Abstract
In higher plants, the PsbS subunit of photosystem II (PSII) plays a crucial role in pH- and xanthophyll-dependent nonphotochemical quenching of excess absorbed light energy, thus contributing to the defense mechanism against photoinhibition. We determined the amino acid sequence of Zea mays PsbS and produced an antibody that recognizes with high specificity a region of the protein located in the stroma-exposed loop between the second and third putative helices. By means of this antiserum, the thylakoid membranes of various higher plant species revealed the presence of a 42-kDa protein band, indicating the formation of a dimer of the 21-kDa PsbS protein. Crosslinking experiments and immunoblotting with other antisera seem to exclude the formation of a heterodimer with other PSII protein components. The PsbS monomer/dimer ratio in isolated thylakoid membranes was found to vary with luminal pH in a reversible manner, the monomer being the prevalent form at acidic and the dimer at alkaline pH. In intact chloroplasts and whole plants, dimer-to-monomer conversion is reversibly induced by light, known to cause luminal acidification. Sucrose-gradient centrifugation revealed a prevalent association of the PsbS monomer and dimer with light-harvesting complex and PSII core complexes, respectively. The finding of the existence of a light-induced change in the quaternary structure of the PsbS subunit may contribute to understanding the mechanism of PsbS action during nonphotochemical quenching.
Higher plants and algae are oxygenic photoautotrophic organisms that require light for life. Light intensity in the natural environment may vary over several orders of magnitude in a time scale from seconds to days. A large light-harvesting antenna complex is necessary for efficient capture when light is limited, but it may become a problem when light is in excess. In fact, excitation of photosystem II (PSII) at a rate exceeding its capacity for photochemical conversion results in a loss of photosynthetic activity associated with photooxidative damage and, in extreme conditions, pigment bleaching and death of the organism (1, 2). Indeed, plants have evolved a number of responses to changes in light intensity. Nonradiative dissipation of excitation energy in the light-harvesting system of PSII in response to excess light is the most important and prompt reaction. This protective mechanism is most commonly measured as the nonphotochemical quenching (NPQ) of chlorophyll fluorescence (3-5). The signal that triggers most of the dissipative response is energization of the thylakoid membrane, through the formation of a pH gradient (6). This energy-dependent quenching has therefore been called qE. Acidification of the lumen under light has at least two important consequences associated with the formation of qE: activation of violaxanthin deepoxidase in the lumen (7, 8) and protonation of the lumen-exposed domains of one or more PSII proteins. Violaxanthin deepoxidase is an enzyme of the xanthophyll cycle that catalyzes the conversion of violaxanthin to zeaxanthin via antheraxanthin (9). There is considerable evidence that the xantophyll cycle is involved in the formation of qE (10, 11).
PsbS is a member of the Lhc superfamily, first isolated in PSII membranes of spinach as a 22-kDa protein (12), having four putative transmembrane helices, unlike the other Lhc proteins, which have three (13, 14). Although Funk et al. (15) reported isolation of PsbS as a chlorophyll-protein complex, the matter remains unclear because (i) PsbS does not contain the conserved histidine residues that coordinate chlorophyll molecules in the other Lhc members; (ii) at variance with other Lhc apoproteins, PsbS is stable in the absence of chlorophyll (16); and (iii) in vitro reconstitution in the same conditions used for other Lhc proteins failed with PsbS (17). Instead, PsbS is able to bind zeaxanthin in vitro, and this binding seems to be the origin of the absorption change at 535 nm associated with the formation of qE (18). In fact, PsbS has been shown to be directly involved in NPQ, because the Arabidopsis npq4-1 mutant that lacks PsbS loses most of its capacity to form qE, although its light-harvesting and photosynthetic efficiencies are normal (19, 20). PsbS protein has been hypothesized to function as a sensor of luminal pH (19), and this view was confirmed by recent experiments showing that protonation of some lumen-exposed residues of PsbS is closely correlated to the formation of qE (21).
Several authors have proposed that a pH-induced conformational change of PsbS is the triggering event for the formation of qE, but this change has not yet been identified nor has a clear mechanism for concerted action producing qE been demonstrated. In this article, we report a quaternary structural change of PsbS induced by its protonation and by light.
Materials and Methods
Isolation, Sequencing, and Subcloning of Maize PsbS cDNA. Total DNA from a λ-Zap II cDNA library of Zea mays (22) was amplified by PCR with synthetic oligonucleotides 5′-GTNGARGAYGGNATHTTYGG-3′ (forward) and 5′-TCRTCNGTDATRAAYTTNCC-3′ (reverse), designed on conserved amino acid sequences VEDGIFG and GKFITDE. Based on the sequence of the resulting major band, specific nested oligonucleotides 5′-ACCTGGAGACGGGCATC-3′ (forward) and 5′-TTGGTGAACCCGAACAGC-3′ (reverse) were synthesized and further used in separate PCR amplifications with universal primers T7 (reverse) and T3 (forward). Sequencing was performed completely on both strands with the BigDye terminator system on an ABI PRISM 3700 DNA sequencer (Applied Biosystems) at Interdipartimental Research Center for Innovative Biotechnologies's sequencing service (University of Padua). The entire cDNA sequence was finally reconstituted by using unique internal (BstEII) or adjacent (BstXI and XhoI) restriction sites. Conventional procedures were used for cloning steps and computer analysis. SalI and HindIII sites of cDNA were exploited for cloning the 3′ half of the coding region into expression vector pQE31 (Qiagen, Valencia, CA). Synthetic oligonucleotides 5′-ACCGCGGCGGATCCGTCGACGAGG-3′, 5′-ACCAAGTCGAACGGATCCTTCGTGGGGCG-3′, and 5′-TCAACATCGGATCCGGGGTGCCCATCAACG-3′ (introducing BamHI sites) were used in separate PCR amplifications as forward primers with reverse primer 5′-GCAGCAGCAGCCGAAT TCCTAT TCCTCTTCCTCGC-3′ (introducing EcoRI site) for amplification of the DNA fragments cloned into expression vector pGEX-4T-1 (Amersham Biosciences).
Production of Antigen and Antiserum. Induction of His- or GST-tagged fusion proteins was performed according to the manufacturer's recommended procedures. Recombinant antigen (≈200 μg/liter of 4-h culture) was obtained from cleared lysate of sonicated cells in 50 mM NaH2PO4 (pH 8), 300 mM NaCl, 10 mM imidazole, and 8 M urea by affinity purification on Ninitrilotriacetic acid resin (Qiagen), and it was used to immunize rabbits by s.c. injections, with poly(A)-poly(U) as adjuvant and following standard routes.
Gel Electrophoresis and Western Blotting. Proteins were separated in SDS/12% PAGE in the presence of 6 M urea (23). Gels were either stained with Coomassie blue or silver or transblotted onto poly(vinylidene difluoride) (PVDF) membrane and decorated with anti-PsbS antibodies at 1:10,000 or 1:20,000 dilution for 1.5 h. All antibodies used were from our laboratory. Horseradish peroxidase-coupled anti-rabbit IgG (Kirkegaard & Perry Laboratories) was used as a secondary antibody, and blots were developed by using the enhanced chemiluminescence system (Pierce). All reported experiments were repeated at least three times and gave similar results.
Thylakoid Membrane Preparation. Thylakoids were isolated as described (24). Briefly, maize leaves were homogenized in 0.33 M sorbitol, 50 mM Tricine (pH 7.8), 5 mM MgCl2, and 10 mM NaCl. After filtering, the homogenate was centrifuged at 4,500 × g for 2 min, and the pellet was resuspended in 50 mM Tricine (pH 7.8), 5 mM MgCl2, and 10 mM NaCl. After centrifugation at 4,500 × g for 10 min, the resulting thylakoids were resuspended in 100 mM sorbitol, 50 mM Tricine (pH 7.8), 5 mM MgCl2, and 10 mM NaCl. Chlorophyll concentration was calculated according to the method of Arnon (25).
Crosslinking. 3,3′-Dithiodipropionic acid di(N-hydroxysuccinimide ester) (DTSP) was used at 0.015% (wt/vol) for thylakoids, diluted to 300 μg/ml chlorophyll concentration in a medium containing 20 mM Tricine (pH 7.5) and 0.3% n-dodecyl β-d-maltoside (DM), and kept in the dark. The reaction was stopped after 30 min by the addition of 10 mM Tris (pH 7.5) and 1 mM EDTA (26).
Sucrose-Gradient Centrifugation. Thylakoids were resuspended in sucrose-free buffer and solubilized with 1% (wt/vol) DM for 5 min on ice. After centrifugation, the supernatant equivalent to 60 μg of chlorophyll was loaded on the top of a 4-ml 0.1-1 M sucrose gradient. In addition to sucrose, the solution contained 10 mM Tricine, pH 7.8 (or 10 mM acetate buffer, pH 4.0) and 0.03% DM. Gradients were centrifuged for 6 h at 4°C at 56,000 rpm (TST 60.4 rotor, Kontron, Zurich) and then fractionated into 150-μl aliquots. Fractions were subjected to SDS/PAGE and immunoblotting.
Immunoprecipitation. Thylakoids (500 μg/ml chlorophyll) were solubilized with 0.3% (wt/vol) DM for 20 min and centrifuged for 3 min at 13,000 × g. One hundred microliters of supernatant was gently shaken with 50 μl of serum overnight at 4°C in the presence of 4 μl of protease inhibitor mixture (Sigma). The mixture was incubated with 30 μl of Protein A Sepharose beads (Amersham Biosciences) for 40 min, centrifuged for 30 sec (13,000 × g), and washed three times with PBS (140 mM NaCl/2.7 mM KCl/10.1 mM Na2HPO4/1.8 mM KH2PO4, pH 7.4). Then, 10-μl beads, treated with standard buffer (SB; 9% SDS/30% glycerol/125 mM Tris, pH 6.8/0.1 M dithiotreitol), were loaded on SDS/PAGE.
Preparation of Intact Chloroplasts. Intact chloroplasts were isolated by modification of the method described by Cerovic and Plesnicar (27). Briefly, maize leaves were homogenized with 340 mM sorbitol, 0.4 mM KCl, 0.04 mM EDTA, and 2 mM Hepes (pH 7.8), filtered twice, and centrifuged for 10 sec at 3,500 × g. Pellets were resuspended in 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 1 mM MnCl2, 50 mM Hepes (pH 7.0), and 0.2% BSA. Purity was checked by inverted microscope.
Results
Cloning and sequencing of the full-length maize cDNA for PsbS protein were performed as described in Materials and Methods. The GenBank accession no. AY291061 was assigned to the cDNA sequence. Comparison of the deduced amino acid sequence with the six PsbS sequences already present in the GenBank catalog (Oryza sativa, Nicotiana tabacum, Solanum sogarandinum, Spinacia oleracea, Arabidopsis thaliana, and Lycopersicon esculentum) confirmed extensive homology (80% to 83% identity in the mature protein). The two glutamate residues (E123 and E227), which are considered essential for the role of PsbS in NPQ (21), were conserved. Differences from the protein sequence deduced from overlapping expressed sequence tags of Z. mays (GenBank accession no. AY104328) were limited to the putative N-terminal chloroplast transit peptide and consisted of the absence of the first seven amino acids (MAAQSML) and the insertion of an alanine residue at position 35. The former difference may be attributed to premature termination of reverse transcription during preparation of the cDNA library (22) and the latter either to a naturally occurring polymorphism or an error in EST sequencing and/or clustering of overlapping sequences.
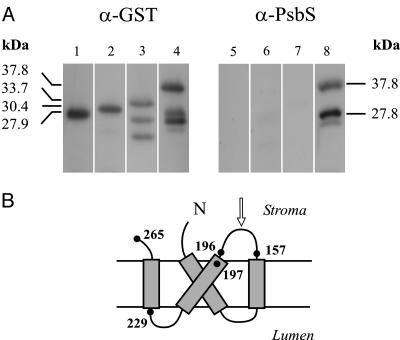
The C-terminal half of the mature protein, from V157 to E265, was expressed in Escherichia coli with a His-tag to favor purification. The recombinant polypeptide was used to produce a polyclonal antiserum. Its specificity was characterized by detection of a different portion of the original antigen expressed in fusion with the GST tag (amino acids V157 to E265, F197 to E265, and G229 to E265, corresponding to theoretical molecular masses of 37.8, 33.7, and 30.4 kDa, respectively). Although all fusion polypeptides were correctly detected by a commercial anti-GST antibody, only the longest one was recognized by the polyclonal antiserum produced in our laboratory (Fig. 1). This observation indicates that the major epitope recognized by our antiserum lies in the sequence comprised between amino acids V157 and L196, corresponding to the stroma-exposed loop between helices 2 and 3 of the PsbS protein (Fig. 1B). Furthermore, in the lanes corresponding to the longest GST-PsbS fusion (Fig. 1 A, lanes 4 and 8), both anti-GST and anti-PsbS sera detected a peptide fragment that may be an aborted translation product resulting from improper codon usage. The position of the first rare codon for E. coli (CCC) in the corresponding portion of the chimerical gene gives a calculated molecular mass of 27.7 kDa, which matches the position of the lower band in the blot. This peptide evidently exposes the major epitope more than the complete fusion and, indeed, is much better detected by our antiserum.
Fig. 1.
Characterization of anti-PsbS serum. (A) Immunoblotting analysis of total extracts of E. coli cells expressing GST alone or GST-PsbS fusion proteins, detected by commercial anti-GST polyclonal antibodies (Left) or anti-PsbS serum produced against recombinant His-tagged PsbS (Right). Shown are GST protein (lanes 1 and 5), GST-G229-E265 fusion (lanes 2 and 6), GST-F197-E265 fusion (lanes 3 and 7), and GST-V157-E265 fusion (lanes 4 and 8). Molecular masses are calculated from amino acid sequences, as deduced from recombinant genes. Two bands detected by anti-GST antibodies under GST-F197-E265 fusion polypeptide (lane 3) are possible products of proteolytic digestion. See text for identification of bands in lanes 4 and 8. (B) Proposed topology of PsbS polypeptide in thylakoid membrane. Number of amino acids defining boundaries of peptides expressed in fusion with GST are shown. Arrow indicates the amino acid loop containing the mapped epitope.
The amino acid sequence corresponding to the mapped epitope was compared with both other known PsbS gene products and, in a BLAST-restricted research, other available maize sequences. As expected, this analysis revealed wide homology among PsbS proteins (71% to 80% identity); instead, the similarity with other maize chloroplast proteins, particularly the related light-harvesting complexes (LHCs), was very small in this region.
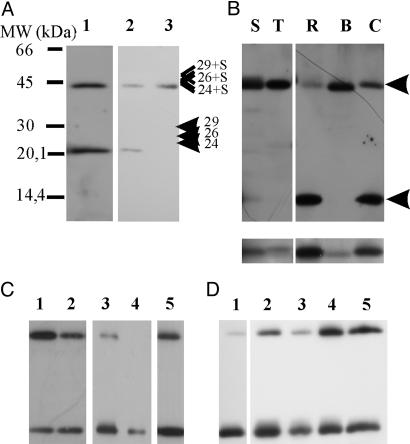
When the antiserum produced was used to reveal the presence and size of PsbS in the thylakoid membrane of Z. mays, two bands were observed in Western blots at 21 and 42 kDa, suggesting the presence of a dimeric form of the protein (Fig. 2A, lane 1). In agreement with this hypothesis is the result shown in Fig. 2 A, lanes 2 and 3: when the bifunctional disulfide reagent, DTSP, was added to thylakoid suspension kept in the dark, monomeric PsbS could no longer be detected, whereas an increase in the intensity of the 42-kDa band was observed.
Fig. 2.
PsbS anti-serum recognizes a 42-kDa band in various plant species. (A) Western blot of maize thylakoid developed with anti-PsbS at 1:20,000 dilution (lane 1). Control (lane 2) and DTSP-treated (lane 3) thylakoids are shown. Positions of CP24, CP26, and CP29 (as determined from Western blots by using antibodies against these proteins) are indicated together with calculated positions of possible heterodimers. (B) Thylakoids of spinach (S), tobacco (T), rice (R), barley (B), and carrot (C) were loaded. (Lower) Bands in 20-kDa range of same samples but solubilized for longer. Apparent masses of bands, calculated by mean of a calibration curve obtained with mass markers, were as follows: S and R, 20.6 and 41.3 kDa; T, 21.0 and 42.1 kDa; B, 20.1 and 40.5 kDa; and C, 20.8 and 41.8 kDa. Arrows indicate the positions of upper and lower bands. All lanes were loaded with 5 μg of chlorophyll. (C) Western blot of thylakoids solubilized with 1.3% (lane 1) and 3% (lane 2) SDS. Thylakoids (lane 3), BBY (lane 4), and grana membrane preparations (lane 5) from maize were solubilized with standard buffer yielding 3% final SDS concentration in the sample. (D) Thylakoids of maize solubilized with standard buffer (lane 1) or with 0.5% (wt/vol) of each detergent: l-α-lysophosphatidylcholine (lane 2), 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS; lane 3), 1-O-n-octyl-β-d-glucopyranoside (lane 4), and DM (lane 5). After solubilization for 15 min, samples were loaded on an SDS/PAGE containing 0.5% SDS in the absence of standard buffer.
Because the occurrence of a PsbS dimer had never been reported before, we checked for its presence in various species. In all species examined, the antiserum, used at 1:20,000 dilution, detected two bands with the expected molecular masses (Fig. 2B) but with different relative abundances. Harsher solubilization of samples from various species increased the intensity of the 21-kDa band, even in those species in which it was barely detectable (Fig. 2B Lower). Accordingly, in a separate set of experiments performed on thylakoids from maize, we observed that the relative intensity of the 42-kDa band varied, depending on the quantity of SDS present in the solubilization buffer (Fig. 2C, lanes 1 and 2). The sensitivity of the dimer to solubilization condition is indicated also by the following observations. The dimer was not present in PSII-enriched membranes obtained by solubilization with Triton X-100 (Fig. 2C, lane 4), whereas it was observable in PSII membranes obtained with digitonin (Fig. 2C, lane 5) (28). To better evaluate the direct effect of various detergents on the relative amount of the extracted dimer and monomer, maize thylakoids, solubilized with four different detergents at 0.5% (wt/vol), were loaded on a gel that contained 0.5% SDS but without SDS in the solubilization buffer (Fig. 2D). For comparison, thylakoids treated with the standard solubilization buffer were loaded on the same gel (Fig. 2D, lane 1). 1-O-n-octyl-β-d-glucopyranoside and DM were most effective in extracting the dimeric form.
In all species the apparent masses of the upper bands were, within experimental error, twice as great as the masses of the lower bands (see Fig. 2 A and B), suggesting the homodimeric nature of the high band. However, aggregation of PsbS with a different protein of similar apparent molecular mass could not be excluded. In Western blots, with antibodies against the known components of PSII with masses similar to that of PsbS (i.e., CP24, CP26, LHCII, and PsbP), in no case were bands with high molecular mass observed, but only with CP24 antiserum. However, the latter antiserum was found to crossreact with PsbS and, in addition, the sum of the apparent masses of PsbS (21.0 kDa) and CP24 (23.5 kDa) was significantly higher than that of the upper band observed in the immunoblot with anti-CP24 (see also Fig. 2 A). Various attempts to determine the amino acid sequences of the 21- and 42-kDa bands failed, in agreement with PsbS being blocked to sequence analysis (13, 14). In the next experiment, thylakoids were equilibrated at different pH values and left in the dark for 10 min before solubilization and analysis. As shown in Fig. 3A, the monomer/dimer ratio depended on pH, the monomer form being favored at acidic pH. The effect of pH on the extent of dimerization was found to be at least partially reversible (data not shown).
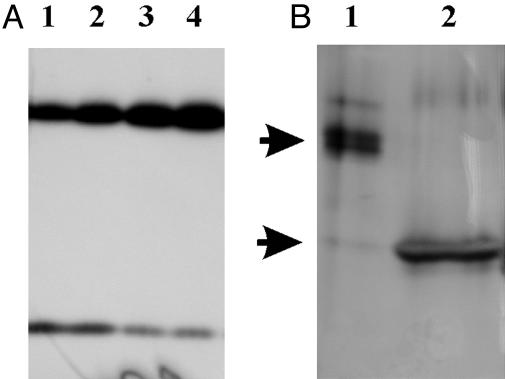
Fig. 3.
pH-induced variation of the dimer/monomer ratio. (A) Maize thylakoids from the same preparation were resuspended at pH 4.8 (lane 1), 6.0 (lane 2), 7.1 (lane 3), and 8.0 (lane 4) (20 mM Hepes/5 mM MgCl2/0.03% DM) and loaded on SDS/PAGE (5 μg of chlorophyll per lane). (B) Bands at 42 (lane 1) and 21 (lane 2) kDa were cut from SDS/PAGE, treated with a solution at pH 3.0 for 30 min (see text), loaded on SDS/PAGE, and silver stained. Arrows indicate 42- and 21-kDa regions.
Taking advantage of the pH dependence of the monomer/dimer equilibrium, we performed a further experiment to check once again the homodimer nature of the 42-kDa band. Thylakoid membranes were treated at pH 9.0 and loaded on SDS/PAGE. After runs, part of the gel was cut out, transblotted, and decorated with anti-PsbS antibody. In parallel, the rest of the gel was stained, and the bands corresponding to the dimer and monomer (as judged from Western blots) were cut from the stained gel, treated at pH 3.0, and loaded on a second gel. Subsequent silver staining revealed the presence of a single band in the 20-kDa zone in the acid-treated dimer sample (Fig. 3B), corresponding to monomeric PsbS, as confirmed by Western blotting (data not shown). This result is expected if the 42-kDa band was a PsbS homodimer.
The addition of two uncoupling agents, dinitrophenol or carbonyl cyanide 4-trifluoromethoxyphenylhydrazone (FCCP), did not change the pH-dependence of the monomer/dimer ratio (data not shown), as expected if our isolated thylakoids were permeable to protons. When intact chloroplasts, freshly isolated at pH 6.0, were resuspended in a medium at pH 9.0 and left in dark, the quantity of the dimeric form significantly increased (data not shown). Thus, in vitro, the monomer/dimer equilibrium for PsbS depends on pH. The following experiment was aimed at establishing whether pH-dependent structural change is also relevant in physiological conditions. Intact chloroplasts, isolated from maize plants grown in normal light conditions (150 μmol of photons m-2·s-1), were analyzed for the relative amounts of PsbS monomer and dimer on exposure to varying light conditions. The results shown in Fig. 4A demonstrate that the dimer-to-monomer ratio decreases with increasing light intensity. Light-induced monomerization is already complete within 10 min [at least at 350 μmol of photons m-2·s-1, a light intensity sufficient to induce NPQ in intact spinach chloroplasts (29) and luminal acidification in maize chloroplasts, measured as described in ref. 30 (data not shown)]. Both 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; 20 mM), a herbicide that blocks electron transfer, and the uncoupler carbonyl cyanide 4-trifluoromethoxyphenylhydrazone (FCCP; 10 μM), as expected, prevented light-induced monomerization but only partially (Fig. 4 B and C, respectively). This observation suggests that in intact chloroplasts, monomerization does depend on acidic luminal pH, but other factors may play a role as well. The process is fully reversible, in that the equilibrium value does not depend on whether the sample has been preilluminated or not. Similar results were obtained with intact plants of maize: after 6 h of adaptation to high light, the dimeric form of PsbS almost completely disappeared (Fig. 4D).
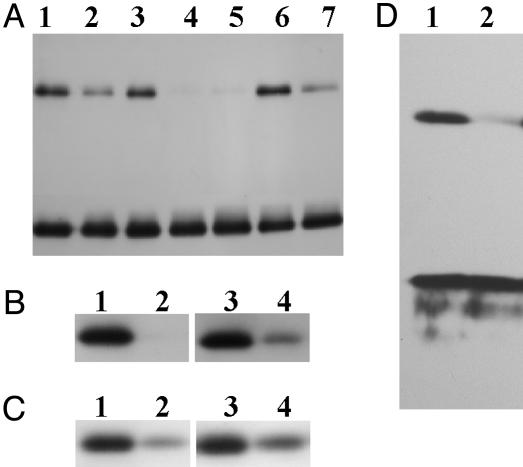
Fig. 4.
pH and light-induced changes in quaternary structure of PsbS in intact chloroplasts and whole plants. (A) Intact maize chloroplasts were kept on ice in the dark (lane 1) or left at room temperature and illuminated for 10 min at 150 (normal light; lane 2), 0 (lane 3), or 350 (high-light; lane 4) μmol of photons m-2·s-1. Sample of lane 5 was the same as that of lane 4 but illuminated for 20 min. One aliquot of high-light-treated sample (lane 4) was reequilibrated in the dark (lane 6) or at 150 μmol of photons m-2·s-1 (normal light; lane 7). (B) Thylakoids left in dark (lanes 1 and 3) or treated with high light (lanes 2 and 4) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 20 mM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). (C) Thylakoids were left in the dark (lanes 1 and 3) or treated with normal light (lanes 2 and 4) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 10 μM carbonyl cyanide 4-trifluoromethoxyphenylhydrazone (FCCP). (D) Thylakoids isolated from maize plants kept in the dark (lane 1) or treated with high light for 6 h (lane 2). All Western blots shown were decorated with anti-PsbS antibodies.
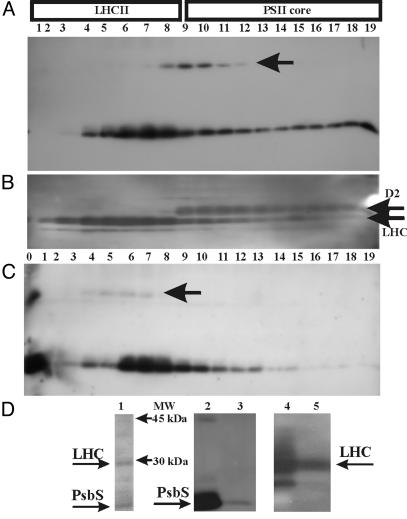
PsbS is believed to be located in the interface region between LHCII and the PSII core (17, 31, 32). For more information on this point, thylakoids were slightly solubilized with 1% DM for 5 min and loaded on a continuous sucrose gradient. In agreement with our previous work, four green bands separated on centrifugation, corresponding, from top to bottom, to monomeric and trimeric LHCII, monomeric PSII, and a mixture of dimeric PSII and PSI (24). The gradient was fractionated, and the resulting fractions were assayed for the presence of PsbS. As shown in Fig. 5A, the PsbS dimer was present only in fractions 8-12, whereas the monomer was present prevalently in fractions 5-9 but also in the lower gradient fractions. Identity of fractions was checked by using anti-LHC and anti-D2 antibodies (Fig. 5B). It may be concluded that the dimer corresponding to the 42-kDa band comigrates mainly with the PSII core. When the same gradient was run at acidic pH (pH 4.0), as expected, most of the dimeric form underwent monomerization, and the monomers migrated preferentially to the LHC region (Fig. 5C). The remaining dimers also were found in the LHC region. To exclude a casual comigration of PsbS with the LHC region, we performed immunoprecipitation with our anti-PsbS serum by using solubilized thylakoid membrane as starting material. Silver staining reveals two prevalent bands (Fig. 5D, lane 1) in the immunoprecipitate, corresponding to PsbS and LHC proteins as identified by Western blots (Fig. 5D, lanes 2 and 3 and lanes 4 and 5, respectively). The fact that our anti-PsbS antibody, which does not recognize other proteins in Western blots, pulls down LHC proteins during immunoprecipitation strongly suggests a close interaction between PsbS and LHC proteins.
Fig. 5.
(A-C) Distribution of PsbS dimers and monomers in a sucrose gradient of solubilized thylakoid membrane. Fractions of sucrose gradients (150 μl), performed at pH 7 (A)orpH4(C), were loaded on SDS/PAGE (lanes 1-19 from top to bottom) and analyzed by Western blotting. Lane 0 in C contains whole thylakoids for reference. Blots were immunodecorated with 1:10,000 anti-PsbS (A and C) and then stripped and redecorated with anti-D2 and anti-LHCII antibodies (B). In this case, the same pattern was observed at both pH values. Arrows indicate the position of the dimer. Gradients at varying pH values were obtained from the same preparation of thylakoids and processed together throughout the experiment. (D) Silver-stained SDS/PAGE of immunoprecipitate (lane 1), Western blots of thylakoids (lanes 2 and 4) and immunoprecipitate (lanes 3 and 5) decorated with anti-PsbS (lanes 2 and 3) and anti-LHC (lanes 4 and 5) antibodies.
Discussion
In this article, we show the existence of a physiologically relevant dimeric form of the PsbS subunit. The dimer/monomer ratio in thylakoid membranes depends on pH, with acidic luminal pH favoring the monomer. Accordingly, after short-term illumination of isolated chloroplasts, complete monomerization of PsbS is induced. Low luminal pH, protonation of PsbS, and xanthophyll deepoxidation are three essential elements for NPQ to occur (5, 18, 33). Binding of protons and probably of zeaxanthin to PsbS and eventually to other PSII proteins is thought to trigger a conformational change, resulting in the formation of a quenching complex (34). Our results suggest that the hypothesized conformational change induced by protonation involves monomerization of PsbS.
The fact that the 42-kDa band recognized by our antibody actually corresponds to a PsbS homodimer is strongly indicated by several observations. First, a high molecular weight band was identified in all species examined by our specific anti-PsbS serum and, in all cases, its apparent molecular mass, within the errors of the method, was twice as great as that of the corresponding lower band. A second point in favor of the homodimer is the result of the crosslinking experiment. Given the length of the spacer arm (11 Å) in DTSP, it was reasonable to expect a linkage with a near-neighbor protein. The exact location of PsbS in the PSII supercomplex is not known (35), but a point at the interface between PSII core and LHCII proteins, in the region of minor antenna proteins CP29, CP26, and CP24, has been proposed (17, 32). We definitely may exclude the formation of a heterodimer with CP26 and CP29, because the 42-kDa band is not detected by the respective antibodies (data not shown). Our anti-CP24 crossreacts with PsbS and therefore cannot be used to exclude a heterodimer PsbS-CP24. However, the only observed crosslinking product has an apparent molecular mass of 42 kDa, identical to that of the band observed in nontreated thylakoids, which seems incompatible with the apparent molecular mass expected for a PsbS-CP24 heterodimer. A third point is the fact that partial monomerization of the 42-kDa band induced by acid pH treatment yields a single protein band at 21 kDa in a silver-stained gel. Finally, the presence of a dimeric PsbS is consistent with the previously reported stoichiometry of two PsbS subunits per PSII reaction center in spinach thylakoids (15).
The pH dependence of the dimer/monomer equilibrium has been demonstrated both in vitro for isolated thylakoid membranes and in a more integrated system, i.e., intact chloroplasts, and in vivo for whole plants. The clear effect of pH in the range of 5-8 is observable in isolated thylakoid membranes (Fig. 3A) and in intact chloroplasts (data not shown). The finding that the same type of phenomenon is induced by light in intact chloroplasts and whole plants (Fig. 4 A and D) allows us to conclude that dimer/monomer pH-dependent equilibrium is a physiological process that operates in vivo as a consequence of light-induced pH changes in the thylakoid lumen. However, other factors beside acidification are probably involved (Fig. 4 B and C).
Protonation of two glutamic acid residues in the lumen-exposed loops of PsbS have been shown to be essential for the role played by PsbS in qE (21). We have shown that, on acidification of the lumen, PsbS dimers reversibly dissociate into monomers. Although the exact pK value for monomerization was not established, in our experiments monomerization was complete at pH 4. This observation is compatible with the dissociation equilibrium operating in the pH range of 5-6, which is the estimated value for luminal pH in high-light conditions (36).
The question arises of what kind of force keeps together the two subunits in a way that they can, at least partially, resist treatment with SDS? Solubilization of thylakoids and chloroplasts before loading on SDS/PAGE was performed in a reducing environment excluding the presence of disulfide bridges. PsbS is a highly hydrophobic protein with distinct zones; the second and fourth putative helices are characterized by extreme hydrophobicity because of the presence of several phenylalanine residues (17, 18). These regions may well be responsible for a strong interaction resistant to solubilization by SDS. A similar situation has been described for the tetrameric potassium channels of the bacterial membrane. In this case, although the subunits are not covalently linked to each other, bands corresponding to dimers, trimers, and tetramers can clearly be seen in SDS/PAGE (37). Glycophorin A, the major component of human erythrocyte membranes, also forms SDS-resistant dimers through very specific helix-helix contacts within the membrane-embedded region of the molecule (38). Three-dimensional structural studies on this protein reveal that segments rich in Val, Ile, Thr, and Leu are critical for dimer formation (39). Both the predicted second and fourth helices of PsbS are very rich in these amino acids. In our case, the interaction was strong enough to detect the dimer in SDS/PAGE, to an extent, however, that depends on solubilization conditions (Fig. 2 C and D).
Our experiments show that high-light treatment induces monomerization of PsbS, suggesting that it is this form of the protein that plays a role in NPQ. Monomerization may expose the hydrophobic phenylalanine ring on PsbS, which has been proposed to be a binding site for zeaxanthin, at least in vitro (18). It is worth recalling that zeaxanthin, forming at low luminal pH and found either free in the lipid phase (40) or bound to Lhcb proteins (41, 42), does not bind to PsbS in vitro in a pH-dependent manner (18). However, constitutive accumulation of zeaxanthin in a Dunaiella salina mutant does not bring about permanent quenching nor affect photosynthesis or sensitivity to irradiance stress, and it has been proposed that zeaxanthin is involved in photoprotection after PSII photodamage (43). A recent hypothesis envisions dynamic translocation of pigments and different domain localization for violaxanthin and zeaxan-thin during the reversible xantophyll cycle (43).
A matter of great interest with some debate is the location of PsbS. According to a current hypothesis, PsbS may have more than one location in the PSII supercomplex (17, 32, 35). Analysis of our sucrose-gradient experiment fits this view. Thus, at neutral pH, PsbS monomers are found to migrate mostly to the LHC region, whereas the dimers are found in the fractions corresponding to the PSII core complexes. However, when sucrose-gradient fractionation was performed at acid pH, not only was the quantity of dimers greatly reduced, as expected, but they were also no longer associated with the PSII core but rather with LHC complexes, where PsbS monomers also are located. In this experiment, we chose to mimic physiologically occurring luminal pH by equilibrating thylakoids at pH 4, because the same experiment performed on thylakoids derived from high-light treated plants would have failed because of prolonged centrifugation time (6 h) in the dark. A direct association of LHC proteins with the monomeric form of PsbS also is indicated by the coimmunoprecipitation (Fig. 5D).
On the basis of the results reported above, we propose that a change in the quaternary conformation of PsbS is an important event in the molecular mechanism by which this protein is involved in producing qE. Thus, the monomer, which we believe is the relevant form of the protein for NPQ, is associated with the LHCII moiety, where it may carry out its function in photoprotection. This result fits the known fact that NPQ has never been reported to occur in isolated PSII membranes (BBY particles), PSII cores, or reaction center preparations. Our results suggest a possible mechanism by which PsbS behaves as a component of the signal-transducing device capable of sensing luminal pH, undergoing conformational change, and inducing the development of a quenched state.
Acknowledgments
We thank G. Walton for revision of the English text. This work was supported by the Italian Ministry for University and Research under projects Programma Nazionale per la Ricerca and Fondo Investimenti per la Ricerca di Base. A European Molecular Biology Organization Young Investigator Program grant to I.S. is gratefully acknowledged.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PSII, photosystem II; NPQ, nonphotochemical quenching; qE, energy-dependent quenching; DTSP, 3,3′-dithiodipropionic acid di(N-hydroxysuccinimide ester); DM, n-dodecyl β-d-maltoside; LHC, light-harvesting complex.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY291061).
References
- 1.Barber, J. & Andersson, B. (1992) Trends Biochem. Sci. 17, 61-66. [DOI] [PubMed] [Google Scholar]
- 2.Aro, E. M., Virgin, I. & Andersson, B. (1993) Biochim. Biophys. Acta 1143, 113-134. [DOI] [PubMed] [Google Scholar]
- 3.Horton, P., Ruban, A. V. & Walters, R. G. (1996) Annu. Rev. Plant Physiol. Mol. Biol. 47, 655-684. [DOI] [PubMed] [Google Scholar]
- 4.Niyogi, K. K. (1999) Annu. Rev. Plant Physiol. Mol. Biol. 50, 333-359. [DOI] [PubMed] [Google Scholar]
- 5.Muller, P., Li, X. P. & Niyogi, K. K. (2001) Plant Physiol. 125, 1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton, P. & Hague, A. (1988) Biochim. Biophys. Acta 932, 107-115. [Google Scholar]
- 7.Demmig-Adams, B. (1990) Biochim. Biophys. Acta 1020, 1-24. [Google Scholar]
- 8.Yamamoto, H. Y., Bugos, R. C. & Hieber, A. D. (1999) in The Photochemistry of Carotenoids, eds. Frank, H. A., Young, A. J., Britton, G. & Cogdell, R. J. (Kluwer, Dordrecht, The Netherlands), pp. 293-303.
- 9.Bugos, R. C. & Yamamoto, H. Y. (1996) Proc. Natl. Acad. Sci. USA 93, 6320-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demmig-Adams, B. & Adams, W. W., III (1996) Planta 198, 460-470. [Google Scholar]
- 11.Niyogi, K. K., Grossman, A. R. & Bjorkman, O. (1998) Plant Cell 10, 1121-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljunberg, U., Akerlund, H. E. & Andersson, B. (1986) Eur. J. Biochem. 158, 477-482. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S., Sandunsky, P., Bowlby, N. R., Aebersold, R., Green, B. R., Vlahakis, S., Yocum, C. F. & Pichersky, E. (1992) FEBS Lett. 314, 67-71. [DOI] [PubMed] [Google Scholar]
- 14.Wedel, N., Klein, R., Ljungberg, U., Andersson, B. & Herrmann, R. G. (1992) FEBS Lett. 314, 61-66. [DOI] [PubMed] [Google Scholar]
- 15.Funk, C., Schröder, W. P., Napiwotzki, A., Tjius, S., Renger, G. & Andersson, B. (1995) Biochemistry 34, 11133-11141. [DOI] [PubMed] [Google Scholar]
- 16.Funk, C., Adamska, I., Green, B. R., Andersson, B. & Renger, G. (1995) J. Biol. Chem., 270, 30141-30147. [DOI] [PubMed] [Google Scholar]
- 17.Dominici, P., Caffarri, S., Armenante, F., Ceoldo, S., Crimi, M. & Bassi, R. (2002) J. Biol. Chem. 277, 22750-22758. [DOI] [PubMed] [Google Scholar]
- 18.Aspinall-O'Dea, M., Wentworth, M., Pascal, A., Robert, B., Ruban, A. & Horton, P. (2002) Proc. Natl. Acad. Sci. USA 99, 16331-16335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X. P., Bjorkman, O., Shih, C., Grossman, A. R., Rosenquist, M., Jansson, S. & Niyogi, K. K. (2000) Nature 403, 391-395. [DOI] [PubMed] [Google Scholar]
- 20.Peterson, R. B. & Avir, E. A. (2000) Planta 210, 205-214. [DOI] [PubMed] [Google Scholar]
- 21.Li, X. P., Phippard, A., Pasari, J. & Niyogi, K. K. (2002) Funct. Plant Biol. 29, 1131-1139. [DOI] [PubMed] [Google Scholar]
- 22.Bergantino, E., Sandonà, D., Cugini, D. & Bassi, R. (1998) Plant Mol. Biol. 36, 11-22. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 24.Barbato, R., Bergo, E., Szabò, I., Dalla Vecchia, F. & Giacometti, G. M. (2000) J. Biol. Chem. 275, 10976-10982. [DOI] [PubMed] [Google Scholar]
- 25.Arnon, D. J. (1949) Plant Physiol. 24, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansson, S., Andersen, B. & Scheller, H. V. (1996) Plant Physiol. 112, 409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerovic, Z. G. & Plesnicar, M. (1984) Biochem. J. 223, 543-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyle, D. J., Kuang, T. Y., Watson, J. L. & Arntzen, C. J. (1984) Biochim. Biophys. Acta 765, 89-96. [Google Scholar]
- 29.Noctor, G., Rees, D., Young, A. & Horton, P. (1991) Biochim. Biophys. Acta 1057, 320-330. [Google Scholar]
- 30.Noctor, G. & Horton, P. (1990) Biochim. Biophys. Acta 1016, 228-234. [Google Scholar]
- 31.Thidholm, E., Lindstrom, V., Tissier, C., Robinson, C., Schroder, W. P. & Funk, C. (2002) FEBS Lett. 513, 217-222. [DOI] [PubMed] [Google Scholar]
- 32.Nield, J., Funk, C. & Barber, J. (2000) Philos. Trans. R. Soc. London B 355, 1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demmig-Adams, B. & Adams, W. M. (1996) Trends Plant Sci. 1, 21-26. [Google Scholar]
- 34.Ruban, A. V., Pascal, A. A., Robert, B. & Horton, P. (2002) J. Biol. Chem. 277, 7785-7789. [DOI] [PubMed] [Google Scholar]
- 35.Nield, J., Orlova, E., Morris, E. P., Gowen, B., van Heel, M. & Barber, J. (2000) Nat. Struct. Biol., 7, 44-47. [DOI] [PubMed] [Google Scholar]
- 36.Li, X. P., Gilmore, A. M. & Niyogi, K. K. (2002) J. Biol. Chem. 277, 33590-33597. [DOI] [PubMed] [Google Scholar]
- 37.Ungar, D., Barth, A., Haase, W., Kauzinger, A., Lewitzki, E., Ruiz, T., Reilander, H. & Michel, H. (2001) Eur. J. Biochem. 268, 5386-5396. [DOI] [PubMed] [Google Scholar]
- 38.Lemmon, M. A., Flanagan, J. M., Treutlein, H. R., Zgang, J. & Engelman, D. M. (1992) Biochemistry 31, 12719-19725. [DOI] [PubMed] [Google Scholar]
- 39.MacKenzie, K. R., Prestegard, J. H. & Engelman, D. M. (1997) Science 276, 131-133. [DOI] [PubMed] [Google Scholar]
- 40.Havaux, M. & Niyogi, K. K. (1999) Proc. Natl. Acad. Sci. USA 96, 8762-8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bassi, R., Pineau, B., Dainese, P. & Marquardt, J. (1993) Eur. J. Biochem. 212, 297-303. [DOI] [PubMed] [Google Scholar]
- 42.Ruban, A. V., Young, A. J. & Horton, P. (1996) Biochemistry 35, 674-678. [DOI] [PubMed] [Google Scholar]
- 43.Jin, E. S., Yokthongwattana, K., Polle, J. E. W. & Melis, A. (2003) Plant Physiol. 132, 352-364. [DOI] [PMC free article] [PubMed] [Google Scholar]