Abstract
Hyperpolarization-activated, cyclic nucleotide-gated cation currents, termed If or Ih, are generated by four members of the hyperpolarization-activated, cyclic nucleotide-gated cation (HCN) channel family. These currents have been proposed to contribute to several functions including pacemaker activity in heart and brain, control of resting potential, and neuronal plasticity. Transcripts of the HCN4 isoform have been found in cardiomyocytes and neurons, but the physiological role of this channel is unknown. Here we show that HCN4 is essential for the proper function of the developing cardiac conduction system. In wild-type embryos, HCN4 is highly expressed in the cardiac region where the early sinoatrial node develops. Mice lacking HCN4 channels globally, as well as mice with a selective deletion of HCN4 in cardiomyocytes, died between embryonic days 9.5 and 11.5. On average, If in cardiomyocytes from mutant embryos is reduced by 85%. Hearts from HCN4-deficient embryos contracted significantly slower compared with wild type and could not be stimulated by cAMP. In both wild-type and HCN4-/- mice, cardiac cells with “primitive” pacemaker action potentials could be found. However, cardiac cells with “mature” pacemaker potentials, observed in wild-type embryos starting at day 9.0, were not detected in HCN4-deficient embryos. Thus, HCN4 channels are essential for the proper generation of pacemaker potentials in the emerging sinoatrial node.
Four genes encoding hyperpolarization-activated, cyclic nucleotide-gated cation (HCN) channels have been identified and functionally expressed. All four HCN channels carry an inward current with the typical features of a current termed Ih in the brain and If in the heart (1–4). These currents have been implicated in a wide range of physiological functions including pacemaking activity of spontaneously firing brain and heart cells, control of resting membrane potential, response to sour taste, neuronal plasticity, and dendritic integration (reviewed in refs. 5–7).
HCN4 is the predominant HCN transcript in the adult sinoatrial node (8–11). If and HCN transcripts have also been identified in mouse embryonic hearts with HCN4 being the prevalent type at early stages (12). Little is known about the specific contributions of the individual HCN isoforms to the function of the heart. One of the main, but still controversially discussed hypotheses is that If is one of the major currents contributing to the spontaneous diastolic depolarization of pacemaker cells and thereby to sinus node rhythm (5, 13). Furthermore, the β-adrenergic up-regulation of sinus node rhythm has been attributed to the binding of cAMP to HCN channels resulting in an enhanced If. However, it has been shown that diastolic depolarization is generated by multiple ionic currents with complex interactions (reviewed in ref. 14). The importance of If has also been questioned because activation thresholds of If vary and conflicting results were obtained with If blockers (15–18).
Direct evidence demonstrating the functional significance of HCN4 channels is lacking. To determine the physiological role of HCN4 channels, we generated mice deficient for this HCN isoform. Here we report that pacemaker cells with a mature, sinoatrial node-like pacemaker potential do not develop in these mice.
Materials and Methods
The generation of mice globally deficient for HCN4 channels is described in Supporting Methods, which is published as supporting information on the PNAS web site.
Cardiomyocyte-Specific Deletion of HCN4. HCN4+/- mice were crossed with transgenic MLC2a-Cre animals (MLC2atg/0) (4) to obtain double-heterozygous mice (HCN4+/-; MLC2atg/0). These mice were mated with animals homozygous for the floxed HCN4 locus (HCN4L2/L2) to generate offspring with a cardiomyocytespecific deletion of HCN4 (HCN4L2/-; MLC2atg/0).
Whole-Mount lacZ Staining of Embryos. Details are given in Supporting Methods.
In Situ Hybridization and Immunohistochemistry. Embryos were fixed in 4% paraformaldehyde, embedded in paraffin wax, and cut into 10-μm sections. In situ hybridization was done as described (11). For immunohistochemistry, endogenous peroxidase activity was quenched by using H2O2/methanol. Antigen retrieval was accomplished by microwaving slides in citrate buffer (0.2 M Na2HPO4/0.1 M trisodium citrate, pH 4.5) for 40 sec. Sections were preincubated in blocking buffer (TBS/0.1% Tween 20/10% normal goat serum) and HCN4 antibody was applied in this buffer overnight at 4°C. Bound antibodies were detected by using VECTASTAIN ABC Kit (Vector Laboratories) and 3,3-diaminobenzidine/H2O2.
Generation of HCN4 Antibody and Western Blot. A rabbit polyclonal antibody was raised against a peptide (TA A PQR EPGARSEPVRSK) from the carboxyl terminus of murine HCN4 and affinity-purified by using a peptide-Sepharose column. Microsomal membranes from HEK293 cells transfected with HCN4 cDNA were prepared essentially as described (19). Ten wild-type embryonic day (E) 11.5 hearts were pooled and homogenized in lysis buffer (50 mM Tris, pH 7.4/1 mM EDTA/protease inhibitor mixture). Proteins were separated on 7% SDS/PAGE gels, blotted, and probed with HCN4 antibody by using a chemiluminescence detection system.
RT-PCR. For whole-embryo RT-PCR, total RNA was isolated by using TRIzol reagent (GIBCO/BRL). First-strand cDNA was synthesized with SuperScript II (GIBCO/BRL). For single-cell RT-PCR, after recordings, single cells were collected intact with patch pipettes, frozen on dry ice, and used as template in the One-Step RT-PCR system (Qiagen). HCN1-4 and GAPDH cDNAs were amplified by using intron-spanning primer pairs to avoid amplification of genomic DNA. Primer sequences are given in Table 1, which is published as supporting information on the PNAS web site.
Isolation and Culture of Embryonic Hearts and Cardiomyocytes. Whole hearts for heart rate determination and isolated cardiomyocytes for electrophysiological recordings were prepared essentially as described (20, 21). Details and composition of solutions are given in Supporting Methods.
Electrophysiology. Twenty-four hours after isolation, If in the whole-cell configuration at 23 ± 1°C (22) and action potentials (APs) in the perforated patch configuration at 37 ± 1°C (23) were recorded from beating cardiomyocytes. In addition, HCN currents from HEK293 cells, transiently transfected with cDNAs encoding murine HCN1, HCN3, or HCN4 channels (11), were measured and analyzed under the same conditions as the native If. Solutions, instruments, and analyzing methods are described in detail in Supporting Methods.
Values are given as means. Error bars in the figures represent SEM, and “±” in the text represents the range of mean. n is the number of experiments. Statistical differences were determined by using Student's unpaired t test, and P values <0.05 were considered significant.
Results
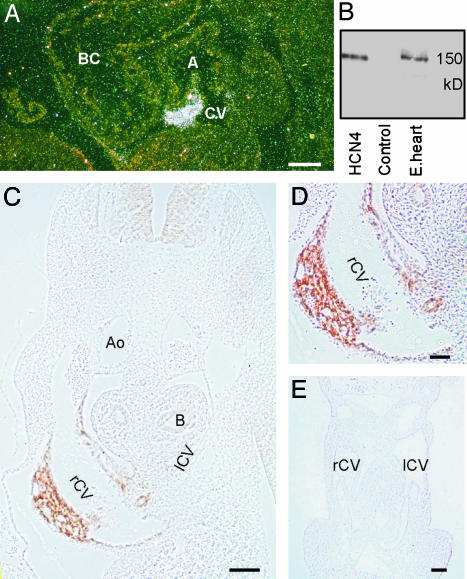
Expression of HCN4 in Mouse Embryos. We determined the expression of HCN4 in murine embryos by in situ hybridization and immunohistochemistry. At E10.5, HCN4 transcripts are highly localized to a specific cardiac region, the wall of the common cardinal vein entering the sinus venosus (Fig. 1A). Compared with the heart, around one order of magnitude lower expression of HCN4 mRNA was detected in the developing central nervous system (not shown).
Fig. 1.
Expression of HCN4 mRNA and protein in embryos at E10.5. (A) In situ hybridization. A dark-field micrograph of a sagittal section from a wild-type E10.5 embryo labeled with an HCN4-specific riboprobe is shown. A, common atrial chamber; BC, bulbus cordis; CV, common cardinal vein. (B) Specificity of the anti-HCN4 antibody. Western blots of membranes from HEK293 cells transfected with either HCN4 or empty vector (Control) and a protein extract from E11.5 embryo hearts (E.heart) are shown. Blots were decorated with the anti-HCN4 antibody. (C) Immunohistochemical detection of HCN4. A transverse section through a wild-type E10.5 embryo labeled with the anti-HCN4 antibody is shown. Ao, Aorta; B, bronchus; rCV and lCV, right and left common cardinal veins. (D) Higher-magnification image of the section shown in C demonstrating labeling of HCN4 in the wall of the right common cardinal vein. (E) Transverse section through the heart and common cardinal veins of an E10.5 HCN4-/- embryo labeled with the anti-HCN4 antibody. Staining of the embryo is completely absent. (Scale bars: A, C, and E, 100 μm; D, 50 μm.)
To investigate the expression of HCN4 at the protein level, we generated an HCN4-specific antibody. The antibody recognizes specifically HCN4 protein as a single 150-kDa band in membranes from HCN4-transfected HEK293 cells and in an extract prepared from E11.5 embryonic hearts (Fig. 1B). This antibody extensively labeled HCN4 protein in the wall of the right common cardinal vein on sections of E10.5 wild-type embryos (Fig. 1 C and D). The staining extends to the left common cardinal vein and the opening of these veins in the sinus venosus. We observed no labeling in similar sections prepared from E10.5 HCN4-/- embryos (Fig. 1E), confirming the successful generation of an HCN4-/- mutant described below.
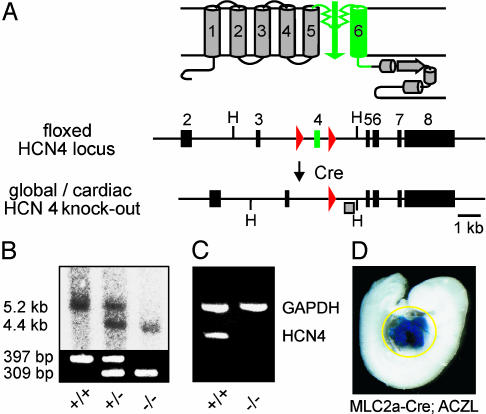
Mice Lacking HCN4 Channels Die During Embryonic Development but Do Not Show Structural Abnormalities. We generated mice globally deficient in HCN4 channels by deleting exon 4 of the HCN4 gene that encodes the pore region and transmembrane segment S6 of the channel (Fig. 2A; and see Fig. 6, which is published as supporting information on the PNAS web site). Heterozygous HCN4+/- animals were viable and bred normally, and were indistinguishable from wild-type littermates. However, no homozygous HCN4-/- pups were born from heterozygous matings. Analysis of timed matings revealed that until E9.5, normally developed HCN4-/- as well as HCN4+/- and HCN4+/+ embryos were present at a Mendelian ratio (n = 20, 42, and 21, respectively; pooled from 10 pregnant mice). At E10.5, a portion of the HCN4-/- embryos were dead (3 of 20). From E11.5 to E14.5, only dead, reabsorbed HCN4-/- embryos were found. These results suggested that HCN4-/- embryos died between E9.5 and E11.5. In addition to PCR and Southern blot analysis (Fig. 2B), RT-PCR using total RNA from whole E9.5 embryos demonstrated the deletion of the HCN4 gene (Fig. 2C). We did not identify structural abnormalities in HCN4-/- embryos at E9.5 and E10.5 (Fig. 7 A and B, which is published as supporting information on the PNAS web site). In particular, serial histological sections revealed no obvious defect in the cardiac structure of mutant embryos (Fig. 7 C and D).
Fig. 2.
Generation of HCN4-deficient mice. (A) Gene targeting strategy. (Upper) Schematic diagram of HCN4 channel structure. The six putative transmembrane segments S1–S6 are numbered. (Lower) Floxed HCN4 locus and knockout allele after Cre-mediated deletion of exon 4. Exons are indicated by boxes. Exon 4 (green) encodes pore and S6 segment of the channel. loxP sites are represented by red triangles. H, HindIII. (B) Genomic analysis of mice lacking HCN4 channels globally. (Upper) Southern blot of HindIII-digested DNA prepared from whole E9.5 embryos. The blot was hybridized with the probe (shaded box) indicated in A, leading to 5.2- and 4.4-kb fragments representing wild-type and HCN4-deletion allele, respectively. (Lower) PCR resulting in a 397-bp product for wild type and a 309-bp product for the null allele. (C) RT-PCR analysis of mice globally deficient in HCN4 channels. Total RNA was prepared from whole HCN4+/+ and HCN4-/- E9.5 embryos. HCN4, HCN4 amplification product, primer pair A/B (refer to Table 1); GAPDH, internal control, amplified in the same reaction as HCN4. (D) Whole-mount X-gal stain of an ACZL β-galtg/0; MLC2a-Cretg/0, double-transgenic embryo. The yellow circle outlines the X-gal-stained cardiac region.
Cardiac-Specific Deletion of HCN4 Results in the Same Embryonic Lethality as Global Deletion. Because HCN4 channels are not only expressed in the embryonic heart but also, albeit at lower expression levels, in the developing central nervous system, we generated mice lacking HCN4 channels specifically in the heart (Fig. 2 A and D). We used transgenic MLC2a-Cre mice that excise loxP-flanked DNA segments selectively in cardiac myocytes (4). Cre recombinase activity during development was analyzed by X-gal staining of embryos double transgenic for MLC2a-Cre and a β-gal reporter gene (ACZL, 24). E9.5 (Fig. 2D) and E10.5 embryos (not shown) show intense staining and, accordingly, efficient recombination selectively in the heart. Embryos with a cardiac-specific deletion of HCN4 channels (n = 12) died during the same developmental period as embryos lacking HCN4 globally, i.e., between E9.5 and E11.5. The control embryos (n = 57) from these litters developed normally. These results strongly support the notion that the death of HCN4-deficient embryos is related to a cardiac rather than a neuronal dysfunction.
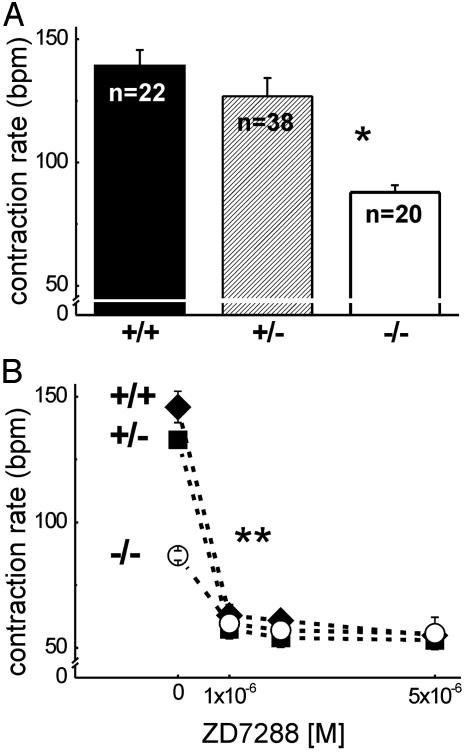
HCN4-Deficient Embryos Have a Slower Heart Rate. Because we found no structural abnormalities in the HCN4-/- embryonic hearts accounting for their premature death, we examined the function of these hearts. Isolated wild-type, heterozygous, and HCN4-deficient embryonic hearts were regularly contracting without obvious arrhythmias (see Movies 1–3, which are published as supporting information on the PNAS web site). However, the basal contraction rate of E9.5 HCN4-/- embryonic hearts (Fig. 3A) was significantly reduced compared with both HCN4+/+ and HCN4+/- embryos of the same age (88 ± 14 bpm for HCN4-/- vs. 139 ± 22 and 127 ± 21 bpm for HCN4+/+ and HCN4+/-, respectively; P < 0.001).
Fig. 3.
Contraction rates of isolated embryonic hearts, E9.5. (A) Basal contraction rates in beats per minute (bpm) of HCN4 wild-type (+/+), heterozygous (+/-), and homozygous knockout (-/-) hearts. The number (n) of counted hearts is indicated. *, P < 0.001 vs. HCN4+/+ or HCN4+/-. Representative video recordings of isolated beating hearts can be found in Movies 1–3. (B) Modulation of contraction rates by ZD7288, which was added to the medium after a 10-min prerun period (0 M ZD7288) in which the basal contraction rate was determined. The number of counted hearts was 10 (HCN4+/+,♦), 9 (HCN4+/-, ▪), and 8 (HCN4-/-, ○). **, P < 0.05, modulated vs. basal rate for all genotypes. Values are mean ± SEM.
Application of ZD7288, a specific If blocker (25), reduced the contraction rates of HCN4+/+, HCN4+/-, and HCN4-/- hearts significantly (Fig. 3B). The effect was much more pronounced in the wild-type and heterozygous hearts, where 1 × 10-6 ZD7288 reduced the mean heart rate from 146 to 63 bpm (n = 10) and 133 to 57 bpm (n = 9), respectively, compared with a reduction from 87 to 60 bpm (n = 8) in the HCN4 knockout hearts. In contrast to heart rates under control conditions, the contraction rates after application of ZD7288 no longer differed significantly. The same result was observed after application of 2 × 10-3 M cesium or 1 × 10-7 M DK-AH269, another specific If blocker (26) (data not shown). The effects obtained with If blockers suggested that there may be a component of If in HCN4-/- embryonic cardiomyocytes not generated by HCN4 channels.
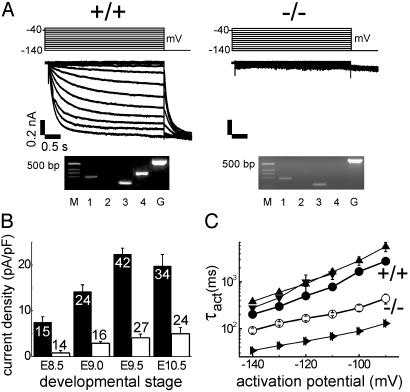
HCN4-/- Cardiomyocytes Show a Significant Reduction of If. If was analyzed in isolated cardiomyocytes of E8.5, E9.0, E9.5, and E10.5 embryos by whole-cell patch clamp recordings. If densities in small, round, beating cardiomyocytes from wild-type embryos increased significantly between E8.5 and E9.5 from a mean of 7.5–22.4 pA/pF (n = 15 and 42; Fig. 4 A Top and B) at a hyperpolarizing pulse to -140 mV. No further increase of the current was observed on E10.5 (19.8 pA/pF, n = 34). In HCN4 knockout cardiomyocytes, only a very small If could be detected at E8.5 (0.8 pA/pF, n = 14), which increased to mean densities of 2.9, 4.1, and 4.9 pA/pF at E9.0, E9.5, and E10.5, respectively (Fig. 4B). This corresponds to a 75–90% reduction of the If density compared with wild-type cardiomyocytes. The same reduction of If was observed in the heart-specific HCN4-/- mice (e.g., in E9.5 cardiomyocytes: 29.6 ± 14.4 pA/pF; wild type, n = 12 vs. 4.9 ± 3.4 pA/pF; knockout, n = 10). There was no significant difference in cell size determined by cell capacitance between HCN4 knockout and wild-type cardiomyocytes of the same type and developmental stage.
Fig. 4.
If in isolated embryonic cardiomyocytes. (A)If traces of HCN4+/+ (Left) and HCN4-/- (Right) cardiomyocytes, at E9.5, recorded on hyperpolarizing steps from a holding potential of -40 mV to the range -140 to 30 mV. Time and current amplitude scales are the same for both. Above the current traces are pulse protocols. Below the current traces are single-cell RT-PCR from these cells. M, DNA size marker; 1, 2, 3, and 4, HCN1, HCN2, HCN3, and HCN4 amplification products; G, GAPDH. Primers for HCN4 and GAPDH are the same as in Fig. 2C. (B) If densities of HCN4+/+ (filled columns) and HCN4-/- (open columns) cardiomyocytes at developmental stages E8.5, E9.0, E9.5, and E10.5. The number of evaluated cells is indicated. (C) Voltage-dependent activation time constants (τact) for If of HCN4+/+ •) and HCN4-/- (○) cardiomyocytes (E9.5) compared with HCN4 (▴), HCN3 (▾), and HCN1 (▴) currents measured from transfected HEK293 cells. Note log10 of τact-ordinate. The number of evaluated cells was 12–34. Values in B and C are mean ± SEM.
cAMP application did not increase the absolute If amplitude in wild-type or knockout cardiomyocytes when the channels were already maximally activated by a voltage step to -140 mV. However, the voltage-dependent activation curve of the wildtype If was shifted significantly to more positive potentials: V1/2 was -94.0 ± 3.2 mV without and -83.7 ± 4.7 mV with cAMP (n = 20) at E9.5. This effect was not observed for the HCN4 knockout If.
HCN4 Is the Major but Not Exclusive HCN Isoform in Embryonic Cardiomyocytes. To clarify the molecular constituents of If in embryonic wild-type and knockout cardiomyocytes, we combined patch-clamp recordings with semiquantitative single-cell RT-PCR. Fig. 4A shows example If traces of HCN4+/+ and HCN4-/- E9.5 cardiomyocytes together with their corresponding RT-PCR results. Wild-type cardiomyocytes with a prominent If had a high expression of HCN4, whereas no HCN4 mRNA was amplified from HCN4-/- embryos. Lower amounts of HCN1 and HCN3 could be amplified from both genotypes. Together with the electrophysiological characteristics of If described above and below, these results indicate that HCN4 is the major component of If in embryonic cardiomyocytes. An additional RT-PCR strategy, described in Supporting Methods, demonstrated that the wild-type and knockout cells compared in Fig. 4 are equivalent cell types, i.e., the HCN4 gene is transcribed in both, but HCN4-/- cells only produce mutated HCN4 mRNA that does not lead to translation of a functional HCN4 protein (Figs. 8 and 9, which are published as supporting information on the PNAS web site).
Comparison of the activation kinetics of wild-type and knockout If supports the finding that more than one HCN-isoform generates the native If (Fig. 4C). Compared with the cloned murine HCN4, HCN3, and HCN1 channels expressed in HEK293 cells, the native embryonic wild-type If at E9.5 is activated slightly faster than HCN4 or HCN3 currents but considerably slower than HCN1 (e.g., mean τact at -140 mV: 202 ms for wild-type cardiomyocytes compared with 379 ms for HCN4, 276 ms for HCN3, and 34 ms for HCN1). This comparison strengthens the notion that the native If may be produced by several HCN isoforms with HCN4 being the main type. In contrast, the residual If in HCN4-/- cardiomyocytes has a mean τact of 91 ms at -140 mV; this is much faster than the wild-type If or the expressed HCN3 and HCN4 currents but slower than HCN1. These results are in accordance with the single-cell RT-PCR data and suggest that the residual If in HCN4 knockout cardiomyocytes is generated by HCN1 and HCN3.
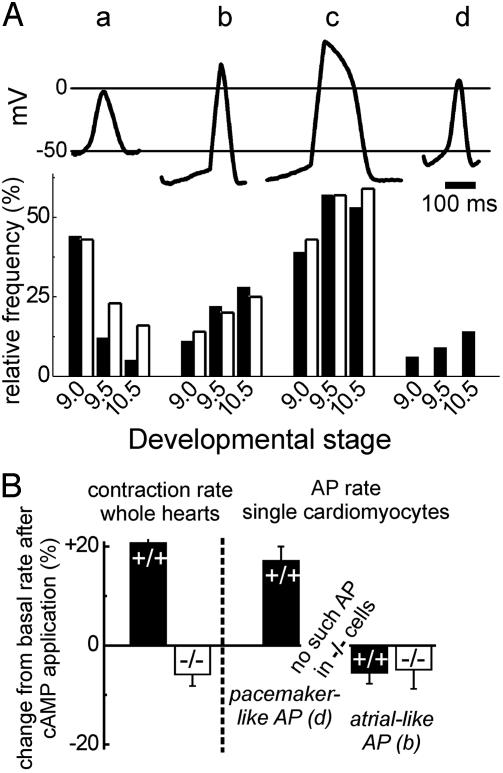
No Pacemaker-Like APs in HCN4-/- Embryonic Cardiomyocytes. AP recordings in beating E8.0–E10.5 cardiomyocytes revealed four different types of APs (Fig. 5A). Three of these types, namely the primitive, the atrial-like (or intermediate), and the ventricularlike AP, were recorded similarly from wild-type and HCN4-/- cardiomyocytes. The almost exclusive type of the early heart (E8.0–E8.5), the primitive (early pacemaker) AP, is characterized by relatively narrow membrane potential fluctuations between -50 and 0 mV (Fig. 5A, a). At later stages, i.e., E9.5–E10.5, this AP occurred slightly more frequently in HCN4-/- than in wild-type embryos, e.g., in 16% of cells compared with 5% in wild-type embryos at E10.5 (Fig. 5A Lower). No differences were found in the relative frequencies of atrial-like (or intermediate) and ventricular-like APs between wild-type and knockout embryos at any stage. The basic characteristics of these APs were not significantly different between HCN4+/+ and HCN4-/- cardiomyocytes. For example, the diastolic depolarization rate (DDR) and mean diastolic potential (MDP) of atrial-like/intermediate APs at E10.5 were 59.3 ± 19.9 mV/s and -65.0 ± 10.1 mV in knockout cells (n = 19) and 73.4 ± 10.5 mV/s and -70.1 ± 7.0 mV in wild-type cells (n = 18). Both atrialand ventricular-like APs show high upstroke velocities between 70 and 90 V/s, overshoots up to +30 mV, and a plateau phase.
Fig. 5.
APs in isolated embryonic cardiomyocytes, E9.5. (A Upper) AP types found in a cardiomyocyte preparation from a wild-type E9.5 embryonic heart. a, primitive (early pacemaker); b, atrial-like/intermediate; c, ventricular-like; d, mature pacemaker/sinus node-like. (A Lower) Relative frequency of the AP types shown above in cardiomyocytes isolated from HCN4+/+ (filled columns) and HCN4-/- (open columns) embryonic hearts of the developmental stages E9.0, E9.5, and E10.5. The number of evaluated cells for HCN4+/+/HCN4-/- was 72/56, 101/60, and 64/76 for E9.0, 9.5, and 10.5, respectively. Note that no mature pacemaker-like APs were detected in HCN4-/- cell preparations. (B) Steady state modulation of contraction rates of whole hearts (left side) and AP rates of single cardiomyocytes (right side) by cAMP. Displayed is the change of the rates relative to the baseline rate 8 min after the addition of 10-4 M 8-Br-cAMP to the bath. Filled columns represent HCN4+/+ hearts/cardiomyocytes, and open columns represent HCN4-/- hearts/cardiomyocytes. The number of experiments was between 6 and 14. Values in B are mean ± SEM.
As early as E9.0, cells with a “mature” pacemaker-like AP were found in wild-type hearts (Fig. 5A, d) having only marginal overshoots up to +5 mV reached by upstroke velocities of 10–20 V/s, a mean MDP of -55.8 ± 5.1 mV, and a DDR of 118.5 ± 19 mV/s. This depolarization rate is much faster than that of the atrial- and ventricular-like APs. The values of the pacemaker-like APs are very similar to the pacemaker potentials of adult sinoatrial node cells. The small percentage of these pacemaker cells relative to all cells is in accordance with the in situ hybridization and immunohistochemistry data (Fig. 1) showing HCN4 transcripts and immunoreactivity in ≈5–10% of the cardiac tissue based on analysis of serially sectioned embryos.
In marked contrast to the wild-type heart, in all analyzed HCN4-/- cells (E8.5–E10.5, n = 192) not a single pacemakerlike potential could be recorded. This lack of pacemaker-like AP in cells proven to transcribe the HCN4 gene by single cell RT-PCR (see Supporting Methods) suggests that HCN4 plays a major role in generating the genuine pacemaker potential.
Heart Rate and Action Potential Rate in HCN4-/- Embryos Cannot Be Accelerated by cAMP. cAMP accelerates the heart rate by increasing the AP firing rate of adult sinoatrial node pacemaker cells. Therefore, we tested whether cAMP modulated the contraction rate of the embryonic heart and its pacemaker cells. For the comparison of heart and AP rates we used the relative reaction to cAMP because the AP rate of single cardiomyocytes was generally higher than the heart rate, possibly because of the missing feedback of adjacent cells in isolated cell preparations. The contraction rate of isolated wild-type E9.5 hearts increased after the application of cAMP by 20% after 8 min (Fig. 5B), and this increased pace was maintained for up to 30 min. In contrast, contraction rates of HCN4-/- hearts could not be increased. In addition, after prolonged exposure to cAMP (over 10 min), the heart rate consistently decreased by 5–15%, an effect not observed in wild-type hearts. Similar to the whole heart, the rate of the pacemaker-like AP of wild-type cardiomyocytes increased by almost 20% after application of cAMP. Atrial-like (Fig. 5B) as well as primitive and ventricular-like APs (data not shown) of both HCN4+/+ and HCN4-/- embryos could not be stimulated by cAMP. Surprisingly, the AP rate in these cells was even decreased to a similar extent as the contraction rate of isolated HCN4-/- embryonic hearts. These results support the above finding that there are no “mature” cAMP-regulated pacemaker cells in HCN4 knockout hearts.
Discussion
The murine embryonic heart starts to show regular contractions as early as E8.0 (20). In the adult heart, pacemaker channels like HCN4 are thought to contribute to a regular heart beat by modulating the pacemaker potential in the sinoatrial node, considered as the primary pacemaker region of the heart. Although HCN4 transcripts have been found in embryonic hearts before (12), it was not clear whether a functional sinoatrial node-like region with this pacemaker channel already exists at an early embryonic age. In this communication, the expression of HCN4 channels is demonstrated in a highly specialized region of the developing heart. Both HCN4 mRNA and protein could be detected in the area where the right and left common cardinal veins enter the sinus venosus. This region develops to the area where the right superior vena cava enters the right atrium, referred to as the adult sinoatrial node region.
Mice lacking HCN4 channels either globally or specifically in the heart die during embryonic development between E9.5 and E11.5. During this time, the highly localized cardiac expression of HCN4 was detectable. At the same time, cardiac cells with a prominent, HCN4-based If appear in wild-type embryos. Surprisingly and despite the 75–90% reduction of If, the hearts of HCN4-/- embryos contracted regularly at a reduced rate. Addition of various If blockers further decelerated but did not abolish heart contractions in both HCN4-/- and HCN4+/+ embryos, suggesting that If is involved in pacemaking, even though a basal heart rate can be sustained without If.
Electrophysiological recordings from single cardiomyocytes showed different types of APs appearing at specific developmental stages. These APs resemble the types found during the differentiation of embryonic stem cells to pulsating cardiomyocytes in embryoid bodies (27). Starting at E9.0, cells with differentiated APs from the developing atria and ventricles were identified in both HCN4+/+ and HCN4-/- hearts. These atrial- and ventricular-like APs show similarities to adult atrial and ventricular APs and are probably created by the concerted action of several ionic currents including INa, ICa, IK, IKI, Ito, IK,ACh, and IK,ATP. However, in contrast to adult ventricular cells, both of these embryonic cell types do not yet have a stable resting potential but spontaneously depolarize. Apparently, HCN4 does not contribute significantly to the AP of these cells. This notion does not exclude the possibility that other HCN isoforms, namely HCN1 and HCN3, which were detected at low levels in almost all embryonic cardiomyocytes of both HCN4+/+ and HCN4-/- embryos, could contribute to the spontaneous depolarizations in atrial- and ventricular-like cells.
True pacemaker cells among the embryonic cardiomyocytes start with a “primitive” pacemaker AP that is probably generated by only two ionic currents, ICa,L and IK,to (27), or even just by intracellular Ca2+ oscillations (28). This AP can be found in both HCN4+/+ and HCN4-/- embryos. Primitive APs are sufficient to drive the small, tubal heart at an early age. However, starting already at E9.0, “mature” sinus node-like APs were found in HCN4+/+ embryos but were never detected in HCN4-/- embryos of any age. Thus, besides several Ca2+ and K+ currents, the If carried by HCN4 channels is likely to be involved in the generation of mature pacemaker potentials.
The lack of functional mature pacemaker cells in HCN4-/- embryos was confirmed by the inability to stimulate the contraction rate of single cardiomyocytes or the whole heart by cAMP. Generally, β-adrenergic stimulation increases the heart rate of adult animals. One possible mechanism involves direct modulation of HCN channels by cAMP in pacemaker cells of the sinoatrial node. The HCN4 channel mediates the cAMP-dependent stimulation of wild-type embryonic heart and embryonic “mature” pacemaker cells because the contraction frequency could not be increased in hearts or cells from mice lacking this channel. In accordance with these results, a direct modulation of If by cAMP was only found in wild-type embryonic cardiomyocytes, whereas the residual If from knockout cells was not stimulated.
The functional correlation between the early embryonic death and the inactivation of the HCN4 gene is not obvious. A possible cause is the deficiency to develop functional or mature pacemaker cells. The slow heart rate of knockout hearts together with the prevalence of intermediate and primitive APs support the hypothesis that these APs are insufficient to drive the heart at the necessary rate needed to supply the developing embryo with enough blood. We conclude that the HCN4 channel is essential for the cardiac function of the developing murine embryo because without HCN4, cardiomyocytes cannot generate sinus node-like pacemaker potentials.
Supplementary Material
Acknowledgments
We thank J. Graw (GSF-National Research Center for Environment and Health, Institute of Developmental Genetics) for help with preparation of embryo sections, S. Moosmang (Institut für Pharmakologie, Technische Universität München) for help with HCN4-antibody preparation, and K. R. Chien (Institute of Molecular Medicine, University of California at San Diego, La Jolla) for providing MLC2a-Cre mice. The excellent technical support of A. Vens and A. Thomer is highly appreciated. This work was supported by grants from Deutsche Forschungsgemeinschaft and Fond der Chemischen Industrie.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AP, action potential; En, embryonic day n; HCN, hyperpolarization-activated, cyclic nucleotide-gated cation.
References
- 1.Biel, M., Ludwig, A., Zong, X. & Hofmann, F. (1999) Rev. Physiol. Biochem. Pharmacol. 136, 165-181. [DOI] [PubMed] [Google Scholar]
- 2.Santoro, B. & Tibbs, G. R. (1999) Ann. N.Y. Acad. Sci. 868, 741-764. [DOI] [PubMed] [Google Scholar]
- 3.Kaupp, U. B. & Seifert, R. (2001) Annu. Rev. Physiol. 63, 235-257. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig, A., Budde, T., Stieber, J., Moosmang, S., Wahl, C., Holthoff, K., Langebartels, A., Wotjak, C., Munsch, T., Zong, X., et al. (2003) EMBO J. 22, 216-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiFrancesco, D. (1993) Annu. Rev. Physiol. 55, 455-472. [DOI] [PubMed] [Google Scholar]
- 6.Pape, H. C. (1996) Annu. Rev. Physiol. 58, 299-327. [DOI] [PubMed] [Google Scholar]
- 7.Robinson, R. B. & Siegelbaum, S. A. (2003) Annu. Rev. Physiol. 65, 453-480. [DOI] [PubMed] [Google Scholar]
- 8.Shi, W., Wymore, R., Yu, H., Wu, J., Wymore, R. T., Pan, Z., Robinson, R. B., Dixon, J. E., McKinnon, D. & Cohen, I. S. (1999) Circ. Res. 85, e1-e6. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig, A., Zong, X., Stieber, J., Hullin, R., Hofmann, F. & Biel, M. (1999) EMBO J. 18, 2323-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii, T. M., Takano, M., Xie, L.-H., Noma, A. & Ohmori, H. (1999) J. Biol. Chem. 274, 12835-12839. [DOI] [PubMed] [Google Scholar]
- 11.Moosmang, S., Stieber, J., Zong, X., Biel, M., Hofmann, F. & Ludwig, A. (2001) Eur. J. Biochem. 268, 1646-1652. [DOI] [PubMed] [Google Scholar]
- 12.Yasui, K., Liu, W., Opthof, T., Kada, K., Lee, J.-K., Kamiya, K. & Kodama, I. (2001) Circ. Res. 88, 536-542. [DOI] [PubMed] [Google Scholar]
- 13.Brown, H. F., DiFrancesco, D. & Noble, S. J. (1979) Nature 280, 235-236. [DOI] [PubMed] [Google Scholar]
- 14.Schram, G., Pourrier, M., Melnyk, P. & Nattel, S. (2002) Circ. Res. 90, 939-950. [DOI] [PubMed] [Google Scholar]
- 15.Vassalle, M. (1995) Cardiovasc. Res. 30, 309-310. [PubMed] [Google Scholar]
- 16.Noma, A. (1996) Jpn. Heart J. 37, 673-682. [DOI] [PubMed] [Google Scholar]
- 17.Miake, J., Marban, E. & Nuss, H. B. (2002) Nature 419, 132-133. [DOI] [PubMed] [Google Scholar]
- 18.Chevaleyre, V. & Castillo, P. E. (2002) Proc. Natl. Acad. Sci. USA 99, 9538-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig, A., Flockerzi, V. & Hofmann, F. (1997) J. Neurosci. 17, 1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porter, G. & Rivkees, S. (2001) Am. J. Physiol. 281, R401-R407. [DOI] [PubMed] [Google Scholar]
- 21.An, R. H., Davies, M. P., Doevendans, P. A., Kubulak, S. W., Bangalore, R., Chien, K. R. & Kass, R. S. (1996) Circ. Res. 78, 371-378. [DOI] [PubMed] [Google Scholar]
- 22.Hamill, O. P., Marty, A., Neher, E., Sakmann, B. & Sigworth, F. J. (1981) Pflugers Arch. 391, 85-100. [DOI] [PubMed] [Google Scholar]
- 23.Korn, S. J. & Horn, R. (1989) J. Gen. Physiol. 94, 789-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akagi, K., Sandig, V., Vooijs, M., Van der Valk, M., Giovannini, M., Strauss, M. & Berns, A. (1997) Nucleic Acids Res. 25, 1766-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.BoSmith, R. E., Briggs, I. & Sturgess, N. C. (1993) Br. J. Pharmacol. 110, 343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raes, A., Van de Vijver, G., Goethals, M. & Van Bogaert, P. P. (1998) Br. J. Pharmacol. 125, 741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hescheler, J., Fleischmann, B. K., Lentini, S., Maltsev, V. A., Rohwedel, J., Wobus, A. M. & Addicks, K. (1997) Cardiovasc. Res. 36, 149-162. [DOI] [PubMed] [Google Scholar]
- 28.Viatchenko-Karpinski, S., Fleischmann, B. K., Liu, Q., Sauer, H., Gryshchenko, G. J. & Hescheler, J. (1999) Proc. Natl. Acad. Sci. USA 96, 8259-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.