Abstract
Chlorophyll (chl) breakdown during senescence is an integral part of plant development and leads to the accumulation of colorless catabolites. The loss of green pigment is due to an oxygenolytic opening of the porphyrin macrocycle of pheophorbide (pheide) a followed by a reduction to yield a fluorescent chl catabolite. This step is comprised of the interaction of two enzymes, pheide a oxygenase (PaO) and red chl catabolite reductase. PaO activity is found only during senescence, hence PaO seems to be a key regulator of chl catabolism. Whereas red chl catabolite reductase has been cloned, the nature of PaO has remained elusive. Here we report on the identification of the PaO gene of Arabidopsis thaliana (AtPaO). AtPaO is a Rieske-type iron–sulfur cluster-containing enzyme that is identical to Arabidopsis accelerated cell death 1 and homologous to lethal leaf spot 1 (LLS1) of maize. Biochemical properties of recombinant AtPaO were identical to PaO isolated from a natural source. Production of fluorescent chl catabolite-1 required ferredoxin as an electron source and both substrates, pheide a and molecular oxygen. By using a maize lls1 mutant, the in vivo function of PaO, i.e., degradation of pheide a during senescence, could be confirmed. Thus, lls1 leaves stayed green during dark incubation and accumulated pheide a that caused a light-dependent lesion mimic phenotype. Whereas proteins were degraded similarly in wild type and lls1, a chl-binding protein was selectively retained in the mutant. PaO expression correlated positively with senescence, but the enzyme appeared to be post-translationally regulated as well.
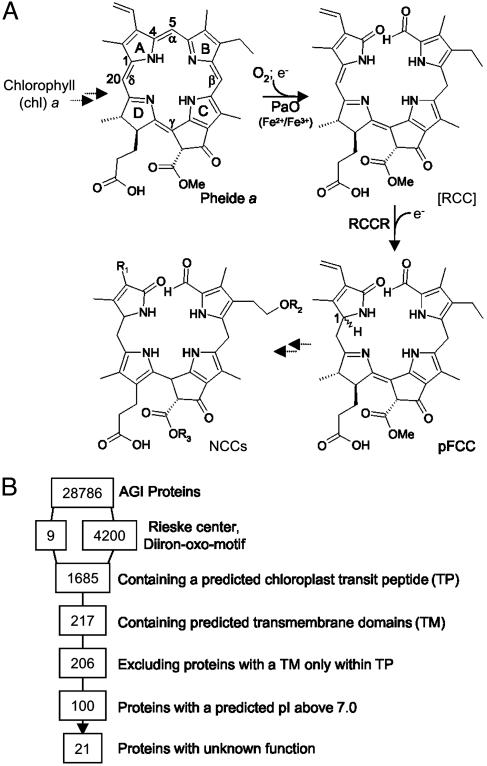
During leaf senescence, chlorophyll (chl) is degraded to colorless linear tetrapyrroles, termed nonfluorescent chl catabolites (NCCs; refs. 1–3). The pathway of chl catabolism (Fig. 1A) includes the occurrence of both colored and colorless intermediates. Thus, in two subsequent reactions catalyzed by chlorophyllase and Mg dechelatase, respectively, phytol and the central Mg atom are removed. Then, the ring structure of pheophorbide (pheide) a (Fig. 1 A) is oxygenolytically opened at the α-mesoposition between C4 and C5 by pheide a oxygenase (PaO). The product, red chl catabolite (RCC), does not accumulate in vivo (4) but is rapidly converted to a primary fluorescent chl catabolite (pFCC) by a stereospecific reduction of the C20/C1 double bond. The source of the responsible enzyme, RCC reductase (RCCR), defines which of two possible C1 isomers, pFCC-1 or -2, occurs (Fig. 1 A). RCCR of Arabidopsis has been shown to produce pFCC-1 (5). Further steps of the chl breakdown pathway involve reactions known from plant detoxification mechanisms (6). FCCs are hydroxylated and in some cases conjugated with a glucosyl or malonyl moiety (7, 8), followed by their export into the vacuole by a primary active ATPase (9). Finally, FCCs are nonenzymically tautomerized to the respective NCCs because of the acidic pH inside the vacuole (10).
Fig. 1.
The pathway of chl catabolism, and identification of possible PaO proteins in A. thaliana.(A) The pathway of chl catabolism. The key reaction is catalyzed by the joint action of PaO and RCCR. Chemical constitutions of chl catabolites are shown. Pyrrole rings (A–D), methine bridges (α-δ), and relevant carbon atoms are labeled. NCCs are species-specifically modified at different side positions (R1-R3). (B) Screening of the ATH1.pep database of the Arabidopsis Genome Initiative (AGI Proteins) for proteins with known properties of PaO.
The biochemistry of chl catabolism has been investigated extensively during the last years (for recent reviews, see refs. 3, 5, 11, and 12). Surprisingly, PaO turned out to be a key regulator of this pathway. Thus, PaO activity is detectable only during senescence (13, 14), whereas activities of other enzymes, such as chlorophyllase and RCCR, are constitutive (15–17). In addition, the reactions catalyzed by PaO and RCCR are responsible for the loss of pigment color. Biochemical evidence suggests that the two enzymes are interacting during catalysis. Thus in vitro, the intermediate, RCC, does not accumulate to substantial amounts in the absence of RCCR, indicating that RCC is metabolically channeled (4). PaO has been demonstrated to be located at the inner envelope of senescing chloroplasts (18). In contrast, RCCR is a stroma protein, suggesting that pheide a to pFCC conversion occurs at the stromal periphery of the inner envelope (4, 19). The recent cloning of RCCR (20) has uncovered a distinct relationship to other plant bilin reductases, all of which are ferredoxin (Fd)-dependent (21). Reduced Fd is also needed as a source of electrons for the PaO/RCCR-catalyzed reaction (13, 19). PaO is a nonheme iron type (14) monooxygenase that introduces one atom of molecular oxygen at the α-methine bridge of pheide a (Fig. 1 A), giving rise to the formyl group attached to pyrrole ring B in pFCC (22). Its activity is restricted to pheide a, with pheide b being a competitive inhibitor. Consequently, all NCCs identified so far are derived from chl a (23). Before entering this degradation pathway, chl b has to be converted to chl a. A respective chlorophyll cycle that is responsible for the interconversion of both types of chl has been described (24, 25). In addition, activity of one of the enzymes involved in chl b to a conversion, chl b reductase, increases during barley leaf senescence (26).
Senescence is the final stage of leaf development, ultimately leading to the death of the entire leaf. It is a highly regulated process that involves an ordered disintegration of chloroplast components, such as thylakoid membranes, along with the remobilization of amino acids from proteins, such as the chl a/b-binding proteins, and the subsequent release of potentially phototoxic chl. It is commonly believed that the function of chl degradation in plants is to avoid this hazard (3). chl in its free form would cause photooxidative damage to the senescing cell, and it has been shown that the accumulation of photoactive intermediates of chl biosynthesis causes a premature cell death phenotype in two chl biosynthetic mutants and transgenic plants (27, 28). This is termed a lesion mimic phenotype that, in contrast to the phenotypically similar hypersensitive response, occurs in the absence of pathogen infection. In addition to chl biosynthetic mutants, Arabidopsis accelerated cell death 2 (acd2) has been isolated that develops a light-dependent lesion mimic phenotype (29). acd2 is deficient in RCCR, and the phenotype has been suggested to be caused by the accumulation of phototoxic RCC (30). Thus, the ability of plants to degrade chl during senescence seems vitally important. Here we describe the molecular identification of PaO. In addition, we show that a mutant that is defective in PaO shows a stay-green phenotype in the dark and accumulates pheide a, which causes light-dependent premature cell death.
Materials and Methods
Plants and Growth Conditions. The maize lls1 mutant, containing the reference allele, was obtained from the Maize Genetics Cooperation Stock Center, University of Illinois at Urbana–Champaign, and was grown for 7–9 wk in a greenhouse. Arabidopsis thaliana ecotype Columbia was grown in soil under short-day conditions at 120 μmol·m-2·s-1. For dark induction of senescence, excised Arabidopsis leaves or lls1 leaf discs (1.0-cm diameter) were incubated on moistened filter paper or floating on tap water for several days, as indicated in Figs. 3, 4, 5.
Fig. 3.
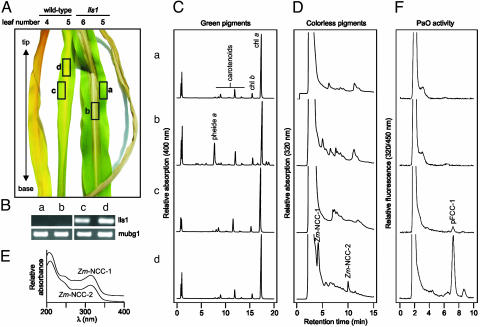
Characterization of lls1 and of wild-type leaf tissue. (A) Visible phenotype of lls1 and wild-type leaves. The boxed areas (a–d) represent the developmental stage of tissues used for the analyses depicted in B–F. (B) Relative RT-PCR analysis of lls1 and wild-type plants. The amount of mRNA encoding polyubiquitin 1 (mubg1) is unchanged. Lls1 mRNA (lls1) is absent in the mutant. (C) HPL chromatograms of green chl pigments. (D) HPLC traces of colorless chl catabolites. Two NCCs, Zm-NCC-1 and -2, accumulate only in senescent wild-type tissue. (E) Absorbance spectra of Zm-NCC-1 and -2. (F) HPLC traces of PaO/RCCR assays. All analyses presented were repeated with qualitatively identical results.
Fig. 4.
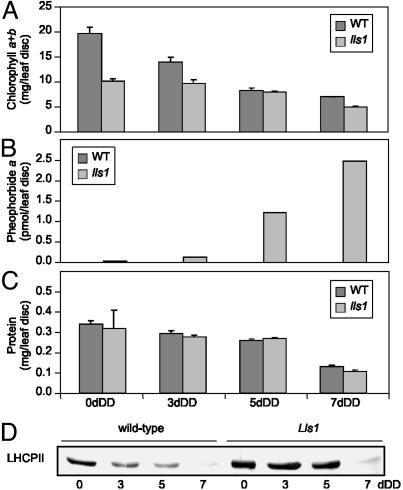
Characterization of maize lls1 and wild-type leaf discs during senescence. To induce senescence, leaf discs were incubated for 0, 3, 5, or 7 d in complete darkness (DD). (A–C) Wild type (WT) is shown with filled bars and lls1 with shaded bars. The values presented are means from one experiment with three replicates. All analyses presented were repeated with similar results. (A) Analysis of chl contents; (B) analysis of pheide a accumulation; (C) analysis of total protein content; (D) immunoblot analysis of light harvesting complex protein II in lls1 and wild type.
Fig. 5.
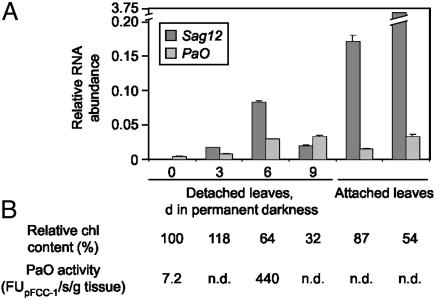
Analysis of AtPaO expression during senescence. (A) For isolation of RNA, either detached leaves incubated in permanent darkness for 0, 3, 6, or 9 d or attached leaves were used. RNA abundances of Sag12, a marker gene for senescence (filled bars), and Acd1/PaO (PaO; shaded bars) were quantified by real-time RT-PCR and normalized to the levels of mRNA encoding the actin 2 protein. Values are means of three replicates. (B) Progression of senescence was monitored by determining the chl content and by measuring PaO activity. n.d., not determined.
Computational Analysis. The BULK PROTEIN ANALYSIS tool at The Arabidopsis Information Resource (TAIR; www.arabidopsis.org) was used to screen the ATH1.pep database (Ver. 4.0) of the Arabidopsis Genome Initiative (31) for the presence of the Rieske motif (PF00350). Proteins containing a diiron-oxo motif (32) were identified with the patmatch tool at TAIR. By using the BULK PROTEIN ANALYSIS tool (TAIR), a subset of 4,209 proteins was analyzed for the following properties: (i) proteins containing a putative plastid-targeting sequence (cTP), (ii) proteins containing putative transmembrane domains (TM), and (iii) prediction of pI. Proteins containing a TM domain only within the predicted cTP were eliminated manually. Screening for known protein function was done with the gene ontology and gene hunter tools at TAIR.
Analysis of Recombinant PaO. Full-length cDNA clones of Acd1 (pda07874) and At4g25650 (pda06002) were obtained from the RIKEN Tsukuba Institute BioResourceCenter (33). For subcloning into pQE30 (Qiagen, Chatsworth, CA), convenient restriction sites were introduced at the ends of the ORFs by PCR. After confirmation by sequencing, the respective constructs were expressed in Escherichia coli strains JM109 and C43 according to standard procedures. Total proteins were extracted as described (20), except that after sonication, extracts were treated with 2% (vol/vol) Triton X-100 (14) to solubilize membrane proteins. After the addition of 10% (vol/vol) glycerol, extracts were frozen in liquid nitrogen and stored at -80°C until use. Expression of proteins was analyzed with immunoblots by using a RGS·His antibody (Qiagen).
PaO Assays. PaO activity was assessed by using a coupled PaO/RCCR assay according to established protocols (4, 14, 20). Briefly, the assay contained, in a total volume of 50 μl, PaO, RCCR, and the following supplements: 2 mM pheide a (isolated according to ref. 20); 10 μg of Fd (Sigma) and a Fd-reducing system [2 mM glucose-6-phosphate; 1 mM NADPH; 50 milliunits of glucose-6-phosphate dehydrogenase; 10 milliunits of Fd-NADPH-oxidoreductase (Sigma)]. As a source of PaO, either E. coli protein extracts (see above) or chloroplast membranes isolated from maize, A. thaliana, or oilseed rape (14) were used. As a source of RCCR, Arabidopsis RCCR expressed in E. coli was used (20). The production of pFCC-1 from pheide a was linear for >1 h of incubation at 24°C and was followed by reversed-phase HPLC (4, 20) by using 40 mM potassium phosphate, pH 7.0/methanol (2:3, vol/vol) as solvent. pFCC-1 eluting after ≈8.0 min under these conditions was quantified as integrated fluorescence units (14). Calculation of the apparent Km was performed according to Lineweaver and Burk (34).
Analysis of chl and chl Catabolites. chl and polar green chl catabolites (pheides and chlorophyllides) were extracted from leaf tissues and separated by reversed-phase HPLC (35). For extraction of NCCs, maize leaf material (eight leaf discs) was homogenized in liquid nitrogen and extracted into 0.4 ml 0.1 M Tris·HCl, pH 8.0/methanol (1:4, vol/vol). After centrifugation, the supernatant was analyzed by HPLC (36). NCCs were eluted with a linear gradient of solvent B (20% (vol/vol) 25 mM potassium phosphate buffer, pH 7.0, and 80% methanol) in solvent A (50 mM potassium phosphate, pH 7.0) as follows: 0% to 100% over 15 min and 100% solvent B for 8 min. All pigments were identified by their absorption spectra and, in addition, green pigments were identified and quantified by using authentic standards (14, 36). chl was determined as described (37).
RNA Isolation and RT-PCR. RNA was prepared by using the Plant RNeasy Kit (Qiagen). After digestion of DNA with RQ1 RNase-free DNase (Promega), 1 μg of RNA was reverse-transcribed with the RETROscript Kit (Ambion, Austin, TX). By using the QuantiTect SYBR Green PCR Kit (Qiagen), quantitative PCR was performed with a LightCycler according to the manufacturer's suggestions (Roche Diagnostics, Rotzrenz, Switzerland). Alternatively, relative RT-PCR analysis was performed according to the manufacturer's protocols (Ambion). Specific primers to the following genes were used: Arabidopsis ACT2 (GenBank accession no. U41998: forward, 5′-TGGAATCCACGAGACAACCTA-3′; reverse, 5′-TTCTGTGAACGATTCCTGGAC-3′); Sag12 (GenBank accession no. U37336: forward, 5′-CGAAGGCGGTTTAATGGATA-3′; reverse, 5′-CACCTCCTTCA AT TCCA ACG-3′); Acd1 (GenBank accession no. NM_114357: forward, 5′-ACGGCATGGTAAGAGTCAGC-3′; reverse, 5′-AAACCAGCAAGAACCAGTCG-3′); maize MubG1 (GenBank accession no. U29159: forward, 5′-GACCCTGACTGGAAAAACCA-3′; reverse, 5′-ACATCGGCAGCTTAAACGAC-3′); Lls1 (GenBank accession no. U77346: forward, 5′-TCGTTGAAATGCTCGTCTTG-3′; reverse, 5′-TCAATGTCAGCATGCACGTA-3′).
Extraction and Analysis of Leaf Protein. Proteins were extracted from leaf tissues as follows. Eight leaf discs were ground in liquid nitrogen, extracted into 0.4 ml of extraction buffer [20 mM sodium phosphate, pH 7.5; 0.1% (vol/vol) 2-mercapto ethanol; 1% (wt/vol) polypyrrolidone], and filtrated through two layers of miracloth (Calbiochem). Samples corresponding to equal amounts of leaf tissue were loaded onto SDS/PAGE gels. An antiserum raised against LHCII protein of Festuca pratensis was used for immunoblot analyses (38).
Results and Discussion
Identification of Possible PaO Genes in the Arabidopsis Genome. The PaO/RCCR catalyzed reaction has been extensively characterized in the past (4, 13, 14, 20, 22). Exploiting PaO properties known from these biochemical analyses, we performed an in silico analysis to narrow down the number of putative genes in the Arabidopsis genome potentially encoding PaO. PaO is a nonheme iron (NHI) containing monooxygenase. In vitro, the activity of PaO is independent of a pterin, α-ketoglutarate, or ascorbate (14), known to be cofactors of different NHI proteins (39). Taken together, this information restricts the molecular nature of PaO to two major types of NHI enzymes, i.e., Rieskeand diiron-oxo-type oxygenases (39). For iron-coordinating clusters of both types of NHI enzymes, conserved motifs have been established that we used to screen the ATH1.pep database of the Arabidopsis Genome Initiative (31). From nine (Rieske) and 4,200 (diiron-oxo) hits obtained, respectively, proteins were selected that met additional properties of PaO, in particular its localization in the chloroplast inner envelope (18). Thus, proteins were selected that contained a putative cTP and putative transmembrane domains (TM). After elimination of proteins containing a putative TM only within the predicted cTP and furthermore of proteins with a predicted pI below 7.0 (envelope proteins generally have a rather alkaline pI; ref. 40), proteins with known function were excluded (Fig. 1B). Among the resulting 21 proteins (Table 1) was Accelerated Cell Death 1 (ACD1, At3g44880) the maize homologue of which Lethal Leaf Spot 1 (LLS1) has been cloned (41).
Table 1. List of 21 PaO candidates in the Arabidopsis genome.
| Locus | Mr | pl | TM | AtDB annotation |
|---|---|---|---|---|
| At1g05750 | 55,761 | 8.40 | 2 | Pentatricopeptide repeat-containing protein |
| At1g11470 | 16,268 | 9.61 | 1 | Hypothetical protein |
| At1g48880 | 55,063 | 8.47 | 1 | Hypothetical protein |
| At1g54320 | 38,937 | 10.00 | 2 | Membrane protein common family |
| At1g67390 | 54,457 | 8.81 | 1 | Hypothetical protein |
| At1g79760 | 33,653 | 9.00 | 2 | Expressed protein |
| At2g07370 | 62,077 | 9.38 | 3 | Hypothetical protein |
| At2g26920 | 71,040 | 7.28 | 1 | Expressed protein |
| At2g40390 | 56,848 | 8.79 | 2 | Expressed protein |
| At2g45870 | 46,262 | 8.83 | 1 | Expressed protein |
| At3g02430 | 24,052 | 7.45 | 4 | Hypothetical protein |
| At3g27180 | 56,369 | 9.34 | 2 | Expressed protein |
| At3g44880 | 60,738 | 7.54 | 2 | Rieske [2Fe-2S] domain-containing protein similar to lethal leaf-spot 1 from Zea mays |
| At3g61320 | 46,522 | 8.11 | 2 | Expressed protein hypothetical protein |
| At4g24860 | 62,210 | 7.08 | 1 | Hypothetical protein |
| At4g25650 | 63,801 | 8.81 | 2 | Rieske [2Fe-2S] domain-containing protein similar to cell death suppressor protein lls1 from Z. mays |
| At5g27010 | 98,190 | 8.58 | 1 | Hypothetical protein, predicted proteins |
| At5g35570 | 72,893 | 7.26 | 1 | Auxin-independent growth promoter-related protein |
| At5g42660 | 53,044 | 10.07 | 1 | Expressed protein |
| At5g63390 | 62,873 | 10.30 | 2 | Auxin-independent growth promoter-related protein |
| At5g64710 | 94,038 | 8.28 | 1 | Expressed protein |
Included are the names of the respective loci; their predicted molecular weight (Mr), pl, and number of TM domains, respectively; and their annotation in the A. thaliana database. TM, transmembrane domain.
Although its biochemical function has not been established, Lls1 was proposed to encode an active suppressor of cell death (41) that plays a role in the removal of a potential photosensitive metabolite (42). ACD1 and its closest relative (At4g25650) occurred in both the Rieske and the diiron-oxo search and appeared to us the most likely candidates for PaO for the following reasons. (i) lls1 and acd1 mutants develop a light-dependent lesion mimic phenotype (41–43) that, by analogy to acd2 mutants (30), might result from the accumulation of intermediates of chl catabolism. (ii) ACD1 and LLS1 are predicted to contain a Rieske-type iron–sulfur cluster and a mononuclear iron-binding site necessary for oxygen activation. (iii) ACD1 has been assigned to the envelope in a plastid proteomic approach (S. Baginsky, personal communication). (iv) Gene expression analyses by using Affymetrix GeneChips demonstrated that Acd1 gene expression is enhanced during Arabidopsis leaf senescence (V. Buchanan-Wollaston and U. Zentgraf, personal communications), and (v) the Arabidopsis genome encodes four ACD1-related proteins (42), one of which (At1g44446) is chl a oxygenase that catalyses the conversion of chl a to b. Interestingly, like in the PaO reaction (22), one atom of molecular O2 is incorporated into the substrate, the electrons being supplied by reduced Fd (24). For these reasons, initial studies focused on ACD1 and its closest relative At4g25650 (Table 1).
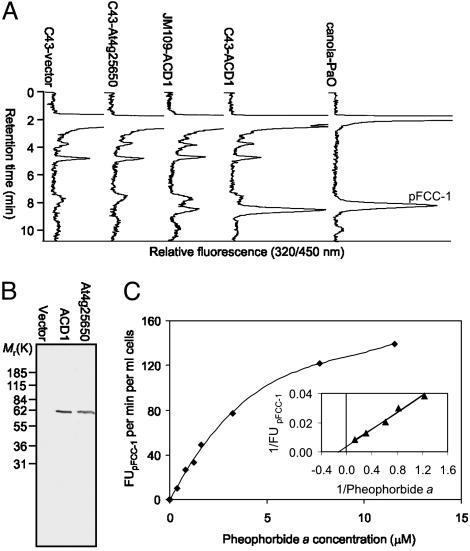
Functional Expression of AtPaO/ACD1 in E. coli. To investigate whether ACD1 or At4g25650 has PaO activity, we expressed cDNA sequences of both in two different E. coli strains. In vitro assays using crude E. coli extracts (Fig. 2A) demonstrated that pFCC-1-forming activity was obtained only from cells expressing ACD1. In addition, activity was higher in E. coli C43 that exhibits improved capacity for the production of eukaryotic membrane proteins (44). Expression of proteins was confirmed by Western blot analysis by using an antibody directed against the His-tag (Fig. 2B). PaO activity in C43-ACD1 cells exhibited requirements identical to those of PaO activity from senescent canola chloroplasts (14). Thus, pFCC-1 formation required RCCR, Fd, a Fd-reducing system, and the substrates pheide a and O2 (data not shown). pFCC-1 formation as a function of pheide a concentrations followed Michaelis–Menten kinetics (Fig. 2C) with an apparent Km of 6.0 μM. As shown for PaO from canola (14), pheide b was not a substrate for heterologously expressed ACD1 (data not shown). In addition, a type 2 RCCR (5) that specifically produces the C1 isomer of pFCC-1 (pFCC-2), together with recombinant PaO, yielded pFCC-2 as well (not shown). Taken together, these data show that ACD1 has PaO activity in vitro.
Fig. 2.
In vitro formation of pFCC-1 by using heterologously expressed ACD1 and kinetics of pFCC-1 formation. (A) HPLC traces of PaO/RCCR assays with protein extracts of E. coli JM109 or C43, transformed with the expression vector harboring either no insert (C43-vector), At4g25650 cDNA (C43-At4g25650), or ACD1 cDNA (JM109-ACD1 and C43-ACD1, respectively). An assay using PaO from senescent canola cotyledons (canola-PaO) was used as an authentic pFCC-1 standard. (B) Immunoblot analysis of E. coli C43 cells transformed with the pQE30 expression vector harboring either no insert (Vector Laboratories), ACD1 cDNA (ACD1), or At4g25650 cDNA (At4g25650). The blot was developed by using an antibody against the His-tag. (C) Nonlinear regression of pFCC-1 formation of C43-ACD1 protein extracts vs. pheide a concentration. (Inset) The Km value was derived from a Lineweaver–Burk plot.
LLS1 Is Required for chl Breakdown. To elucidate whether ACD1 and/or its maize homologue LLS1 are responsible for chl catabolism in vivo as well, we analyzed an lls1 mutant in respect to senescence-related characteristics. The lesions of lls1 have been described to occur in a development-related fashion (41). Thus, lesion formation mostly starts at the tip of a leaf, with the oldest leaf being affected first. Subsequently, lesions run down the leaf blade progressing from the center to the edges. This phenotype very much resembles senescence progression in wild-type leaves (Fig. 3A). The absence of LLS1 RNA from mutant tissues was confirmed by RT-PCR (Fig. 3B). We analyzed tissues indicated by the boxed areas of Fig. 3A for the presence of green chl catabolites (Fig. 3C). In necrotic mutant tissue, pheide a was present at rather high concentrations. No polar chl catabolites accumulated in lls1 green leaf tissue or in the wild-type tissues. Concomitant with the disappearance of chl in wild type, the appearance of NCCs as the final catabolites of chl was observed (Fig. 3D). To our knowledge, NCCs have so far not been characterized from maize. Deduced from their typical absorbance spectrum (Fig. 3E and ref. 1), there are two NCCs, tentatively named Zm-NCC-1 and -2 (for a nomenclature of NCCs, see ref. 36). The structures of these NCCs remain unresolved, but from their retention time on reversed-phase HPLC, they appear to represent unique NCCs not found in other plant species analyzed so far. Zm-NCC-1 and -2 did not accumulate in lls1 (Fig. 3D). Furthermore, we assessed PaO activity in mature green leaf tissues and necrotic and senescent tissue of lls1 and wild type, respectively. Only extracts from senescent wild-type leaves produced substantial amounts of pFCC-1 in the in vitro assay (Fig. 3F).
The obvious correlation of lesion formation in lls1 with leaf senescence in wild type tempted us to use an artificial system for senescence induction, i.e., dark incubation of excised leaf discs. Under these conditions, chl degradation was retarded in lls1, whereas in wild type, chl was degraded in a time-dependent manner (Fig. 4A). NCCs were exclusively found in wild type and their accumulation positively correlated with the loss of chl (data not shown). Again, pheide a but not b or chlorophyllides progressively accumulated in lls1 but not in wild type (Fig. 4B). The same result was obtained in preliminary experiments with an Arabidopsis line harboring a T-DNA insertion in Acd1 (data not shown). Two recessive mutants, F. pratensis Bf 993 and Mendel's “green peas,” have been described that accumulate pheide a on senescence induction. Although the affected genes have not been identified, in both mutants, absence of PaO activity is the biochemical lesion causing a stay-green phenotype (45, 46). In dark-incubated nonyellowing lls1 segments, PaO activity was absent as well (data not shown). Activity of PaO has been shown to be senescence-specifically regulated (Fig. 3F and refs. 13, 14); thus, the absence of PaO activity in lls1 raised the question whether senescence was initiated at all in the mutant. We therefore investigated protein degradation during dark-incubation of leaf discs. Overall protein concentrations decreased similarly in wild type and lls1 (Fig. 4C), but a chl-binding polypeptide, light-harvesting complex protein II, was specifically retained in lls1 (Fig. 4D). Again, the same observation had been made in Bf 993 (47), indicating that in the absence of light, lls1 exhibits a stay-green phenotype, similar to the Bf 993 mutant of F. pratensis. Together, the analysis of senescence-related characteristics of lls1 strongly supports the conclusion that the maize ACD1 orthologue, LLS1, is a PaO that is indispensable for the degradation of chl beyond the level of pheide a. We have shown that Lls1/Acd1 encode PaO enzymes that are involved in senescence-related chl degradation in vivo. Therefore, we designate the respective maize and Arabidopsis genes ZmPaO and AtPaO, respectively.
Both PaO Expression and Activity Are Regulated. Because of the senescence-related activity of PaO (13, 14), we investigated the expression of AtPaO/Acd1 during senescence of Arabidopsis leaves. Expression levels were normalized to the levels of mRNA encoding the actin 2 protein (48). Sag12 encoding a senescence-specific cysteine protease was used as a senescence marker gene (49, 50). AtPaO expression was positively correlated with senescence, but transcripts were also found in presenescent tissue (Fig. 5A). After 6 d of dark incubation, expression was increased 6.2-fold, whereas PaO activity of the same tissue had increased >60-fold (Fig. 4B). This indicated that PaO might also be regulated posttranscriptionally, probably by regulation of its activity. As yet, the nature of this proposed regulation is unknown, but preliminary experiments indicate that a phosphorylation/dephosphorylation mechanism might be involved (data not shown).
Significance of PaO for Cell Death Suppression? The lesion mimic phenotype of lls1 has been suggested to be caused by a chloroplast-derived signal (42). Ion leakage measurements of dark-incubated leaf discs after reexposure to light demonstrated a close relationship between pheide a content and ion leakage (data not shown). Thus, in lls1, cell death occurred faster when leaf discs had accumulated higher amounts of pheide a. This reaction was light-dependent, indicating that photoactivation of pheide a may trigger the formation of reactive oxygen species, which in turn cause cell death. This clearly demonstrates that in lls1, cell death occurs because of the accumulation of a phototoxic compound, pheide a, rather than because of a direct interference with a cell death suppression mechanism (41). The identification of lesion mimic mutants has been described as a powerful tool to unravel programmed cell death pathways in plants (51). Our finding here, along with the isolation of several lesion mimic mutants in which accumulation of intermediates of chl metabolism causes premature cell death (27, 30), indicates that the relationship between the biochemical function of the respective genes and their involvement in a cell death pathway under wild-type conditions remains a complex puzzle yet to be solved.
The biochemical analysis of heterologously expressed ACD1 and the characteristics of the lls1 mutant demonstrate that these proteins are identical to PaO, a key enzyme in the catabolism of chl. The wide distribution of PaO-like genes within photosynthetic organisms indicates their importance. PaO-like genes are present not only in higher plants but also in Chlamydomonas reinhardtii (data not shown) and cyanobacteria (42). In contrast, PaO is not found (data not shown) in the genome of Chlorobium tepidum, an anoxygenic photosynthetic bacterium (52). These bacteria may contain an unrelated oxygen-independent version of PaO. Alternatively, the invention of chl catabolism may have become indispensable for the evolution of oxygenic photosynthetic organisms. This is supported by the finding in several lower plant species of chl degradation products, mainly of the type of RCCs (2, 53, 54). The ability of chl to absorb light and, thus, to enable the conversion of light energy to chemical energy during photosynthesis is vitally important for all life on earth, but it turns into a threat when chl is released from its natural apoprotein environment during uncontrolled senescence.
Acknowledgments
We thank U. Fischer, University of Kaiserslautern, Kaiserslautern, Germany, for support with quantitative RT-PCR analysis; E. Neuhaus for E. coli C43 strain; M. Geissler, University of Zurich, Zurich, for antibodies; H. Thomas, H. Ougham and I. Donnison, Institute of Grassland and Environmental Research, Aberystwyth, U.K., for antibodies and discussion; and L. Bovet, U. Feller, E. Martinoia, and P. Matile for support and discussion. Especially, we thank N. Amrhein for critical reading of the manuscript and for discussion. S.H. was supported by grants from the Swiss National Science Foundation; M.R. was supported by a grant from the Spanish Government.
Abbreviations: chl, chlorophyll; FCC, fluorescent chl catabolite; pFCC, primary FCC; pheide, pheophorbide; PaO, pheide a oxygenase; RCC, red chl catabolite; RCCR, RCC reductase; NCC, non-FCC; Fd, ferredoxin.
References
- 1.Kräutler, B., Jaun, B., Bortlik, K.-H., Schellenberg, M. & Matile, P. (1991) Angew. Chem. Int. Ed. Engl. 30, 1315-1318. [Google Scholar]
- 2.Hörtensteiner, S. (1999) Cell. Mol. Life Sci. 56, 330-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matile, P., Hörtensteiner, S. & Thomas, H. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 67-95. [DOI] [PubMed] [Google Scholar]
- 4.Rodoni, S., Mühlecker, W., Anderl, M., Kräutler, B., Moser, D., Thomas, H., Matile, P. & Hörtensteiner, S. (1997) Plant Physiol. 115, 669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hörtensteiner, S., Rodoni, S., Schellenberg, M., Vicentini, F., Nandi, O. I., Qiu, Y.-L. & Matile, P. (2000) Plant Biol. 2, 63-67. [Google Scholar]
- 6.Kreuz, K., Tommasini, R. & Martinoia, E. (1996) Plant Physiol. 111, 349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mühlecker, W., Kräutler, B., Ginsburg, S. & Matile, P. (1993) Helv. Chim. Acta 76, 2976-2980. [Google Scholar]
- 8.Mühlecker, W. & Kräutler, B. (1996) Plant Physiol. Biochem. 34, 61-75. [Google Scholar]
- 9.Hinder, B., Schellenberg, M., Rodoni, S., Ginsburg, S., Vogt, E., Martinoia, E., Matile, P. & Hörtensteiner, S. (1996) J. Biol. Chem. 271, 27233-27236. [DOI] [PubMed] [Google Scholar]
- 10.Oberhuber, M., Berghold, J., Breuker, K., Hörtensteiner, S. & Kräutler, B. (2003) Proc. Natl. Acad. Sci. USA 100, 6910-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takamiya, K., Tsuchiya, T. & Ohta, H. (2000) Trends Plant Sci. 5, 426-431. [DOI] [PubMed] [Google Scholar]
- 12.Thomas, H., Ougham, H. & Hörtensteiner, S. (2001) in Advances in Botanical Research, ed. Callow, J. A. (Academic, San Diego), Vol. 35, pp. 1-52. [Google Scholar]
- 13.Ginsburg, S., Schellenberg, M. & Matile, P. (1994) Plant Physiol. 105, 545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hörtensteiner, S., Vicentini, F. & Matile, P. (1995) New Phytol. 129, 237-246. [DOI] [PubMed] [Google Scholar]
- 15.Trebitsh, T., Goldschmidt, E. E. & Riov, J. (1993) Proc. Natl. Acad. Sci. USA 90, 9441-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuchiya, T., Ohta, H., Okawa, K., Iwamatsu, A., Shimada, H., Masuda, T. & Takamiya, K. (1999) Proc. Natl. Acad. Sci. USA 96, 15362-15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodoni, S., Vicentini, F., Schellenberg, M., Matile, P. & Hörtensteiner, S. (1997) Plant Physiol. 115, 677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matile, P. & Schellenberg, M. (1996) Plant Physiol. Biochem. 34, 55-59. [Google Scholar]
- 19.Schellenberg, M., Matile, P. & Thomas, H. (1993) Planta 191, 417-420. [Google Scholar]
- 20.Wüthrich, K. L., Bovet, L., Hunziker, P. E., Donnison, I. S. & Hörtensteiner, S. (2000) Plant J. 21, 189-198. [DOI] [PubMed] [Google Scholar]
- 21.Frankenberg, N., Mukougawa, K., Kohchi, T. & Lagarias, J. C. (2001) Plant Cell 13, 965-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hörtensteiner, S., Wüthrich, K. L., Matile, P., Ongania, K.-H. & Kräutler, B. (1998) J. Biol. Chem. 273, 15335-15339. [DOI] [PubMed] [Google Scholar]
- 23.Kräutler, B. & Matile, P. (1999) Acc. Chem. Res. 32, 35-43. [Google Scholar]
- 24.Tanaka, A., Ito, H., Tanaka, R., Tanaka, N. K., Yoshida, K. & Okada, K. (1998) Proc. Natl. Acad. Sci. USA 95, 12719-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, H., Ohysuka, T. & Tanaka, A. (1996) J. Biol. Chem. 271, 1475-1479. [DOI] [PubMed] [Google Scholar]
- 26.Scheumann, V., Schoch, S. & Rüdiger, W. (1999) Planta 209, 364-370. [DOI] [PubMed] [Google Scholar]
- 27.Hu, G., Yalpani, N., Briggs, S. P. & Johal, G. S. (1998) Plant Cell 10, 1095-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruse, E., Mock, H.-P. & Grimm, B. (1995) EMBO J. 14, 3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg, J. T., Guo, A., Klessig, D. F. & Ausubel, F. M. (1994) Cell 77, 551-563. [DOI] [PubMed] [Google Scholar]
- 30.Mach, J. M., Castillo, A. R., Hoogstraten, R. & Greenberg, J. T. (2001) Proc. Natl. Acad. Sci. USA 98, 771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arabidopsis Genome Initiative. (2000) Nature 408, 796-813. [DOI] [PubMed] [Google Scholar]
- 32.Kurtz, D. M. (1997) J. Biol. Inorg. Chem. 2, 159-167. [Google Scholar]
- 33.Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. (2002) Science 296, 141-145. [DOI] [PubMed] [Google Scholar]
- 34.Lineweaver, H. & Burk, D. (1934) J. Am. Chem. Soc. 56, 658-666. [Google Scholar]
- 35.Langmeier, M., Ginsburg, S. & Matile, P. (1993) Physiol. Plant 89, 347-353. [Google Scholar]
- 36.Ginsburg, S. & Matile, P. (1993) Plant Physiol. 102, 521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strain, H. H., Cope, B. T. & Svec, W. A. (1971) Methods Enzymol. 23, 452-487. [Google Scholar]
- 38.Hörtensteiner, S., Chinner, J., Matile, P., Thomas, H. & Donnison, I. S. (2000) Plant Mol. Biol. 42, 439-450. [DOI] [PubMed] [Google Scholar]
- 39.Solomon, E. I., Brunold, T. C., Davis, M. I., Kemsley, J. N., Lee, S.-K., Lehnert, N., Neese, F., Skulan, A. J., Yang, Y.-S. & Zhou, J. (2000) Chem. Rev. 100, 235-349. [DOI] [PubMed] [Google Scholar]
- 40.Ferro, M., Salvi, D., Rivière-Rolland, H., Vermat, T., Seigneurin-Berny, D., Grunwald, D., Garin, J., Joyard, J. & Rolland, N. (2002) Proc. Natl. Acad. Sci. USA 99, 11487-11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray, J., Close, P. S., Briggs, S. P. & Johal, G. S. (1997) Cell 89, 25-31. [DOI] [PubMed] [Google Scholar]
- 42.Gray, J., Janick-Buckner, D., Buckner, B., Close, P. S. & Johal, G. S. (2002) Plant Physiol. 130, 1894-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenberg, J. T. & Ausubel, F. M. (1993) Plant J. 4, 327-341. [DOI] [PubMed] [Google Scholar]
- 44.Miroux, B. & Walker, J. E. (1996) J. Mol. Biol. 260, 289-298. [DOI] [PubMed] [Google Scholar]
- 45.Vicentini, F., Hörtensteiner, S., Schellenberg, M., Thomas, H. & Matile, P. (1995) New Phytol. 129, 247-252. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, H., Schellenberg, M., Vicentini, F. & Matile, P. (1996) Bot. Acta 109, 3-4. [Google Scholar]
- 47.Hilditch, P. I., Thomas, H., Thomas, B. J. & Rogers, L. J. (1989) Planta 177, 265-272. [DOI] [PubMed] [Google Scholar]
- 48.Bovet, L., Eggmann, T., Meylan-Bettex, M., Polier, J., Kammer, P., Marin, E., Feller, U. & Martinoia, E. (2003) Plant Cell Environ. 26, 371-381. [Google Scholar]
- 49.Noh, Y. S. & Amasino, R. M. (1999) Plant Mol. Biol. 41, 181-194. [DOI] [PubMed] [Google Scholar]
- 50.Noh, Y. S. & Amasino, R. M. (1999) Plant Mol. Biol. 41, 195-206. [DOI] [PubMed] [Google Scholar]
- 51.Lorrain, S., Vailleau, F., Balagué, C. & Roby, D. (2003) Trends Plant Sci. 8, 263-271. [DOI] [PubMed] [Google Scholar]
- 52.Eisen, J. A., Nelson, K. E., Paulsen, I. T., Heidelberg, J. F., Wu, M., Dodson, R. J., Deboy, R., Gwinn, M. L., Nelson, W. C., Haft, D. H., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engel, N., Jenny, T. A., Mooser, V. & Gossauer, A. (1991) FEBS Lett. 293, 131-133. [DOI] [PubMed] [Google Scholar]
- 54.Doi, M., Shima, S., Egashira, T., Nakamura, K. & Okayama, S. (1997) J. Plant Physiol. 150, 504-508. [Google Scholar]