubMed] [Google Scholar]
- 33.Jiang H, Su ZZ, Lin JJ, et al. The melanoma differentiation associated gene mda-7 suppresses cancer cell growth. Proc Natl Acad Sci U S A. 1996;93:9160–5. doi: 10.1073/pnas.93.17.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Riordan JM, Abdel-latif MM, Ravi N, et al. Proinflammatory cytokine and nuclear factor kappa-B expression along the inflammation-metaplasia-dysplasia adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005;100:1257–64. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- 35.Fitzgerald RC, Onwuegbusi BA, Bajaj-Elliott M, et al. Diversity in the oesophageal phenotypic response to gastro-oesophageal reflux: immunological determinants. Gut. 2002;50:451–9. doi: 10.1136/gut.50.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moons LM, Kusters JG, van Delft JH, et al. A pro-inflammatory genotype predisposes to Barrett's esophagus. Carcinogenesis. 2008;29:926–931. doi: 10.1093/carcin/bgm241. [DOI] [PubMed] [Google Scholar]
- 37.Wong NA, Wilding J, Bartlett S, et al. CDX1 is an important molecular mediator of Barrett's metaplasia. Proc Natl Acad Sci U S A. 2005;102:7565–70. doi: 10.1073/pnas.0502031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu N, Flaig MJ, Su H, et al. Comprehensive characterization of annexin I alterations in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:6013–6022. doi: 10.1158/1078-0432.CCR-04-0317. [DOI] [PubMed] [Google Scholar]
- 39.Kimchi ET, Posner MC, Park JO. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 40.Paweletz CP, Ornstein DK, Roth MJ, et al. Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res. 2000;60:6293–6297. [PubMed] [Google Scholar]
- 41.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 42.Caudell EG, Mumm JB, Poindexter N, et al. The protein product of the tumor suppressor gene, melanoma differentiation-associated gene 7, exhibits immunostimulatory activity and is designated IL-24. J Immunol. 2002;168:6041–6. doi: 10.4049/jimmunol.168.12.6041. [DOI] [PubMed] [Google Scholar]
- 43.Lebedeva IV, Sarkar D, Su ZZ, et al. Molecular target-based therapy of pancreatic cancer. Cancer Res. 2006;66:2403–13. doi: 10.1158/0008-5472.CAN-05-3510. [DOI] [PubMed] [Google Scholar]
- 44.Aggarwal S, Takada Y, Mhashilkar AM, et al. Melanoma differentiation-associated gene-7/IL-24 gene enhances NF-kappa B activation and suppresses apoptosis induced by TNF. J Immunol. 2004;173:4368–76. doi: 10.4049/jimmunol.173.7.4368. [DOI] [PubMed] [Google Scholar]
- 45.Enzinger PC, Mayer RJ. Esophageal Cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 46.Ramaswamy S. Translating cancer genomics into clinical oncology. N Engl J Med. 2004;350:1814–1816. doi: 10.1056/NEJMp048059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.