Abstract
Visual cortex exhibits smooth retinotopic organization on the macroscopic scale, but it is unknown how receptive fields are organized at the level of neighboring neurons. This information is crucial for discriminating among models of visual cortex. We used in vivo two-photon calcium imaging to independently map ON and OFF receptive field subregions of local populations of layer 2/3 neurons in mouse visual cortex. We found that receptive field subregions are often precisely shared among multiple neighboring neurons. Furthermore, large subregions appear to be assembled from multiple smaller, non-overlapping subregions of other neurons in the same local population. These experiments provide the first characterization of the diversity of receptive fields in a dense local network of visual cortex, and reveal elementary units of receptive field organization. Our results suggest that a limited pool of afferent receptive fields is available to a local population of neurons, and reveal new organizational principles for the neural circuitry of the mouse visual cortex.
The organization of receptive fields (RFs) in the visual cortex1 has been the subject of renewed and intense interest with the advent of new imaging and recording methods. A smooth retinotopic organization on the macroscopic scale has been demonstrated in mice using intrinsic signal optical imaging2, 3, corroborating earlier extracellular recordings4, 5. However, it remains unknown how RFs are organized on the microscopic scale, at the level of local networks of neighboring neurons. In monkeys, which also exhibit smooth retinotopic organization, Hubel and Wiesel6 found that the scatter of RFs for units recorded simultaneously on the same electrode or during the same perpendicular penetration was quite large, approximately equal to the RF size. More recent studies have confirmed that when multiple neurons are recorded simultaneously on the same electrode, they can have very different RFs7, 8 and responses to natural stimuli9. However, these studies could only sample a small number of neurons that could fortuitously be recorded on a single electrode and discriminated. In order to determine the diversity of RFs, and identify any underlying structure, it is necessary to systematically map larger samples in a defined local population.
Exploring the diversity of RFs within a local population can yield important insights into the function of visual cortex. The geometry of retinal ganglion cell mosaics10 can provide a constraint for neurons in visual cortex to form their RFs10-12. If neurons sample from a small pool of unique afferent RFs, then local populations of cortical neurons should exhibit substantial redundancy in representing a few RFs. Alternatively, an ample pool of afferent RFs and a large amount of convergence could permit neighboring cortical neurons to integrate many different unique sets of afferents, thus rendering highly variable RFs and a low level of redundancy.
To address these issues, we used two-photon calcium imaging in vivo to obtain the first high-resolution RF maps for large numbers of neurons in a local population. By using sparse noise visual stimulation, we independently mapped the ON and OFF subregions of multiple neighboring neurons in mouse visual cortex. We discovered new features of RF micro-organization in visual cortex: shared and spanned receptive fields. These results can be used to constrain models of visual cortex, thus deepening our understanding of neocortical circuitry.
Results
Mapping RFs for populations of neurons in visual cortex
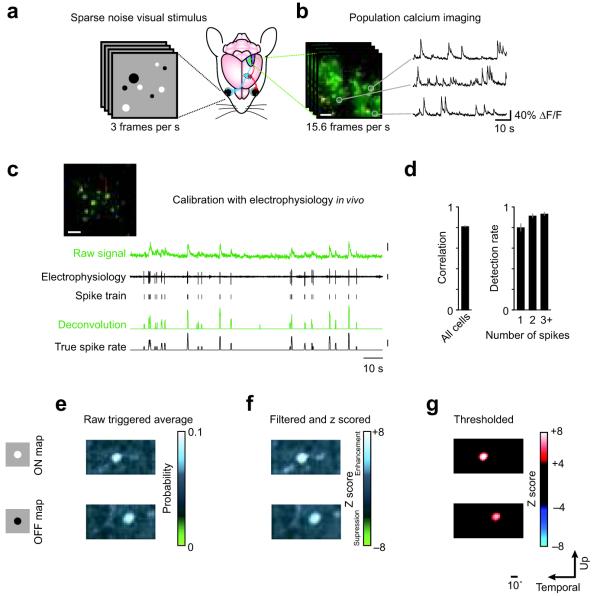
Bolus loading of calcium indicator dyes and two-photon microscopy13, 14 were used to monitor spiking activity in a local population of layer 2/3 neurons spanning a square area (230 μm to a side) in mouse primary visual cortex during sparse noise visual stimulation (Fig. 1a,b). A deconvolution-based algorithm was used to convert calcium signals into estimated spike rates. To calibrate the algorithm, we made loose cell-attached patch-clamp recordings from identified neurons in the population during two-photon calcium imaging and stimulus presentation (Fig. 1c). After calibrating using the electrophysiology data, the algorithm yielded spike rate estimates that were highly correlated to the recorded spike trains (r = 0.81 ± 0.02, n = 19; Fig. 1d), and the false positive rate was low (4.4 ± 1.7%).
Figure 1. Mapping receptive fields with population calcium imaging and sparse noise visual stimuli.
(a) Sparse noise visual stimuli were presented to a mouse during (b) simultaneous population calcium imaging in visual cortex. (c) The calcium signals were deconvolved using parameters obtained from electrophysiology in order to obtain estimated spike rates. Scale bars in b and c: 20 μm; scale bars at right, top to bottom: 20% ΔF/F, 1 mV, 2 spikes/frame. (d) Left, the mean correlation coefficient between deconvolved calcium signals and spike rates obtained from simultaneous on-cell recordings was 0.81 ± 0.02. Right, detection reliability as a function of the number of spikes within one frame. Error bars are ± S.E.M. (e) Using the deconvolved calcium signals as an estimate of spike rate, a triggered average of stimulus frames was computed. Separate ON and OFF maps were generated for the white dots (top) and black dots (bottom), respectively. (f) This estimate was filtered using a Gaussian kernel and z scored using an area of the triggered average away from the RF. (g) This was then thresholded to obtain RFs.
The estimated spike rates were used to construct a spike-triggered average (Fig. 1e) for each neuron in the population. Visual stimuli consisted of random black and white dots on a gray background15. Using the estimated spike rate time courses, we generated two spike-triggered averages, one for the white dots (the ON map) and one for the black dots (the OFF map). Maps were then smoothed using a Gaussian kernel equal in size to the smallest dot used in the stimulus set (1.3°), z scored (Fig. 1f), and thresholded (Fig. 1g). With this method, we obtained maps for 44 ± 4% of imaged neurons (189/434 neurons in 6 mice). RFs for at least 20 neurons (range 21 - 68) were obtained in each animal. The relative ratio of ON and OFF subregions within a local population of neurons varied significantly among animals, but in contrast to macaque visual cortex which is dominated by OFF responses16, overall did not show a consistent bias (mean: 1.08 ± 0.31; range: 0.39 to 2.17).
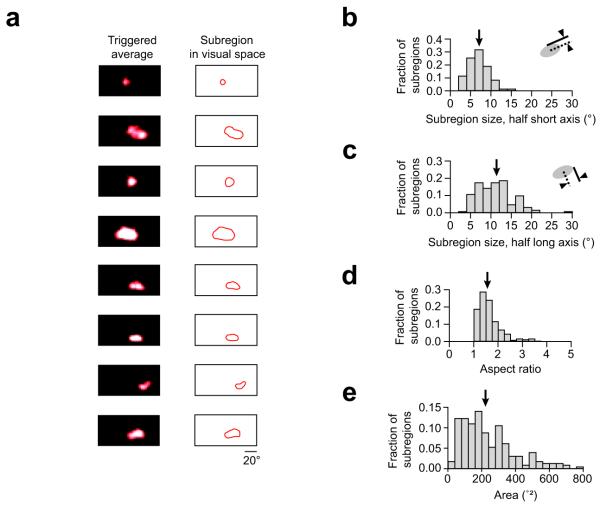
RF subregions varied in size (Fig. 2a) and were often elongated. To quantify RF subregion shape, we fit the subregions with ellipses and computed the half short axis, which was 6.9 ± 1.5° (mean ± S.D., n = 189 RFs from 6 mice; Fig. 2b). This is similar to RF subregions of individual neurons in previous studies of mouse visual cortex obtained using electrophysiological techniques, which generally reported a 5 - 7° range for RF radius17-19. The half long axis of the RF subregions we measured was on average 11.1 ± 4.3° (mean ± S.D.; Fig. 2c), which reflects their often elongated structure. The mean aspect ratio of the elliptical fits was 1.7 ± 0.6 (mean ± S.D.; Fig. 2d). The mean area of the RF subregions was 237 ± 157°2 (mean ± S.D.; Fig. 2e). ON and OFF subregions in individual RFs exhibited low overlap (Supplementary Fig. 1), indicating that most cells were simple cells, consistent with extracellular recordings from mouse V1 (Refs. 18, 20).
Figure 2. Receptive field subregions obtained with population calcium imaging.
(a) To illustrate the diversity of subregion sizes and shapes, a representative set of examples are shown, taken from eight different neurons across three different animals. For each example subregion, the z scored triggered average of stimulus frames (left) and the subregion outline used in subsequent analysis (right) are displayed next to each other. The third and sixth subregions are OFF subregions, the rest are ON subregions. For all subregions mapped (n = 228 subregions in 6 mice) we have plotted the distributions of various geometric parameters. (b) The half short axis length and (c) half long axis length, and (d) aspect ratio of elliptical fits to observed subregions. (e) Distribution of areas of RF subregions. Arrows indicate the mean of each histogram.
Overlap analysis of RF subregions
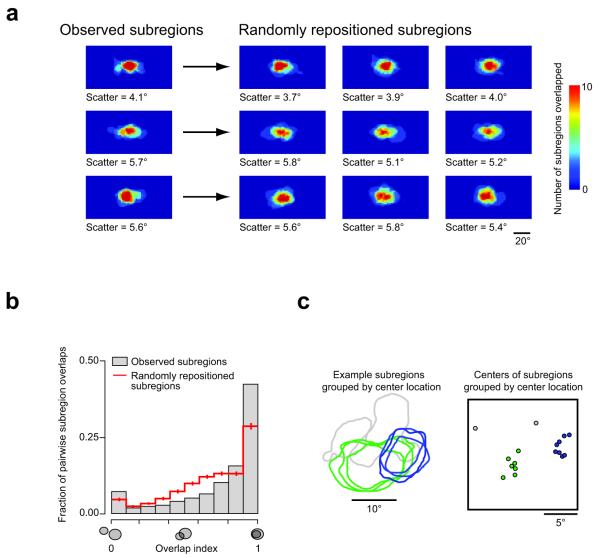
We next examined ensembles of subregions. Ensembles are defined here as all the subregions of a single sign (ON or OFF) from a single population of neurons (20.7 ± 4.8 subregions per ensemble, n = 6 animals). Ensembles of subregions exhibited a scatter of 4.9 ± 0.8° (n = 11 ensembles from 6 animals; Fig. 3a). This scatter is comparable to the subregion size, consistent with findings in primates6. We then examined the degree of overlap in ensembles of subregions. To test whether the amount of overlap observed in the data could result from random positioning, we used a bootstrap method. For each ensemble of subregions, we constructed 1000 new ensembles. These new ensembles used exactly the same population of subregions (no rotations or other distortions), but repositioned them randomly, preserving the subregion scatter observed in the original data (observed: 4.9 ± 0.8°, shuffled: 4.9 ± 0.4°; Fig. 3a). Thus, only the relative positions of subregions were randomized.
Figure 3. Substantial receptive field subregion overlaps occur more frequently than expected for random positioning.
(a) Left, multiple (12 – 15) overlaid subregions (ON, OFF, OFF subregions, respectively) in three different mice are shown. For each point in space, the number of overlapping subregions is coded by color. Scatter is defined as the average deviation of each subregion center from the mean subregion center. Right, the subregions of each ensemble have been randomly repositioned three times. The random repositioning algorithm was designed to not change the scatter (across the full data set, scatter observed: 4.9 ± 0.8°, after repositioning: 4.9 ± 0.4°; mean ± S.E.M.; P = 0.99, Wilcoxon signed-rank test). Note that the random repositioning does not markedly alter the appearance of the ensembles. (b) Pairwise analysis, however, reveals that subregions overlapped more in their observed positions (grey bars) than when randomly repositioned (red line, error bars indicate S.D.; P < 10−5, Kolmogorov-Smirnov test). (c) Example of apparently shared subregions (left) and clustered subregion centers (right) in visual space. The subregion outlines and center markers have been color-coded by hand for clarity. The two gray subregion center markers on the right represent subregions (not shown) that do not appear to be shared with the green and blue groups.
Next, we quantified the pairwise overlap of subregions using a simple metric: the proportion of the smaller of two subregions that overlapped the larger subregion. This metric ranges from 0 (indicating no overlap) to 1 (indicating that one subregion is completely contained within the other subregion). A high proportion of RF subregion pairs exhibited a high degree of overlap (i.e. overlap index close to 1; Fig. 3b). To test whether this unexpectedly high degree of overlap is due to chance, we used the same measure for the randomly repositioned subregions (as shown in Fig. 3a). The randomly repositioned subregions exhibited substantially fewer incidences of high overlap (P < 10−5, two sample Kolmogorov–Smirnov test, Fig. 3b), indicating that the observed substantial overlap between RF subregions is a key, non-random feature of the organization of receptive fields in the mouse visual cortex. The distribution of full RFs (ON + OFF subregions) was similar to that of repositioned subregions, and was again significantly different from the subregions as observed (P < 10−5, two sample Kolmogorov–Smirnov test, Supplementary Fig. 2). Manual inspection of small ensembles of overlapping subregions revealed an apparent trend for subregions to be shared among multiple neurons (Fig. 3c). Therefore, we explicitly tested for the existence of shared subregions.
Shared RF subregions
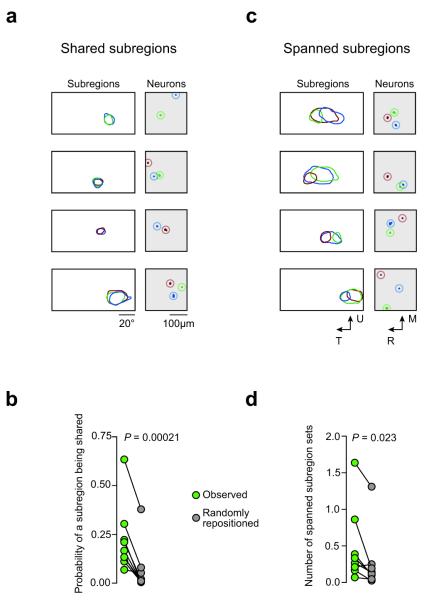
We defined neurons with shared RF subregions as those whose subregions matched in shape, position, and sign (ON or OFF). Such shared RF subregions (Fig. 4a) were observed frequently in all animals, and were often shared across multiple neurons (up to 7). When subregions were repositioned randomly in visual space while preserving scatter (see Fig. 3a), the incidence of shared subregions was significantly reduced (P = 0.00021, paired t-test, n = 11 subregion ensembles from 6 mice; Fig. 4b).
Figure 4. Features of local receptive field organization: shared subregions and spanned subregions.
(a) The left-hand panels show subregions of individual neurons (color-coded) which are shared. The right-hand panels show the relative position of the same neurons in visual cortex (same color code). Four examples are shown. (b) The number of shared subregions in the population is decreased when the subregions are randomly repositioned. Each point represents one ensemble of subregions. (c) In the case of spanned subregions, the subregion of one neuron overlaps two or more non-overlapping subregions from other neurons. Four examples are shown. (d) Again, the incidence of this spatial arrangement decreases when the subregions are randomly repositioned.
In addition to sharing an area of visual space, we determined whether the spatial structure of the spike triggered average within the area is also shared between neurons. We computed the image correlation of the z scored maps and the thresholded RF subregions. We found that the z scored maps also exhibited high degrees of correlation, consistently higher than that of the thresholded RF subregions. This is further evidence of significantly shared RF subregions (Supplementary Fig. 3).
What is the spatial relationship of neurons in visual cortex which share RF subregions? Two-photon population calcium imaging provides us with the exact location of all neurons recorded, allowing us to link their functional properties with their relative locations in the visual cortex. There was only a weak relationship between the distance separating individual neurons in the cortex and the overlap of their subregions in visual space (Supplementary Fig. 4). Therefore, although there is gross retinotopy on the macroscopic scale3, 4, on the scale of neighboring neurons, visual cortex did not exhibit smooth retinotopy.
Correlations among neurons with shared RF subregions
The next question we addressed is whether neurons with shared RF subregions also exhibit correlated activity. We computed activity correlations from the deconvolution-based spike rate estimates rather than from the raw calcium imaging signals themselves in order to avoid overestimating correlation coefficients (Supplementary Figs. 5 and 6). While some neurons with shared RF subregions showed correlated activity, in general neurons that shared RF subregions exhibited low correlations (Supplementary Fig. 5a-d), only slightly higher than the general population (correlation among neurons with shared RF subregions: 0.12 ± 0.12, mean ± SD, n = 149 pairs; correlation among neurons not sharing an RF subregion: 0.085 ± 0.088, mean ± SD, n = 6696 pairs; P = 0.0023, Wilcoxon rank sum test). Overall there was only a weak relationship between the correlation coefficient and the subregion overlap, as quantified by the overlap index (Pearson’s r = 0.13; P < 10−5; Supplementary Fig. 5f). There was a similarly weak relationship between the strength of the cross-correlation and the distance between cell bodies in visual cortex (Pearson’s r = −0.053; P = 0.0021; Supplementary Fig. 5g). Therefore, even when the subregions of two neurons were identical (Supplemental Fig. 5b), individual neurons often responded to different frames of the visual stimulus (Supplementary Fig. 5e).
When we analyzed pairs of cells that both exhibit ON and OFF RF subregions, and shared at least one RF subregion, we found that none of these cell pairs shared both ON and OFF subregions (0/13). To further explore this difference, we examined the overlap and found that among these neurons, their subregions of the opposite sign overlap about 30% less than the shared subregions (P = 0.0097; Supplementary Fig. 7). This tendency for neurons to not share RF subregions of both signs simultaneously could contribute to the low correlations we observe. Additional potential contributors to the low correlations include a low response reliability, differing selectivities to higher order aspects of the stimulus not reflected in the spike triggered average, and active decorrelation due to intracortical circuitry21.
Spanned RF subregions
We also observed evidence for neurons integrating multiple subregions into a single, larger subregion. We refer to this feature as “spanned subregions”: the subregion of one neuron overlaps two or more non-overlapping subregions of the same sign from other neurons (Fig. 4c). Spanned subregions were observed in all animals, but not all ensembles. When subregions were repositioned randomly in visual space while preserving scatter, the incidence of this feature was significantly reduced (P = 0.0234, paired Wilcoxon signed rank test, n = 8 subregion ensembles from 6 mice; Fig. 4d). This indicates that subregions which are not overlapping may themselves be encompassed by a larger spanning subregion that integrates them both.
Spatial offset of ON and OFF RF subregion ensembles
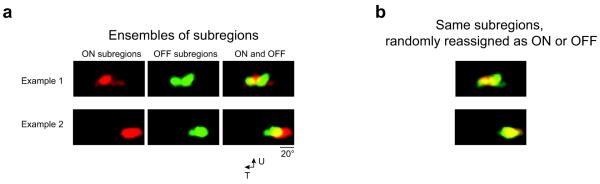
Having determined that subregions of the same sign frequently overlap and thus form clusters in visual space, we next sought to determine the relative relationships between clusters of ON and OFF subregions. To address this issue, the ensemble of subregions for a single local population of neurons was plotted in visual space and color-coded to discriminate ON and OFF subregions. This revealed a clear spatial segregation of ON and OFF subregion populations (Fig. 5a). When subregions were randomly reassigned as ON or OFF subregions (while still maintaining the same total number of ON and OFF subregions), this segregation was not observed (P < 0.02, bootstrap, n = 4 mice; Fig. 5b). Thus, local populations of neurons encode light and dark stimuli in parallel from separate regions of visual space.
Figure 5. ON and OFF subregions are offset with respect to each other in visual space.
(a) The ON subregions in each animal tend to cluster in an area of visual space spatially segregated from the OFF subregions that also cluster together. Data from two animals are shown, with overlap density represented by color intensity (example 1 consists of 15 ON subregions and 18 OFF subregions; example 2 consists of 19 ON subregions and 15 OFF subregions). (b) When the subregions are randomly reassigned as ON or OFF subregions, the spatial segregation is lost. The observed clustering was decreased by randomly reassigning subregions as ON or OFF (P < 0.02, bootstrap method, n = 4 mice).
Discussion
We have mapped RFs in local populations of layer 2/3 neurons in mouse visual cortex using calcium imaging combined with sparse noise visual stimulation. This yielded a cellular-level view of a cortical retinotopic map, revealing independently sampled ON and OFF subregions for individual neurons mapped at high resolution. Within these local populations we found that, although local retinotopy was not smooth, neurons often exhibited highly similar RF subregions, precisely overlapping in visual space, and sharing similar spatial structure. Moreover, large RF subregions often appeared to be composed of two or more smaller, non-overlapping subregions exhibited by other neurons in the same local population. Together, these results suggest that small RF subregions represent elementary units that are shared and integrated by local populations of neurons. This finding indicates that a small pool of afferents strongly influences the organization of RFs in local populations, and places constraints on the functional organization of cortical circuitry10-12, 22.
Population imaging for dense sampling of receptive fields
This study provides the first characterization of the diversity of spatial RFs in local networks of the mammalian visual cortex. Previous studies of visual cortical activity using population calcium imaging used comparatively small numbers of stimuli to reveal the cellular-level cortical organization of orientation selectivity14 23, and ocular dominance24, 25. In our experiments, by combining sparse noise visual stimuli with high speed imaging, we were able to present thousands of visual stimuli and obtain high-resolution RFs from a large fraction of neurons in a local population simultaneously. Such dense sampling is important for constructing accurate models of cortical function15, 26. In particular, our results would have been difficult to obtain using smaller samples of each local population, and highlight the unique advantages of population calcium imaging: excellent isolation of individual neurons and parallel sampling of local populations. Importantly, cross-validation of our results is possible against data obtained with complementary methods. In particular, receptive field sizes and scatter we have observed are consistent with results from electrophysiological studies4, 17-20.
Shared and spanned subregions
The principal finding of our study is that RF subregions of layer 2/3 neurons in mouse visual cortex often precisely overlap with those of other neurons distributed heterogeneously in the same local population. In addition, larger subregions appear to be composed of non-overlapping smaller subregions. These features limit the diversity of RFs and represent a fundamental principle of organization of the visual cortical microcircuit.
The subregions we observed are similar in size to the RFs of retinal ganglion cells27. However, the subregions of cortical neurons are more elongated, with their aspect ratio around 1.6 on average compared to ~1.2 for retinal ganglion cells28, likely reflecting the integration of multiple afferents, possibly upstream of the layer 2/3 populations we have imaged. This integration may occur in the lateral geniculate nucleus, where neurons receive input from 1 to 3 retinal ganglion cells29. While RF properties in mouse visual cortex are quite similar to those observed in the lateral geniculate30, we cannot exclude the possible contribution of further cortical integration upstream from the populations of neurons we imaged.
Neurons that shared subregions were not spatially clustered within visual cortex at the cellular scale, indicating that their dendritic fields did not necessarily need to overlap to produce identical subregions. This distributed processing of visual space stands in contrast to the retina, where individual mosaics of ganglion neurons are coordinated to sample visual space uniformly31. Similarly, neurons with subregions that spanned other subregions did not exhibit a clear pattern of spatial organization within visual cortex. These spanning subregions may occur in neurons downstream in the circuit from those neurons expressing the subregions that they contain, suggesting the possibility of a processing hierarchy of RF subregions. Detailed circuit tracing studies will be required to test this hypothesis.
Pairs of neurons that shared an RF subregion exhibited low correlations in activity, similar to the rest of the population. This suggests that neurons sharing RF subregions remain independent, rather than simply serving as redundant elements in cortical circuitry. There are at least two possible scenarios to consider. First, the neurons sharing an RF subregion may receive input that originated with the same small population of retinal ganglion cells. This naturally explains the high overlap, but does not explain the low activity correlations. There are several potential sources of these low correlations. Low firing rates can limit the peak correlations (see Supplementary Note 1). RF subregions of the opposite (non-shared) sign are another mechanism that will limit correlations, by driving uncorrelated responses when the stimulus is not restricted to the shared RF subregion. Indeed, neurons that share a RF subregion of one sign have less overlapped subregions of the opposite sign (Supplementary Fig. 7). Similarly, neurons may be selective for different higher order features of the visual stimulus that are not captured by the spike-triggered average32, 33. Finally, another potential source for the low correlations in this scenario is active decorrelation due to intracortical circuitry21.
Second, the two cells may receive input that originated from retinal ganglion cells (RGCs) of the same sign and the same region of visual space, but different RGC types with different temporal response profiles to the stimulus. The mouse retina is estimated to contain at least 20 different RGC types, each forming an independent mosaic34-36. Furthermore, given the density of RGCs in the mouse retina and the surface area of visual cortex we image, we estimate about 1270 RGCs contribute to the region of visual space encoded in the portion of visual cortex we image (see Supplementary Note 2). Therefore, independent RGC input with the same sign encoding input for a very similar area of visual space is plausible, and would naturally explain the low activity correlations. However, since each retinal mosaic is positioned independently, the precise overlap seen in our data between different cell types is expected to be uncommon. Hebbian mechanisms could alter receptive fields in the visual pathway and increase the functional overlap in primary visual cortex. It may be helpful to use population imaging to map RFs in developing visual cortex in order to determine whether the substantial overlap of subregions is a feature that is initially present, or only emerges later due to Hebbian mechanisms37.
Spatially offset ensembles of ON and OFF RF subregions
The ON and OFF RF subregions exhibited by local populations of V1 neurons were found to be spatially offset in visual space. The origin of this phenomenon may lie in retinal ganglion cell mosaics (see Supplementary Note 3), but another source could also be responsible. In the optic tract of cats, retinal ganglion cell axons of the same type tend to travel together and form separate retinotopic maps that are slightly out of register with each other38, 39. A similar phenomenon may occur in mice. Although corrective mechanisms may exist to bring these maps back into register, they will have to operate on pathways with very different firing patterns (ON and OFF cells for the same region of visual space). Thus it is easy to imagine how such mechanisms may fail to fully correct the registration error introduced by the fasciculation of axons of the same cell type.
Comparison with other species
Since many principles of visual cortex development and function are shared between mice, cats, and primates40, 41, it will be interesting to determine whether the results presented here may also hold in these other species. In support of this idea, we found that clusters of ON and OFF subregions tend to be spatially segregated in visual space. This is reminiscent of the clustering of ON and OFF geniculate afferents in cat visual cortex42. The segregated processing of light and dark stimuli we found at the local level may also have parallels in humans, given that human test subjects perform more poorly at detecting the interval between opposite polarity lines than same polarity lines, a phenomenon that has been interpreted to be due to independent processing of light and dark stimuli43, 44. However, there are also aspects of the mouse visual system that are different from the visual systems of cats and primates (see Supplementary Note 4). Nevertheless, the principles we have uncovered are potentially applicable to intracortical connections with a low degree of divergence, which are found across a wide range of species.
Functional implications
Our results demonstrate that there is indeed robust functional microorganization in mouse visual cortex which underlies the apparently weak local spatial organization of neurons with similar function previously described across rodent cortex45. This important result provides a useful framework with which we can begin to dissect the cortical microcircuitry. Groups of neurons with shared or spanned RF subregions, despite precisely sharing a portion of their selectivity, act as independent, parallel units in the cortical circuitry.
This result demonstrates a mechanism for generating a high diversity of stimulus selectivities, despite low convergence: local scatter. The precisely shared RF subregions and spanned subregion sets indicate a low degree of convergence. If this low convergence were combined with low scatter retinotopy, more neurons would have shared both ON and OFF subregions and local circuitry would have exhibited much less diversity. By employing a high level of local RF scatter, the diversity of stimulus selectivities is increased. Thus local RF scatter may not be a defect, but rather a mechanism for generating diverse cortical computations.
Visual cortex circuitry can be considered in terms of discrete, self-contained circuits (“columns”), or alternatively as a uniform network with smoothly varying stimulus selectivities. Our finding that the visual cortex contains groups of neurons that perform parallel processing of visual stimuli46 of the same sign in the same area of visual space (at the expense of smooth local retinotopy) may lie somewhere in between these two extremes. Groups of neurons can form discrete cell assemblies47 by virtue of a shared subregion of the same sign, and exhibit a variety of stimulus selectivities due at least in part to their diversity of subregions of the opposite sign, thus forming a parallel processing unit for an area of visual space. However, these units are not mutually exclusive. Each neuron in a group may simultaneously participate in another group due to their subregions of the opposite sign, effectively participating in multiple cell assemblies47. This clear evidence for an intermediate domain of discreteness raises many questions about models of cortical development and function. To address these questions, it will be important to determine the connectivity motifs of groups of neurons that share subregions, and neurons that exhibit spanning subregions. This will ultimately require a combination of dense population imaging as we have performed here, and new approaches for detailed cortical circuit analysis.
Methods
Preparation
All experiments were carried out in accordance with UK Home Office regulations. Adult C57/blk6 mice (1.5 – 3 months of age) were anesthetized with isoflurane (5% for induction, 1 – 3% for surgery) and chlorprothixene (0.5 – 2 mg/kg). During imaging, the isoflurane level was decreased to 0 – 0.5%. A 2 – 3 mm diameter craniotomy was opened over monocular visual cortex. The dura mater was left intact. Bolus loading of neurons with calcium dye was performed using a standard protocol13, 14, 48. A 50 μg aliquot of Oregon Green BAPTA-1 or Fluo-4 (Invitrogen) was dissolved in a 20% Pluronic 127 solution in DMSO. After vortexing, this was diluted 10-fold with a pH 7.4 solution of 150 mM NaCl, 2.5 mM KCl, 10 mM HEPES, and 50 μM sulforhodamine 101 to counterstain astrocytes49. After filtering the solution, a patch pipette (3 – 7 MΩ) was guided under two photon microscopy to a depth of 150 – 250 μm. Pressure of 300 – 600 mbar was applied for 1 – 3 minutes to eject dye solution from the pipette. This stained a spherical volume of 300 – 400 μm in diameter. We imaged 434 neurons in 6 animals and obtained RFs from 189 of these cells (44%).
Imaging
A custom two-photon microscope with a 16x, 0.8 N.A. (Nikon) water immersion objective and a large aperture collection pathway with low-noise PMTs (hand-selected 3896 and 7422-40P PMTs, Hamamatsu) was used for imaging. Images were acquired using ScanImage in conjunction with MATLAB at 15.6 frames per second with 256 × 128 pixels resolution, covering an area of 230 × 230 μm.
Electrophysiology
In order to calibrate the deconvolution routine, on-cell recordings were performed during imaging and stimulus presentation. Standard techniques50 were used to visually guide the pipette (3 – 5 MΩ), which contained HEPES-buffered artificial cerebrospinal fluid and 25 – 50 μM Alexa 594 (Invitrogen), to the target neuron. Recordings were obtained in current clamp mode with no current injection. Signals were filtered at 3 – 5 kHz and acquired at 10 – 50 kHz.
Visual stimulation
Routines written in MATLAB using the Psychophysics Toolbox (Brainard and Pelli) extensions controlled the visual stimulus presentation. Sparse noise visual stimuli consisted of black (2 cd/m2) and white (86 cd/m2) dots on a gray (40 cd/m2) background 15. Dots ranged in size from 1.3° to 8.0° in diameter. Adjacent frames had no overlap in dot locations so that all pixel transitions were to or from gray, never from black to white or vice versa. The visual stimulus LCD screen (ESAW 7 inch VGA TFT, set at 1024× 768 resolution and 60 Hz refresh rate) was shrouded with a cone up to the eye of the animal to prevent contamination of the imaging pathway with light from the visual stimulus. The visual stimulus extended +20 – +124° in azimuth (in one experiment, the span was +20 – +73°) and −10 – +42° in elevation. Stimulus frames (128 × 128 pixels) were smoothly interpolated so that 1 stimulus frame pixel was equivalent to 0.72°2 in visual space. Between 4000 and 6000 stimulus frames were used in each experiment. Total mapping time was 30 – 45 minutes.
Analysis and statistics
In the calcium imaging movies of visual cortex during stimulus presentation, regions of interest were detected using a local pixel-wise neighborhood cross-correlation (Supplementary Fig. 8). For a given movie, the time course for each pixel was compared to those of its eight neighbors. The average peak cross-correlation value was then used to form a local cross-correlation peak image. This image was thresholded using a locally adaptive filter to obtain regions of interest. Subsequently, morphological filters were used to identify somata.
After filtering the fluorescence time courses with a locally adaptive Wiener filter, a deconvolution-based algorithm generated estimated spike rates. The deconvolution kernel was an instantaneous rise followed by an exponential decay. The time course of the exponential decay was selected based on the electrophysiology data to maximize the match between the real and estimated spiking. Once optimized, the same kernel (τdecay = 0.64 s) was used for all time courses. The resulting trace was then thresholded to remove noise; this threshold was adaptive in that it was adjusted based on the standard deviation and skewness of the deconvolved traces. When the threshold was adjusted manually for each cell individually, the resulting cross-correlations between recorded spike trains and estimated spike trains did not change significantly (automatic noise threshold, cross-correlation: 0.81 ± 0.02; manual noise threshold, cross-correlation: 0.84 ± 0.02). When the noise threshold was set using all but 1 data set, and then tested on the remaining data set, the estimated spike trains again did not change significantly (cross-correlation: 0.81 ± 0.02). Detection rates (Fig. 1d) were calculated as the likelihood that any given, single imaging frame event will be detected. It does not factor the accuracy of event amplitude estimation (number of spikes). The cross-correlation measure does depend on the accuracy of the event amplitude estimation, but is insensitive to changes in scale. We restricted our analysis to measures that were also insensitive to scale, namely spike-triggered averages and cross-correlations.
These estimated spike rate time courses were used for reverse correlation with the visual stimulus and for determining cross-correlations between neurons. Raw fluorescence traces were not used for computing cross-correlations due to concerns that they can yield artificially high correlations (due to a variety of causes, including slow decay time courses after events and small fluctuations in laser power). Indeed, when cross-correlations were computed on raw fluorescence traces, the resulting cross-correlation values were higher than that obtained from deconvolved traces (Supplementary Fig. 6).
We did not explicitly differentiate between excitatory and inhibitory neurons. However, our deconvolution scheme relies on the well-characterized calcium signal in response to action potentials, and the action potential-associated calcium signals in interneurons are typically markedly smaller than those of principal neurons, due to the robust calcium buffering mechanisms unique to interneurons. Together with the higher prevalence of pyramidal neurons compared to interneurons in mammalian cortex, it is therefore likely that the population of neurons reported here is dominated by pyramidal neurons.
Reverse correlation was computed using the delay that yielded the highest signal-to-noise ratio in the RF maps across all neurons (mean 255 ms, range 200 – 300 ms). The same delay parameter was used for all neurons within a population. Estimated spike-triggered averages were thresholded at ±4 z score. To further decrease the noise in the RF maps, a morphological filter excluded small features (less than 2.6° in radius). Negative z score areas (where visual stimuli suppressed spiking) were observed, but were rare and typically weak. For simplicity, we focused solely on the positive z score areas for this analysis. For clarity, interpolated RF contours are shown in some figures. All analysis was performed on uninterpolated (pixel-wise) binary RFs.
The overlap index metric was calculated as the proportion of the smaller of two subregions that overlapped the larger subregion. Shared subregions were defined as having ≥ 90% overlap, centers ≤ 1.5° from each other, and ≤ 25% difference in area. Spanned subregion sets were defined as an ensemble of two or more subregions with ≤ 25% overlap and another subregion that overlapped the ensemble ≥ 75%. To measure the incidence of spanned sets, we calculated the number of unique span sets in the population, normalized by the number of subregions; a span set was defined as a set of non-overlapping subregions and a corresponding spanning subregion. The main results of this study were not acutely sensitive to these definitions. We also compared an alternative measure of subregion overlap to the overlap metric described above. This alternative was a subregion correlation (SC) metric that was calculated using the formula:
where srA and srB are binary images of two subregions with backgrounds of zeros. For this metric, 0 indicates completely non-overlapping subregions, increasing values represent greater amounts of subregion overlap, and a value of 1 indicates that the subregions are perfectly identical. Indistinguishable results were obtained using the overlap metric with the thresholds described above, and the subregion correlation metric with a threshold of 0.85. Thus the results presented in this study do not depend on a specific quantification of subregion overlap.
We also computed image correlations using another metric. The two images were each converted into 1-dimensional series of pixels, A and B, similar to two time series. The correlation R was computed as:
Where μx and σx are the mean and standard deviations of series X, and E is the expected value operator. When used to compare z scored maps, the maps were first masked by a two dimensional Gaussian mask with a standard deviation equal to the longest radius of the RF subregion. This minimally affected the z scored maps near the RF subregion, and flattened out the background regions to values near 0. This approach was taken to ensure that the correlation coefficients calculated were dominated by correlations in or near the RF subregion, and not the background noise.
Span sets were found using a tree search algorithm. Neurons with similar, but not necessarily shared, subregions could increase the number of span sets. For example, take the case where neurons A, B, and C have similar subregions; likewise for neurons D, E, and F; and neurons G and H have similar subregions, which span the previous ones. Then the spanning subregions sets would be: ADG, BDG, CDG, AEG, … CFH. A total of 18 unique sets for 8 neurons. Note that, in practice, not all shared subregions will degenerately contribute to the same spanning subregion count; some shared subregions will vary just enough to exclude them, and vice versa for subregions in the spanning subregion count being excluded from the shared subregions. Since shared RF subregions can contribute to high spanning set counts and thus confound a statistical test of the spanning RF subregions, we relaxed the thresholds for the spanning RF subregion test to the point that the statistical significance for shared RF subregions breaks down. Thus, any remaining difference between observed and repositioned subregions was due to the feature of spanning RF subregions. The potential for high counts in this analysis was mitigated by using the non-parametric, paired, two-sided Wilcoxon signed rank test for determining statistical significance.
In order to test whether the observed ensembles of subregions were significantly different from other random spatial configurations, we compared the ensembles to randomly repositioned versions. To construct these, observed subregions were randomly repositioned in visual space. The repositioning did not change subregion shapes or orientation. The repositioning preserved the scatter and spatial spread (Supplementary Fig. 9) of subregion ensembles in visual space, as observed in the original data. To obtain average values for the randomly repositioned versions of ensembles, the repositioning was repeated 1000 times.
The clustering of ON and OFF subregions was quantified by subtracting the sum of the OFF subregions from the sum of ON subregions. In this way, highly overlapping ON and OFF subregions will result in an image that is on average zero, and any clustering will result in positive and negative peaks. The sum of absolute values of all pixels was then used as a measure of clustering. To test significance, the subregions were randomly re-assigned as ON or OFF and the clustering was re-calculated. After 1000 iterations, the mean of the clustering values for the re-assigned data was used to normalize the observed clustering measures.
Unless otherwise noted, all measurements are expressed as mean ± S.E.M.
Supplementary Material
Acknowledgements
We are grateful to Beverley Clark, Matteo Carandini, James Cottam, Martha Havenith, Julian Jack, Benjamin Judkewitz, Peter Latham, Michael London, Alex Mathy, Ikuko Smith and Christian Wilms for helpful discussions and comments on the manuscript. This work was supported by an HFSP Long-Term Fellowship to SLS, and grants by the Wellcome Trust and the Gatsby Charitable Foundation to MH.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hubel DH, Wiesel TN. Receptive fields of single neurones in the cat’s striate cortex. The Journal of physiology. 1959;148:574–591. doi: 10.1113/jphysiol.1959.sp006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith SL, Trachtenberg JT. Experience-dependent binocular competition in the visual cortex begins at eye opening. Nat Neurosci. 2007;10:370–375. doi: 10.1038/nn1844. [DOI] [PubMed] [Google Scholar]
- 3.Kalatsky VA, Stryker MP. New paradigm for optical imaging: temporally encoded maps of intrinsic signal. Neuron. 2003;38:529–545. doi: 10.1016/s0896-6273(03)00286-1. [DOI] [PubMed] [Google Scholar]
- 4.Wagor E, Mangini NJ, Pearlman AL. Retinotopic organization of striate and extrastriate visual cortex in the mouse. J Comp Neurol. 1980;193:187–202. doi: 10.1002/cne.901930113. [DOI] [PubMed] [Google Scholar]
- 5.Drager UC. Receptive fields of single cells and topography in mouse visual cortex. J Comp Neurol. 1975;160:269–290. doi: 10.1002/cne.901600302. [DOI] [PubMed] [Google Scholar]
- 6.Hubel DH, Wiesel TN. Uniformity of monkey striate cortex: a parallel relationship between field size, scatter, and magnification factor. J Comp Neurol. 1974;158:295–305. doi: 10.1002/cne.901580305. [DOI] [PubMed] [Google Scholar]
- 7.DeAngelis GC, Ghose GM, Ohzawa I, Freeman RD. Functional micro-organization of primary visual cortex: receptive field analysis of nearby neurons. J Neurosci. 1999;19:4046–4064. doi: 10.1523/JNEUROSCI.19-10-04046.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reich DS, Mechler F, Victor JD. Independent and redundant information in nearby cortical neurons. Science. 2001;294:2566–2568. doi: 10.1126/science.1065839. [DOI] [PubMed] [Google Scholar]
- 9.Yen SC, Baker J, Gray CM. Heterogeneity in the responses of adjacent neurons to natural stimuli in cat striate cortex. J Neurophysiol. 2007;97:1326–1341. doi: 10.1152/jn.00747.2006. [DOI] [PubMed] [Google Scholar]
- 10.Wassle H, Boycott BB, Illing RB. Morphology and mosaic of on- and off-beta cells in the cat retina and some functional considerations. Proc R Soc Lond B Biol Sci. 1981;212:177–195. doi: 10.1098/rspb.1981.0033. [DOI] [PubMed] [Google Scholar]
- 11.Ringach DL. On the origin of the functional architecture of the cortex. PLoS One. 2007;2:e251. doi: 10.1371/journal.pone.0000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soodak RE. The retinal ganglion cell mosaic defines orientation columns in striate cortex. Proc Natl Acad Sci U S A. 1987;84:3936–3940. doi: 10.1073/pnas.84.11.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 15.Mata ML, Ringach DL. Spatial overlap of ON and OFF subregions and its relation to response modulation ratio in macaque primary visual cortex. J Neurophysiol. 2005;93:919–928. doi: 10.1152/jn.00668.2004. [DOI] [PubMed] [Google Scholar]
- 16.Yeh CI, Xing D, Shapley RM. “Black” responses dominate macaque primary visual cortex v1. J Neurosci. 2009;29:11753–11760. doi: 10.1523/JNEUROSCI.1991-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metin C, Godement P, Imbert M. The primary visual cortex in the mouse: receptive field properties and functional organization. Exp Brain Res. 1988;69:594–612. doi: 10.1007/BF00247312. [DOI] [PubMed] [Google Scholar]
- 18.Mangini NJ, Pearlman AL. Laminar distribution of receptive field properties in the primary visual cortex of the mouse. J Comp Neurol. 1980;193:203–222. doi: 10.1002/cne.901930114. [DOI] [PubMed] [Google Scholar]
- 19.Liu BH, et al. Visual receptive field structure of cortical inhibitory neurons revealed by two-photon imaging guided recording. J Neurosci. 2009;29:10520–10532. doi: 10.1523/JNEUROSCI.1915-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renart A, et al. The asynchronous state in cortical circuits. Science. 2010;327:587–590. doi: 10.1126/science.1179850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringach DL. Haphazard wiring of simple receptive fields and orientation columns in visual cortex. J Neurophysiol. 2004;92:468–476. doi: 10.1152/jn.01202.2003. [DOI] [PubMed] [Google Scholar]
- 23.Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mrsic-Flogel TD, et al. Homeostatic regulation of eye-specific responses in visual cortex during ocular dominance plasticity. Neuron. 2007;54:961–972. doi: 10.1016/j.neuron.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Gandhi SP, Yanagawa Y, Stryker MP. Delayed plasticity of inhibitory neurons in developing visual cortex. Proc Natl Acad Sci U S A. 2008;105:16797–16802. doi: 10.1073/pnas.0806159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seung HS. Reading the book of memory: sparse sampling versus dense mapping of connectomes. Neuron. 2009;62:17–29. doi: 10.1016/j.neuron.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Balkema GW, Jr., Pinto LH. Electrophysiology of retinal ganglion cells in the mouse: a study of a normally pigmented mouse and a congenic hypopigmentation mutant, pearl. J Neurophysiol. 1982;48:968–980. doi: 10.1152/jn.1982.48.4.968. [DOI] [PubMed] [Google Scholar]
- 28.Hammond P. Cat retinal ganglion cells: size and shape of receptive field centres. J Physiol. 1974;242:99–118. doi: 10.1113/jphysiol.1974.sp010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Regehr WG. Developmental remodeling of the retinogeniculate synapse. Neuron. 2000;28:955–966. doi: 10.1016/s0896-6273(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 30.Grubb MS, Thompson ID. Quantitative characterization of visual response properties in the mouse dorsal lateral geniculate nucleus. J Neurophysiol. 2003;90:3594–3607. doi: 10.1152/jn.00699.2003. [DOI] [PubMed] [Google Scholar]
- 31.Gauthier JL, et al. Receptive fields in primate retina are coordinated to sample visual space more uniformly. PLoS Biol. 2009;7:e1000063. doi: 10.1371/journal.pbio.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carandini M, et al. Do we know what the early visual system does? J Neurosci. 2005;25:10577–10597. doi: 10.1523/JNEUROSCI.3726-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olshausen BA, Field DJ. How close are we to understanding v1? Neural Comput. 2005;17:1665–1699. doi: 10.1162/0899766054026639. [DOI] [PubMed] [Google Scholar]
- 34.Huberman AD, et al. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huberman AD, et al. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volgyi B, Chheda S, Bloomfield SA. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. J Comp Neurol. 2009;512:664–687. doi: 10.1002/cne.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song S, Abbott LF. Cortical development and remapping through spike timing-dependent plasticity. Neuron. 2001;32:339–350. doi: 10.1016/s0896-6273(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 38.Torrealba F, Guillery RW, Eysel U, Polley EH, Mason CA. Studies of retinal representations within the cat’s optic tract. J Comp Neurol. 1982;211:377–396. doi: 10.1002/cne.902110405. [DOI] [PubMed] [Google Scholar]
- 39.Torrealba F, Guillery RW, Polley EH, Mason CA. A demonstration of several independent, partially overlapping, retinotopic maps in the optic tract of the cat. Brain Res. 1981;219:428–432. doi: 10.1016/0006-8993(81)90305-x. [DOI] [PubMed] [Google Scholar]
- 40.Nauhaus I, Benucci A, Carandini M, Ringach DL. Neuronal selectivity and local map structure in visual cortex. Neuron. 2008;57:673–679. doi: 10.1016/j.neuron.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon JA. Cellular mechanisms of visual cortical plasticity: a game of cat and mouse. Learn Mem. 1997;4:245–261. doi: 10.1101/lm.4.3.245. [DOI] [PubMed] [Google Scholar]
- 42.Jin JZ, et al. On and off domains of geniculate afferents in cat primary visual cortex. Nat Neurosci. 2008;11:88–94. doi: 10.1038/nn2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi DM, Jiang BC, Klein SA. Spatial interval discrimination with blurred lines: black and white are separate but not equal at multiple spatial scales. Vision Res. 1990;30:1735–1750. doi: 10.1016/0042-6989(90)90156-f. [DOI] [PubMed] [Google Scholar]
- 44.Levi DM, Westheimer G. Spatial-interval discrimination in the human fovea: what delimits the interval? Journal of the Optical Society of America. 1987;4:1304–1313. doi: 10.1364/josaa.4.001304. [DOI] [PubMed] [Google Scholar]
- 45.Ohki K, Reid RC. Specificity and randomness in the visual cortex. Current opinion in neurobiology. 2007;17:401–407. doi: 10.1016/j.conb.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao E, DeAngelis GC, Burkhalter A. Parallel input channels to mouse primary visual cortex. J Neurosci. 2010;30:5912–5926. doi: 10.1523/JNEUROSCI.6456-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hebb DO. The Organization of Behavior, a Neuropsychological Theory. Wiley; 1949. [Google Scholar]
- 48.Garaschuk O, Milos RI, Konnerth A. Targeted bulk-loading of fluorescent indicators for two-photon brain imaging in vivo. Nat Protoc. 2006;1:380–386. doi: 10.1038/nprot.2006.58. [DOI] [PubMed] [Google Scholar]
- 49.Nimmerjahn A, Kirchhoff F, Kerr JN, Helmchen F. Sulforhodamine 101 as a specific marker of astroglia in the neocortex in vivo. Nat Methods. 2004;1:31–37. doi: 10.1038/nmeth706. [DOI] [PubMed] [Google Scholar]
- 50.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.