Abstract
More than 50 leaf rust resistance (Lr) genes against the fungal pathogen Puccinia triticina have been identified in the wheat gene pool, and a large number of them have been extensively used in breeding. Of the 50 Lr genes, all are known only from their phenotype and/or map position except for Lr21, which was cloned recently. For many years, the problems of molecular work in the large (1.6 × 1010 bp), highly repetitive (80%), and hexaploid bread wheat (Triticum aestivum L.) genome have hampered map-based cloning. Here, we report the isolation of the Lr gene Lr10 from hexaploid wheat by using a combination of subgenome map-based cloning and haplotype studies in the genus Triticum. Lr10 is a single-copy gene on chromosome 1AS. It encodes a CC-NBS-LRR type of protein with an N-terminal domain, which is under diversifying selection. When overexpressed in transgenic wheat plants, Lr10 confers enhanced resistance to leaf rust. Lr10 has similarities to RPM1 in Arabidopsis thaliana and to resistance gene analogs in rice and barley, but is not closely related to other wheat Lr genes based on Southern analysis. We conclude that map-based cloning of genes of agronomic importance in hexaploid wheat is now feasible, opening perspectives for molecular bread wheat improvement trough transgenic strategies and diagnostic allele detection.
During the past decade, a number of disease resistance genes have been isolated from model plant species such as Arabidopsis thaliana and rice or from diploid crop plants such as tomato and barley (for recent reviews see refs. 1 and 2). In many cases, gene isolation was performed through map-based cloning. This process requires the development of high-density genetic maps and the possibility to perform chromosome walking on large genomic fragments. Until recently, such positional cloning has been limited to small genomes, and it has remained very difficult in large (>5,000 Mb) and repetitive (>80%) genomes such as those of barley and wheat. Mlo, the first barley disease resistance gene isolated by map-based cloning (3), was identified through the use of a yeast artificial chromosome (YAC) library and the subsequent construction of a bacterial artificial chromosome (BAC) library from the YAC clone spanning the resistance locus. More recently, the construction of a BAC library from the barley cultivar Morex (4) has greatly facilitated the map-based isolation of the powdery mildew resistance genes Mla1 and Mla6 (5, 6) as well as the stem rust resistance gene Rpg1 (7). In wheat, BAC libraries have been constructed from the diploid species Triticum monococcum (8) and Ae. tauschii (9), whose genomes are related to the A and D genome of hexaploid wheat, respectively. The T. monococcum DV92 library has been used to isolate VRN1, a gene controlling vernalization response in T. monococcum (10), as well as a candidate gene for the Q gene, which confers free-threshing character to domesticated wheat (11). Very recently, Huang et al. (12) have isolated the Lr21 resistance gene introgressed on chromosome 1DS by using a cosmid library from the Aegilops tauschii Lr21 donor line.
Many genes of agronomic interest, including >50 leaf rust (Lr) disease resistance genes, have been characterized by genetic analysis in hexaploid wheat. The Lr10 gene originates from hexaploid wheat and is located on chromosome 1AS (13). In the absence of BAC or cosmid libraries from a wheat variety containing Lr10, cloning of the complete Lr10 resistance locus was not possible from hexaploid wheat. Therefore, we have developed a subgenome chromosome walking strategy in which genetic mapping was performed in a hexaploid wheat population segregating for the Lr10 resistance, and chromosome walking was performed by using BAC clones from the diploid T. monococcum DV92 (14). In one step of chromosome walking, a T. monococcum DV92 physical contig of 280 kb spanning the Lr10 locus in hexaploid bread wheat was established. Sequencing of 211 kb from this contig revealed the presence of two resistance gene analogs (RGAs), rga1 and rga2, in a region that showed complete linkage to Lr10 in Triticum aestivum (14, 15).
Here, two genes (T10rga1 and T10rga2-1A) orthologous to the T. monococcum genes rga1 and rga2 were isolated from the Lr10 donor line ThatcherLr10. Haplotype studies in the wheat gene pool showed that these two genes are the best possible candidates for Lr10. Mutational analysis and stable transformation of the candidate genes demonstrated that T10rga1 is the Lr10 resistance gene. Our data show that map-based cloning of genes of agronomic interest is feasible from hexaploid wheat by using a combination of subgenome chromosome walking and haplotype studies.
Materials and Methods
Mutant Screening. Three thousand seeds of the parental line ThatcherLr10 were treated with 0.35% ethyl methanesulfonate (EMS). M2 seedlings (52,000) were then artificially infected with the leaf rust (Puccinia triticina) isolate, (AvrLr10) 89–201 CBTB(TX), (13) by using a large-scale infection procedure modified from Schachermayr et al. (16). Thirty-three putative mutants were grown to the next generation and were reassessed for susceptibility by artificial infection. Five susceptible fertile mutants were crossed with the susceptible cv. Frisal. Three of the mutants (EMS_19, EMS_25, and EMS_31) resulted in susceptible F1 progeny, suggesting that the mutation is in the Lr10 gene.
PCR Amplification of the T10rga1 and T10rga2-1A Genes and RT-PCR. Three overlapping fragments spanning the T10rga1 gene were amplified by PCR on 50 ng of genomic DNA extracted from the three EMS mutants and from the hexaploid wheat line Canadian3842 by using the following primer combinations: ThLr10_T (5′-CTGAGTGAGCATGAGCAAC-3′) and ThLr10_P (5′-TGGAATTGAGACAGTACAC-3′); ThLr10_E (5′-AGC CCTAATATGGCAACC-3′) and ThLr10_H (5′-TGTAGAAC CGTGCCT TAC-3′); and ThLr10_G (5′-GCTCT TCTA ACGGGGATC-3′) and ThLr10_J (5′-CATCTCTTGAA AGCTCC-3′). Four overlapping fragments spanning T10rga2-1A were amplified by PCR using the specific primer combinations: Rga2_F (5′-GATGGAGACGACGGTGCT-3′) and Rga2_U (5′-CAACTGCTTGTGATCTGGT-3′); Rga2_V (5′-GAAGCCGGATTATAGTGTCA-3′) and Rga2_W (5′-CTGCCCAGCTAAGTTCTTG-3′); Rga2_X (5′-CAATTGT GATGAACTCCTCA-3′) and Rga2_N (5′-AGGTGACA GATAGATTCAC-3′); and Rga2_K (5′-CTTCTGCGAG TGCTGGAC-3′) and Rga2_E (5′-TTCATAGCTCATTG CATC-3′). A second PCR amplification was performed for the regions where point mutations were identified to confirm that they did not result from Taq polymerase errors. RT-PCRs were performed on 3 μg of poly(A)+ RNA by using the primer combinations ThLr10_D (5′-GTCAAGATCCCGTATCAG-3′)/ThLr10_H or ThLr10_G/ThLr10_H for T10rga1 and the specific primers Rga2_V/Rga2_W for T10rga2-1A.
Biolistic Transformation of the Susceptible Wheat Bobwhite S 98 56. A 4.4-kb genomic fragment containing the entire T10rga1 coding region, 21-bp upstream sequence and 380-bp downstream sequence, was cloned under the control of the maize ubiquitin promoter by using the strategy described by Clausen et al. (17) to generate the plasmid pUbi_T10rga1. pUbi_T10rga2-1A, which contains a 3-kb T10rga2-1A full-length cDNA with 65 bp upstream and 203 bp downstream of the coding sequence was generated by using a similar strategy. A total of 350 immature embryos of the Bobwhite accession SH 98 56 (18) were cotransformed with pUBi_T10rga1, pUbi_T10rga2-1A, and a plasmid containing the selectable phosphomannose isomerase marker (19) by using the PDS-1000/He biolistic particle delivery System (Bio-Rad). Regeneration and selection of the transformed plants were performed as described (18, 19). Eight independent T0 transgenic lines containing either the two transgenes or only one of the transgenes were obtained. T1 transgenic plants were artificially infected with the leaf rust isolate TCB/TD. The number of T10rga1 transgene integration events in the T1 transgenic plants was analyzed by Southern hybridization (13). Expression of the T10rga1 transgene was analyzed by Northern analysis (13) with 20 μg of total RNA extracted from the T1 transgenic plants.
Substitution Rate Analysis. Nucleotide sequences from the rga1 sequences of T. aestivum cvs. ThatcherLr10 and Canadian3842 as well as from Triticum durum cv. Langdon and T. monococcum DV92 were aligned by using the pileup program of the gcg software. The nucleotide alignment was adjusted manually with the program lineup by using the amino acid sequence alignment as a guide to keep a codon-by-codon alignment. The rate of nonsynonymous (Ka) vs. synonymous (Ks) nucleotide substitutions per 100 sites (Ka/Ks) was computed with the program DIVERGE, which is based on the algorithm developed by Li (20) and uses Kimura's two-parameter method (21) for analysis. The different domains (N-terminal, NBS, spacer, and LRR) were chosen according to the domains defined by Meyers et al. (22) for NBS-LRR-encoding genes. A Student t test (sas package, SAS Institute, Cary, NC) was used to test for the significance of differences between the Ka/Ks mean values of the N-terminal and the LRR domains.
Phylogenetic Analysis. Alignment of the amino acid sequences for the genes was performed by using the clustalx program (23). A neighbor-joining (NJ) method was then applied to produce a phylogenetic tree. The relative degree of branch support was determined within the NJ framework by using the bootstrap procedure (24). The original data set was resampled 1,000 times. The LR21 sequence was used as an outgroup.
Results
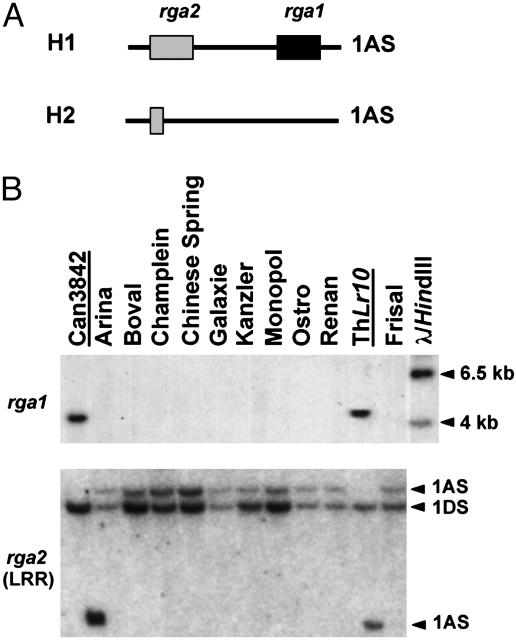
Isolation of Two Candidate Genes for Lr10 in Hexaploid Wheat. Two RGAs, named rga1 and rga2, have been previously identified in a genomic region of T. monococcum DV92 spanning the Lr10 resistance locus in T. aestivum, ThatcherLr10 (14, 15). To test whether these genes are the best candidate genes for Lr10, we have analyzed the Lr10 locus at the molecular level in the wheat gene pool (25). Southern analysis of 113 wild and cultivated diploid and polyploid wheat lines has revealed the presence of two characteristic haplotypes on chromosome 1AS. Haplotype H1 is defined by the presence of the two full-length rga1 and rga2 genes, whereas in haplotype H2, rga1 is absent and only a truncated LRR domain of rga2 can be detected (ref. 25 and Fig. 1A). In a survey of 56 hexaploid European wheat breeding lines, only eight lines had the H1 haplotype, indicating that the H2 haplotype is predominant in T. aestivum (Fig. 1B). Moreover, all of the lines of the H2 haplotype showed identical hybridization patterns at the Lr10 locus; i.e., they lack rga1 and a single fragment hybridizes with the 3′ end of the LRR domain of rga2 on chromosome 1A (Fig. 1B). These data suggest that there is very little variability within the H1 and H2 haplotypes and that there are possibly no other haplotypes at the Lr10 locus in the hexaploid wheat gene pool. Therefore, molecular analysis of these two haplotypes should allow the identification of the complete set of Lr10 candidate genes.
Fig. 1.
Two haplotypes defined by the presence or absence of two candidates for the Lr10 resistance gene on chromosome 1AS are present in the hexaploid wheat gene pool. (A) Schematic representation of the H1 and H2 haplotypes at the Lr10 locus. The candidate genes rga1 and rga2 are present on chromosome 1AS in lines with the H1 haplotype (e.g., ThatcherLr10), but not in lines with the H2 haplotype (e.g., Frisal). (B) The H2 haplotype is predominant and is very conserved in the wheat gene pool. Southern hybridization with HindIII (Upper)- and DraI(Lower)-digested genomic DNA isolated from a subset of 56 hexaploid European wheat breeding lines. Hybridizations were performed with rga1 (Upper) and the LRR domain of rga2 (Lower) as probes. Lines with an H1 haplotype are underlined. The fragments corresponding to T10rga2-1A on chromosome 1AS and T10rga2-1D on chromosome 1DS (25) are indicated with arrowheads.
With the exception of the variety Canadian3842 (Can3842), the H1 lines, including ThatcherLr10, were resistant to leaf rust isolates avirulent on Lr10 (AvrLr10) (data not shown). To identify all candidate genes possibly present at the Lr10 locus in ThatcherLr10, we have characterized the H1 and H2 haplotypes in more detail at the molecular level. Two BAC contigs were established at the Lr10 locus in the tetraploid T. durum cv. Langdon (H1 haplotype) and in the T. aestivum cv. Renan (H2 haplotype) (unpublished data). Low-pass sequencing of the two BACs revealed the presence of the chromosome condensation factor, nodulin-like, and actin genes, which were already identified at the Lr10 locus in T. monococcum DV92 (H1 haplotype) (15). It also confirmed the complete deletion of the rga1 sequence and the presence of a truncated LRR domain of rga2 in the Renan H2 haplotype (data not shown). These studies did not provide additional candidate genes to rga1 and rga2 in the physical interval between markers flanking Lr10 in hexaploid wheat. Therefore, we conclude that rga1 and rga2 of haplotype H1 are the only promising candidate orthologs for the T. aestivum Lr gene Lr10.
The T. monococcum rga1 and rga2 genes were used as probes to isolate the orthologous genes T10rga1 and T10rga2 from a λ library of the resistant hexaploid wheat variety ThatcherLr10. The T10rga1 gene (GenBank accession no. AY270157) has a length of 3,935 bp and contains one intron of 1,171 bp, which, as in most cereal RGAs, is located in the NBS domain at the N terminus of the kinase-2 motif (26). The gene encodes a coiled coil (CC), nucleotide-binding site (NBS), leucine-rich repeat (LRR) (CNL) protein of 919 amino acids with 14 imperfect LRRs at the C terminus (Fig. 2). The length of the CC, NBS, spacer, and LRR domains, as well as the type of amino acid residues contained in the different motives, indicate that T10RGA1 is related to the CNL-C type of proteins (22). By Southern hybridization, it was found that T10rga1 is a single-copy gene on chromosome 1AS in ThLr10. For rga2, two homoeologous genes, T10rga2-1A and T10rga2-1D, were identified on chromosomes 1A and 1D, respectively. The T10rga2-1A gene (GenBank accession no. AY270159) is 4,756 bp long and has two introns of 944 and 310 bp. It encodes a CC-NBS-LRR protein of 1,169 amino acids that is unrelated to T10RGA1. Linkage analysis in a population of 3,120 F2 plants showed that T10rga1 and T10rga2-1A are both completely linked to Lr10. RT-PCR analysis demonstrated that both genes are expressed in the resistant variety ThatcherLr10 (see Fig. 5A, which is published as supporting information on the PNAS web site) and are not induced on leaf rust infection (Fig. 5B).
Fig. 2.
Amino acid sequence of the Lr10 gene. The CC, NBS, spacer, and LRR domains are indicated. Amino acids belonging to characteristic motives in each domain are bold. In the NBS domain they are in the following order: P-loop, RNBS-A, kinase2, RNBS-B, RNBS-C, GLPL, RNBS-D, and MHDV. The four domains that have been used in the Ka/Ks analysis are separated from each other by an empty line. The spacer sequence is indicated in italics and the aliphatic (a) residues in the consensus (xxaxaxx) region of the LRR domain are boxed in yellow. The amino acid residues that are modified in the three EMS mutant genes are highlighted with red boxes with the number of the mutant above them.
Three Independent Mutations Affecting T10rga1 Lead to Susceptibility to Leaf Rust Carrying AvrLr10. Three independent lr10 mutants (T10_EMS19, T10_EMS25, and T10_EMS31) were identified and characterized at the molecular level. The T10rga1 and T10rga2-1A genes were amplified by PCR from the mutants, and their sequences were compared with the sequences of ThatcherLr10. In all mutants, point mutations were detected in T10rga1 but not T10rga2-1A. In T10rga1_EMS19, a C-to-T transition at position 151 resulted in a change of the last leucine residue in the putative CC domain into a phenylalanine residue (Fig. 2). In T10rga1_EMS25, a G-to-A transition at position 608 changed a glycine into an arginine residue in the third conserved glycine of the P-loop motif in the NBS (Fig. 2). A similar transition was detected at position 3,461 in the LRR domain of the T10rga1_EMS31 sequence. It introduced a stop codon instead of a tryptophan residue at the end of the ninth LRR in the LRR domain (Fig. 2). Thus, three independent mutations affect the T10rga1 gene at different positions and result in a loss of resistance against leaf rust, demonstrating that T10rga1 is the Lr10 gene. In addition, these data indicate that the last five LRR are required for Lr10 function and underline the essential role of the leucine and glycine residues in the CC and P-loop motives, respectively.
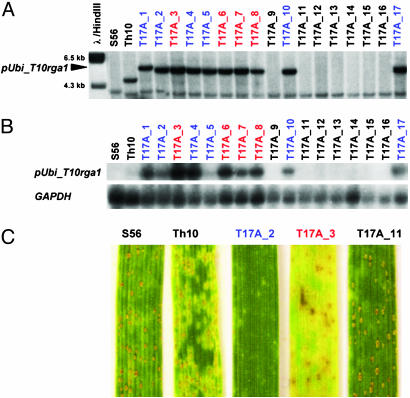
Transgenic Wheat Plants Overexpressing T10rga1 Show Increased Resistance to Leaf Rust Avirulent on Lr10. To test whether T10rga1 is sufficient to confer rust resistance to wheat plants, plasmids containing the T10rga1 (pUbi_T10rga1) and T10rga2-1A (pUbi_T10rga2-1A) genes under control of the maize ubiquitin promoter were cotransformed into the susceptible wheat accession Bobwhite SH 98 56 (18), which has the H2 haplotype. Artificial infection and molecular analysis performed on T1 progeny plants identified two families expressing resistance to leaf rust avirulent on Lr10. In both families (T14 and T17), cosegregation was found between the resistance phenotype and the pUbi_T10rga1 transgene (Fig. 3A). pUbi_T10rga2-1A was present in all of the pUbi_T10rga1 transgenic plants of the T17 family but was not detected in plants of the T14 family. All resistant plants from the segregating T1 families expressed pUbi_T10rga1, as shown by Northern analysis (Fig. 3B), confirming that T10rga1 is the Lr10 gene. Other T1 families, which all contained multiple transgene insertions and rearrangements, were susceptible suggesting gene silencing or incomplete transgene integration (data not shown). The level of expression in the transgenic resistant plants was 8–25 times higher than in ThatcherLr10 (Fig. 3B). Interestingly, in the plants overexpressing T10rga1, the infection type differed from the moderate resistance (small to middle size uredinias surrounded by chlorosis) conferred by the endogenous Lr10 (Fig. 3C). The resistant transgenic plants either showed hypersensitive flecks or developed strong chlorosis and necrotic spots upon leaf rust infection, and no uredinia were formed (Fig. 3C). These data suggest that overexpression of the Lr10 gene enhances leaf rust resistance.
Fig. 3.
Transgenic wheat seedlings overexpressing T10rga1 show enhanced resistance to leaf rust. (A) Southern hybridization of HindIII-digested genomic DNA extracted from Bobwhite SH 98 56 (S56), ThatcherLr10 (Th10), and 17 transgenic T1 plants of the T17 family (T17A_x) with T10rga1 as a probe. The arrowhead indicates the 5.6-kb fragment, which is expected from a HindIII digest of the pUbi_T10rga1 construct. (B) Northern blot of total RNA extracted from the same seedlings hybridized with T10rga1 as a probe. The same blot was hybridized with the housekeeping GAPDH gene as a control. The relative intensity of the hybridization signals in the transgenics vs. wild-type plants was estimated with the Cyclone gene array system (Perkin–Elmer, Boston). Transgenic plants showing chlorotic hypersensitive resistance reaction (i.e., T17A_2) are blue, those with a necrotic phenotype (i.e., T17A_3) are red, while susceptible T1 plants are black. (C) Phenotypes of transgenic plants overexpressing Lr10 compared with the resistant ThacherLr10 and the susceptible Bobwhite SH 98 56, 10 days after artificial infection with the leaf rust isolate TCB/TD AvrLr10.
Diversifying Selection Acts on the N-Terminal Region of the CNL-C Lr10 Gene Product. Comparison of allelic sequences has shown that diversifying selection acts on the LRR encoding domain of many plant disease resistance genes (27). To study whether a particular selective pressure acts on the Lr10 gene, we have compared the sequences of the Lr10 alleles of the two hexaploid wheat lines ThatcherLr10 (T10rga1, GenBank accession no. AY270157) and Can3842 (Can_rga1, GenBank accession no. AY270158), the T. monococcum DV92 (rga1, GenBank accession no. AF326781), as well as a gene amplified from the tetraploid T. durum var. Langdon (Td_rga1, unpublished data). The two latter lines are resistant to leaf rust carrying AvrLr10, but we do not know whether this resistance is conferred by the Lr10 allele, because other resistance genes are possibly present. In contrast, Can3842 is the only susceptible hexaploid wheat line identified so far with an H1 haplotype. Can_rga1 is expressed (see Fig. 6, which is published as supporting information on the PNAS web site), indicating that the susceptibility of Can3842 to AvrLr10 is not due to a lack of gene expression but might result from a loss of recognition between the CAN_RGA1 protein and AVRLr10. The protein sequences encoded by the four genes share, on average, 93% similarity over the entire sequence. Interestingly, conservation is lower in the first 426 amino acids of the LR10 protein (89%) than in the remaining 493 amino acids (96.5%), which mostly form the LRR domain (see Fig. 7, which is published as supporting information on the PNAS web site). Estimation of the number of synonymous (Ks) and nonsynonymous (Ka) nucleotide substitution per site showed that Ks is identical to Ka, indicating selection on the gene as a whole (Table 1). Analysis of the different domains of LR10 (N-terminal, NBS, spacer, and LRR; Fig. 7) revealed that with a Ka/Ks ratio of 0.39, the LRR region is not under positive selection compared with the N-terminal region of the protein, which has a ratio >1 (1.48; Table 1). A statistical t test of the Ka/Ks mean values for the N-terminal and LRR domains showed that the difference is significant (P = 0.0005). The Ka/Ks ratio remained <1 for the intervening residues as well as for the exposed residues (xxaxaxx) of the LRR (data not shown). We conclude that the 5′ end of the Lr10 gene is under diversifying selection.
Table 1. Average rates of nucleotide substitutions per 100 sites among four alleles (T10rga1, Can_rga1, Td_rga1, and rga1) of the Lr10 gene.
| Complete gene | N-terminal | NBS | Spacer | LRR | |
|---|---|---|---|---|---|
| Ka | 3.71 | 8.10 | 3.70 | 1.85 | 1.58 |
| Ks | 3.69 | 5.89 | 3.33 | 3.18 | 2.88 |
| Ka/Ks | 0.98 | 1.48 | 0.99 | 0.83 | 0.39 |
The coding sequences for the N-terminal (amino acids 1–197), NBS (208–502), spacer (503–533), and LRR (534–919) domains were analyzed separately. The Ka/Ks ratio was calculated by averaging the ratio for each comparison.
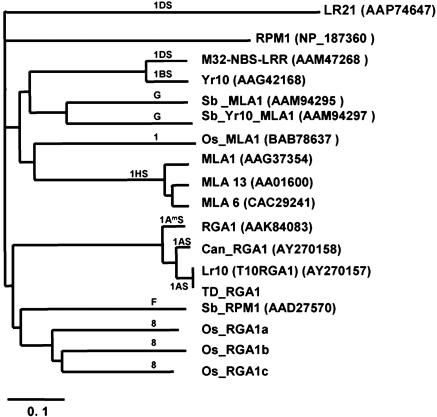
Lr10 Is Related to Other Fungal Disease Resistance Genes in Grasses. To analyze whether Lr10 is similar to other plant disease resistance genes or RGAs, the Lr10 sequence was compared against the databases and against the Lr21 resistance gene (12). More than 50% similarity was found at the amino acid level over the entire sequence with RGAs from rice, barley, sorghum, and wheat, whereas similarity with the LR21 sequence was very low (41%) and restricted to 336 amino acids of the NBS domain. Phylogenetic analysis showed that the wheat LR10 sequence is closely related to RGAs encoded by three predicted genes (Os_RGA1a, Os_RGA1b, and Os_RGA1c) located on a single BAC (AP005158) of chromosome 8 in rice as well as with a sorghum RPM1-like gene product (Sb_RPM1) located on chromosome F (Fig. 4). There was more similarity with the rice and sorghum sequences than with wheat disease RGAs such as M32-NBS-LRR, which segregates with Lr21 at the telomeric end of chromosome 1D in Ae. tauschii (28) or with the putative Yr10 stripe rust resistance gene on chromosome 1BS. Significant similarity was also found with the powdery mildew resistance genes Mla1, Mla 6, and Mla13, which are located on chromosome 1HS in barley (5, 6) as well as with two proteins (Sb_MLA1, Sb_YR10/MLA1) encoded by Mla-like genes on chromosome G in sorghum (ref. 29 and Fig. 4). Finally, LR10 shows significant similarity to the A. thaliana RPM1 sequence (43% over 875 amino acids), which also belongs to the CNL-C class of disease resistance genes and was used as an outgroup in the phylogenetic analysis (Fig. 4). We conclude from this analysis that Lr10 belongs to an ancient class of CNL resistance genes of which members are conserved in the grass genomes. Finally, the Lr10 gene was used as a probe on Southern blots of 24 near isogenic lines of the cultivar Thatcher containing different Lr genes and alleles (Lr1 to Lr34; ref. 30). None of the lines including ThatcherLr21 showed a hybridizing fragment (data not shown), indicating that the other leaf rust genes do not belong to the same NBS-LRR family as Lr10.
Fig. 4.
NJ phylogenetic tree of LR10, LR21, and products of RGAs from different grass species. Except for Td_RGA1 (unpublished results) and the rice homologs on chromosome 8, which were annotated from a BAC sequence (GenBank accession no. AP005158) in this work, the accession numbers of the proteins are given next to the gene names. Genes that did not have names in the database were named after the species of origin (OS, rice; Sb, sorghum) and the homology defined by the authors in the database annotation. The chromosomal location of the genes is indicated on the branch of the phylogenetic tree.
Discussion
Map-Based Cloning in Hexaploid Wheat. The construction of the first BAC library from the diploid wheat T. monococcum DV92 (8) has allowed the recent map-based isolation of VRN1 directly from T. monococcum (10) and the identification of candidate gene orthologs for the hexaploid wheat genes Lr10 (14) and Q (11) by using subgenome map-based cloning. The T. monococcum DV92 line is not known to carry Lr10. Therefore, our work demonstrates that it is possible to isolate a gene from hexaploid bread wheat by using available BAC resources from diploid relatives that do not necessarily contain the target gene. Very recently, the Lr21 resistance gene, which originates from Ae. tauschii, has been isolated (12). In this case, map-based isolation of the gene was performed without chromosome walking as the restriction fragment-length polymorphism probe KSUD14, which is part of the Lr21 gene, could be used directly to screen a cosmid library from the Ae. tauschii Lr21 donor line (12).
The subgenome map-based cloning of Lr10 has succeeded, because T. monococcum DV92 has the same haplotype (H1) as the target hexaploid wheat variety ThatcherLr10 and, therefore, colinearity between the two species was very high at the Lr10 locus. A number of recent studies performed at the interspecific level (31–33), as well as between inbred lines (34), indicate that many rearrangements involving genes have occurred at orthologous loci during plant evolution. For this reason, the isolation of agronomically important genes from hexaploid wheat through subgenome map-based cloning strategies should be performed together with a detailed haplotype characterization of the target locus in the wheat gene pool to identify the most appropriate genomic tools.
Comparison of the Lr10 sequence with Lr21, the only other Lr gene isolated so far (12), showed no significant similarity between the two NBS-LRR sequences, suggesting that the different wheat Lr genes are not very closely related to each other. Consequently, each Lr gene must probably be isolated independently. High-genome coverage BAC libraries have been very recently constructed from tetraploid (35) and hexaploid bread wheat (B. Chalhoub, personal communication). However, given the amount of clones required, i.e., ≈1 million BAC clones for a 6× time coverage of the hexaploid wheat genome, such libraries cannot be constructed for every wheat genotype of interest. In contrast, it should be possible to develop methods for rapidly constructing nonarrayed BAC libraries from a number of diploid wheat genotypes at low cost. We have shown here that libraries from diploid wheat relatives are efficient tools for the map-based isolation of genes from hexaploid wheat.
Lr10 Encodes a CNL-C Protein with an N-Terminal Domain Under Diversifying Selection. The nucleotide substitution pattern in Lr10 alleles varies across the gene with a higher frequency of nonsynonymous substitutions in the 5′ end of the gene, compared with the region encoding the LRR domain, which is highly conserved. This pattern contrasts with the majority of LRR-containing disease resistance proteins for which allelic sequence comparisons and Ka/Ks analysis have demonstrated that the LRR domain is under diversifying selection and plays a role in resistance specificity (27). Only in few cases, such as for L alleles in flax, the N-terminal TIR domain has also been shown to be involved in determining resistance specificity (36). Here, we have compared four alleles of Lr10, and it was not possible to correlate variations observed in the amino acid sequences of the diversifying N-terminal domain with the resistance phenotypes. However, with the Lr10 sequence information and our knowledge about the haplotype composition in the wheat gene pool (25), we have now the possibility to isolate additional Lr10 alleles from wild relatives of wheat. Complementation tests using these alleles will provide essential information if they act as Lr genes and will ultimately help to define the role of the N-terminal region in LR10 resistance specificity. Finally, analysis of the EMS lr10 mutants has demonstrated that the last five LRRs are required for Lr10 function and has confirmed the important role of the leucine and glycine residues in the CC and P-loop motives, respectively. Similar loss of resistance has been observed in mutants of the flax rust resistance M gene lacking 426 bp encoding part of the LRR domain (37), whereas mutational analysis performed in the tobacco N (38) and in the A. thaliana RPM1 (39) genes have highlighted the importance of an intact P-loop motif for resistance gene function.
Lr10 Is Conserved in Grass Species, and It Shows Similarities to RPM1 in A. thaliana. LR10 has significant sequence similarity with the Arabidopsis RPM1 protein. Both genes belong to the same class of CNL-C resistance genes, suggesting that they originate from a common ancestor. Lr10 homologs have been identified in barley, sorghum, and rice. Interestingly, there was more similarity between LR10, a sorghum RPM1-like protein sequence, and the three protein sequences predicted from genes located on rice chromosome 8 than with any other wheat RGAs, suggesting conservation of the RPM1/Lr10 type of resistance genes in these species. Except for the barley Mla genes, none of these grass homologs has been identified as a disease resistance gene or has been mapped at disease resistance loci. The positions of the barley and wheat homologs on the genetic maps of the homoeologous chromosomes 1 did not suggest orthology between these genes and Lr10. However, we have identified by hybridization a homolog of Lr10 in barley, which maps at the telomeric region of chromosome 1HS (unpublished data), where the Rph4 gene is located (40). Mapping of this gene in a population segregating for Rph4 needs to be performed to study whether both wheat and barley rust genes are orthologous. Orthology between resistance genes in grasses is not expected from previous analyses (41), which have shown rapid reorganization of resistance gene loci between related grass species. However, it is possible that in species that are very closely related some orthologous relationships are conserved, as found by Pan et al. (42) for four resistance gene homolog loci in tomato and potato.
Overexpression of Lr10 Enhances Resistance to Leaf Rust Carrying Avrlr10. In ThatcherLr10, the Lr10 gene provides only moderate resistance, is constitutively expressed, and is not induced on leaf rust infection. Plant disease resistance genes are only rarely induced by pathogen attack and are usually expressed at low levels (2), suggesting that too high expression might have negative effects. This result is supported by the finding that in the A. thaliana epigenetic variant bal1, which overexpresses an NBS-LRR gene, developmental abnormalities such as late flowering, dwarfing, and altered floral structures were described (43). Interestingly, we have observed that some resistant transgenic plants overexpressing Lr10 are smaller and have fewer tillers compared with nontransgenic plants (unpublished data). Possible correlations between the level of T10rga1 expression and developmental alterations will be analyzed in the next generations. The overexpression of Lr10 resulted in enhanced resistance with a complete prevention of rust sporulation compared with ThatcherLr10. In addition, a necrotic phenotype, which has not been described before for Lr10 (30), has been observed in some transgenic resistant plants. Similar to transgenic barley plants expressing the stem rust resistance gene Rpg1 (44), it was not possible to strictly correlate the level of Lr10 transgene expression with the resistance types observed in the transgenic plants. Analysis in the next generation will provide additional information about the genotype of the T1 plants and will allow the comparison of rust fungal growth in the two resistant types. Thus, our data suggest that overexpression of a disease resistance gene can improve resistance. This finding has also been shown in the cases of the Pto and Prf genes in tomato (45, 46), and more recently for the Rpg1 resistance gene in barley (44).
In the near future, additional wheat disease resistance genes will be isolated by using similar strategies as in this work. Their characterization will provide a better understanding of the molecular basis of disease resistance in wheat and will allow, in the long term, the development of genomics-guided transgene strategies (47), such as the combination of resistance specificities, the overexpression of resistance genes, and the use of the cloned genes as “perfect” markers for molecular breeding. These strategies should ultimately lead to improved resistance of wheat against fungal diseases.
Supplementary Material
Acknowledgments
We thank Dr. A. Pellegrineschi and Dr. R. Singh (Centro Internacional de Mejoramiento de Maiz y Trigo, Mexico) for providing us with seeds of Bobwhite SH 98 56 and for the leaf rust isolate TCB/TD. This work was supported by Swiss National Science Foundation Grant 3100-065114 and a grant from the Indo-Swiss Collaboration in Biotechnology.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; RGA, resistance gene analog; EMS, ethyl methanesulfonate.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY270157–AY270159).
References
- 1.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826-833. [DOI] [PubMed] [Google Scholar]
- 2.Hulbert, S. H., Webb, C. A., Smith, S. M. & Sun, Q. (2001) Annu. Rev. Phytopathol. 39, 285-312. [DOI] [PubMed] [Google Scholar]
- 3.Buschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., vanDaelen, R., vanderLee, T., Diergaarde, P., Groenendijk, J., et al. (1997) Cell 88, 695-705. [DOI] [PubMed] [Google Scholar]
- 4.Yu, Y., Tomkins, J. P., Waugh, R., Frisch, D. A., Kudrna, D., Kleinhofs, A., Brueggeman, R. S., Muehlbauer, G. J., Wise, R. P. & Wing, R. A. (2000) Theor. Appl. Genet. 101, 1093-1099. [Google Scholar]
- 5.Halterman, D., Zhou, F. S., Wei, F. S., Wise, R. P. & Schulze-Lefert, P. (2001) Plant J. 25, 335-348. [DOI] [PubMed] [Google Scholar]
- 6.Zhou, F. S., Kurth, J. C., Wei, F. S., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R. & Schulze-Lefert, P. (2001) Plant Cell 13, 337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brueggeman, R., Rostoks, N., Kudrna, D., Kilian, A., Han, F., Chen, J., Druka, A., Steffenson, B. & Kleinhofs, A. (2002) Proc. Natl. Acad. Sci. USA 99, 9328-9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lijavetzky, D., Muzzi, G., Wicker, T., Keller, B., Wing, R. & Dubcovsky, J. (1999) Genome 42, 1176-1182. [PubMed] [Google Scholar]
- 9.Moullet, O., Zhang, H. B. & Lagudah, E. S. (1999) Theor. Appl. Genet. 99, 305-313. [Google Scholar]
- 10.Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T. & Dubcovsky, J. (2003) Proc. Natl. Acad. Sci. USA 100, 6263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faris, J. D., Fellers, J. P., Brooks, S. A. & Gill, B. S. (2003) Genetics 164, 311-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, L., Brooks, S. A., Li, W., Fellers, J. P., Trick, H. N. & Gill, B. S. (2003) Genetics 164, 655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feuillet, C., Schachermayr, G. & Keller, B. (1997) Plant J. 11, 45-52. [DOI] [PubMed] [Google Scholar]
- 14.Stein, N., Feuillet, C., Wicker, T., Schlagenhauf, E. & Keller, B. (2000) Proc. Natl. Acad. Sci. USA 97, 13436-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wicker, T., Stein, N., Albar, L., Feuillet, C., Schlagenhauf, E. & Keller, B. (2001) Plant J. 26, 307-316. [DOI] [PubMed] [Google Scholar]
- 16.Schachermayr, G. M., Messmer, M. M., Feuillet, C., Winzeler, H., Winzeler, M. & Keller, B. (1995) Theor. Appl. Genet. 90, 982-990. [DOI] [PubMed] [Google Scholar]
- 17.Clausen, M., Krauter, R., Schachermayr, G., Potrykus, I. & Sautter, C. (2000) Nat. Biotechnol. 18, 446-449. [DOI] [PubMed] [Google Scholar]
- 18.Pellegrineschi, A., Noguera, L. M., Skovmand, B., Brito, R. M., Velazquez, L., Salgado, M. M., Hernandez, R., Warburton, M. & Hoisington, D. (2002) Genome 45, 421-430. [DOI] [PubMed] [Google Scholar]
- 19.Wright, M., Dawson, J., Dunder, E., Suttie, J., Reed, J., Kramer, C., Chang, Y., Novitzky, R., Wang, H. & Artim-Moore, L. (2001) Plant Cell Rep. 20, 429-436. [DOI] [PubMed] [Google Scholar]
- 20.Li, W. H. (1993) J. Mol. Evol. 36, 96-99. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, M. (1980) J. Mol. Evol. 16, 111-120. [DOI] [PubMed] [Google Scholar]
- 22.Meyers, B. C., Kozik, A., Griego, A., Kuang, H. & Michelmore, R. W. (2003) Plant Cell 15, 809-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein, J. (1985) Evolution (Lawrence, Kans.) 39, 783-791. [DOI] [PubMed] [Google Scholar]
- 25.Scherrer, B., Keller, B. & Feuillet, C. (2002) Funct. Integr. Genomics 2, 40-50. [DOI] [PubMed] [Google Scholar]
- 26.Bai, J. F., Pennill, L. A., Ning, J. C., Lee, S. W., Ramalingam, J., Webb, C. A., Zhao, B. Y., Sun, Q., Nelson, J. C., Leach, J. E., et al. (2002) Genome Res. 12, 1871-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergelson, J., Kreitman, M., Stahl, E. A. & Tian, D. (2001) Science 292, 2281-2285. [DOI] [PubMed] [Google Scholar]
- 28.Spielmeyer, W., Sharp, P. J. & Lagudah, E. S. (2003) Crop Sci. 43, 333-336. [Google Scholar]
- 29.Song, R., Llaca, V. & Messing, J. (2002) Genome Res. 12, 1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIntosh, R. A., Wellings, C. R. & Park, R. F. (1995) in Wheat Rusts: an Atlas of Resistance Genes, eds. McIntosh, R. A., Wellings, C. R. & Park, R. F. (CSIRO, Melbourne; Kluwer, Dordrecht, The Netherlands).
- 31.Bennetzen, J. L. & Ramakrishna, W. (2002) Plant Mol. Biol. 48, 821-827. [DOI] [PubMed] [Google Scholar]
- 32.Brunner, S., Keller, B. & Feuillet, C. (2003) Genetics 164, 673-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wicker, T., Yahiaoui, N., Guyot, R., Schlagenhauf, E., Liu, Z.-D., Dubcovsky, J. & Keller, B. (2003) Plant Cell 15, 1186-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu, H. H. & Dooner, H. K. (2002) Proc. Natl. Acad. Sci. USA 99, 9573-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cenci, A., Chantret, N., Xy, K., Gu, Y., Anderson, O. D., Fahima, T., Distelfeld, A. & Dubcovsky, J. (2003) Theor. Appl. Genet., 107, 931-939. [DOI] [PubMed] [Google Scholar]
- 36.Luck, J. E., Lawrence, G. J., Dodds, P. N., Shepherd, K. W. & Ellis, J. G. (2000) Plant Cell 12, 1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson, P. A., Lawrence, G. J., Morrish, B. C., Ayliffe, M. A., Finnegan, E. J. & Ellis, J. G. (1997) Plant Cell 9, 641-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinesh-Kumar, S. P., Tham, W. H. & Baker, B. J. (2000) Proc. Natl. Acad. Sci. USA 97, 14789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tornero, P., Chao, R. A., Luthin, W. N., Goff, S. A. & Dangl, J. L. (2002) Plant Cell 14, 435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin, Y., Cui, G. H., Steffenson, B. J. & Franckowiak, J. D. (1996) Phytopathology 86, 887-890. [Google Scholar]
- 41.Leister, D., Kurth, J., Laurie, D. A., Yano, M., Sasaki, T., Devos, K., Graner, A. & Schulze-Lefert, P. (1998) Proc. Natl. Acad. Sci. USA 95, 370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, Q., Liu, Y.-S., Budai-Hadrian, O., Sela, M., Carmel-Goren, L., Zamir, D. & Fluhr, R. (2000) Genetics 155, 309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stokes, T. L., Kunkel, B. N. & Richards, E. J. (2002) Genes Dev. 16, 171-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath, H., Rostoks, N., Brueggeman, R., Steffenson, B., von Wettstein, D. & Kleinhofs, A. (2003) Proc. Natl. Acad. Sci. USA 100, 364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oldroyd, G. E. D. & Staskawicz, B. J. (1998) Proc. Natl. Acad. Sci. USA 95, 10300-10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, X. Y., Xie, M. T., Kim, Y. J., Zhou, J. M., Klessig, D. F. & Martin, G. B. (1999) Plant Cell 11, 15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauss, S. H. (2003) Science 300, 61-62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.