Abstract
Granulin epithelin precursor (GEP) is a new growth factor that functions in brain development, chondrogenesis, tissue regeneration, tumorigenesis, and inflammation. The goal of this study was to study whether GEP was critical for odontogenesis and amelogenesis both in vivo and in vitro. The in situ hybridization and immunohistochemistry data showed that GEP was expressed in both odontoblast and ameloblast cells postnatally. Knockdown of GEP by crossing U6-ploxPneo-GEP and Sox2-Cre transgenic mice led to a reduction of dentin thickness, an increase in predentin thickness, and a reduction in mineral content in enamel. The in vitro application of recombinant GEP up-regulated molecular markers important for odontogenesis (DMP1, DSPP, and ALP) and amelogenesis (ameloblastin, amelogenin and enamelin). In conclusion, both the in vivo and the in vivo data support an important role of GEP in tooth formation during postnatal development.
Keywords: GEP, tooth development, dentin, enamel, siRNA
INTRODUCTION
GEP, also known as PC-cell-derived growth factor, progranulin, acrogranin, or GP80, was first identified as a growth factor from the conditioned tissue culture media collected from a mouse epithelin precursor of a highly tumorigenic cell line 1. High levels of GEP expression were found in several human cancers such as breast cancer, clear-cell renal carcinoma, invasive ovarian carcinoma, glioblastoma, adipocytic teratoma, and multiple myeloma 2-5. GEP was highly expressed in rapidly cycling epithelial cells such as cells in the immune system and in the nerve system 1-4, 6, as well as in cartilage cells 7.
GEP is a 593-amino-acid secreted glycoprotein with an apparent molecular weight of 80 kDa, which acts as an autocrine growth factor. GEP contains seven-and-a-half repeats of a cysteine-rich motif (CX5-6CX5CCX8CCX6CCXDX2HCCPX4CX5-6C) in the order P-G-F-B-A-C-D-E, where A-G are full repeats and P is the half-motif. The C-terminal region of the consensus sequence contains the conserved sequence CCXDX2HCCP which is likely linked to its regulatory function.
It is well documented that GEP, as growth factor, plays multiple functions in control of cell cycle 8, mesothelial differentiation 9, sexual differentiation of the brain 10, macrophage development 11, response to rheumatoid arthritis and osteoarthritis 12, as well as in wound responses and tissue repair 13, 14.
To address GEP function in chondrogenesis, we recently showed that GEP stimulates chondrocyte differentiation from mesenchymal stem cells in vitro and endochondral ossification ex vivo 15. Importantly, we demonstrated that GEP is a key downstream molecule of BMP2, and that GEP-knockdown mice display skeleton defects 7.
Many studies have indicated that various factors are involved in the formation of dentin and enamel during tooth development 16-26, although there has been no report on the role of GEP in odontogenesis and amelogenesis. In this study, we attempted to study GEP expression pattern in odontoblast and ameloblast cells. Next, we investigated whether GEP plays a role in tooth development in vivo using GEP knock-down mice. We also studied the molecular mechanisms by which GEP controlled odontoblast and ameloblast function in vitro. Our data documented an important role of GEP in controls of tooth development both in vivo and in vitro.
EXPERIMENTAL PROCEDURES
Generation of siGEP Knockdown Transgenic Mice
Based on the technique developed by Deng's laboratory 27, we recently generated the U6-ploxPneo-GEP-RNAi knockdown (KD) transgenic mice where a 19 base pair of 5'-GCCTATCCAAGAACTACAC-3' oligo (silencing GEP) and its antisense oligo with a loop was under the control of the U6 promoter (a ubiquitous promoter) 7. Because a loxP-flanked neomycin cassette is inserted into this promoter to block the promoter activity, there is no expression of RNAi in the absence of Cre. In this study, two independent founders were used for crossing with Sox2 Cre mice (Stock Number: 004783 from Jax mice database) to remove the neo cassette in order to activate expressions of GEP RNAi. The transgenic lines were genotyped using PCR with the following pair of primers (5′-CGAAGTTATCTAGAGTCGAC-3′ and 5′-AAACAAGGCTTTTCTCCAAGG-3′), which amplify ~100 bp from the U6 promoter and the connecting neo gene. The wild-type controls and GEP KD mice were sacrificed at various stages of development, including E17.5, newborn, postnatal day 10, and day 21. All animal studies were in accordance with the guidelines and approved by the IACUC committee of Baylor College of Dentistry.
Alizarin Red/Alcian Blue Staining of the Skeleton
The newborns from wild-type and the GEP KD littermates were skinned, eviscerated, and fixed for more than 1 day in 95% ethanol. They were then stained with Alizarin red (0.09%) and Alcian blue (0.05%) for photography described previously 28-29.
Histology
Teeth were fixed in freshly prepared 4% paraformaldehyde in phosphate-buffered saline (pH 7.4), decalcified, and embedded in paraffin by standard histological procedures as previously described 30. The tissue blocks were cut into 4-µm-thick, mesio-distal, serial sections and mounted on glass slides. Sections were used for H&E staining, in situ hybridization and immunohistochemistry (GEP and DSPP). For Von kossas staining, the samples (newborn) were embedded in paraffin without decalcification.
In Situ Hybridization
The mouse GEP antisense RNA (cRNA) was used for in situ hybridization as described previously 7, 30. Briefly, the digoxigenin (DIG)-labeled mouse GEP cRNA probe was prepared by using an RNA Labeling Kit (Roche, Indianapolis, IN). The hybridization temperature was set at 55℃ and the washing temperature was set at 70℃ to inactivate endogenous alkaline phosphatase. DIG-labeled nucleic acids were detected in an enzyme-linked immunoassay with a specific anti-DIG-AP antibody conjugate and an improved substrate that gives rise to a red signal (Vector, Burlingame, CA).
High-Resolution Tooth Radiography and μ -CT
After dissection, teeth were X-rayed using a Faxitron radiographic inspection unit (Model 8050-020, Field Emmission Corporation, Inc.) with digital image capture capabilities. Using a Scanco μ -CT 35 (SCANCO Medical AG, Switzerland), μ-CT analyses included a high resolution scan of the lower jaw for overall assessment of the tooth shape and structure .
Cell Culture and Cell Proliferation
A preodontoblast or an odontoblast cell line 31 or an ameloblast cell line (LS8, a gift from Dr. Malcolm L. Snead from University of Southern California) was plated into a 6-well plate at a density of 1.2X105 cells per well. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mmol/L L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cell proliferation assay was performed by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] method. Briefly, Cells were seeded into 96-well plates at 5 x104 cells per well. When the cells were at 70% confluences, recombinant GEP protein was added at concentrations of 50 ng/ml, 100 ng/ml, and 200 ng/ml. The media was changed every other day throughout the course of the experiment. The mitogenic effect of recombinant GEP protein was assessed on day one, three, and five after initial treatment by MTT cell proliferation assay kit (ATCC, No. 30-1010K, Manassas, VA). The optimal density was determined at a wavelength of 490 nm. Cells without addition of GEP served as a control group.
RNA Isolation and Quantitative PCR analyses
To examine the effects of the recombinant GEP protein on gene expression, the preodontoblast or ameloblast cells were adapted to serum free medium for 24 hours followed by additions of GEP at 200 ng/ml. Twenty-four hours later, these cells were harvested for RNA isolation and quantitation analysis of gene expression by real time RT-PCR. RNA was isolated using Trizol reagent (Invitrogen, San Diego, CA) according to the manufacturer's protocol. After purification, 1 μg total RNA was treated with Turbo DNase (Ambion) and reverse transcribed into first-strand cDNA using a high-capacity cDNA reverse transcription kit (Invitrogen, San Diego, CA). Using 20 μl SYBR GREEN PCR, reactions were performed in a 96 well optical reaction plate formatted in the 7300 Sequence Detection System (Applied Biosystems) with the following PCR conditions: 40 cycles, 95 °C for 15 s, 60 °C for 1 minute. The transcript of GAPDH mRNA was employed as an internal control for RNA quality. For each gene, three independent RT-PCR reactions from the same reverse transcription sample were also performed. The presence of a single specific PCR product was verified by melting curve analysis, confirmed on an agarose gel, and further sequenced by the Applied Biosystems sequencing system (Applied Biosystems Inc). The genes analyzed were DMP1, DSPP, ALP, Ameloblastin, Enamelin and Amelogenin. GAPDH was used as the internal control.
Statistical Analyses
We computed statistical significance with independent-samples t-test using SPSS 12.0. A P value of < 0.05 was considered statistically significant.
RESULTS
GEP Expression Patterns in Teeth during Embryonic and Postnatal Development
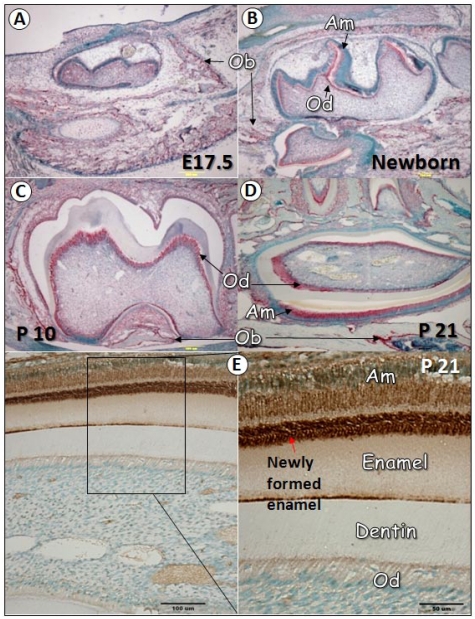
To characterize the temporal and spatial expression pattern of GEP during tooth development, we performed in situ hybridization at multiple time points, including E17.5, newborn, postnatal days 10 (P10), and 21 (P21). As revealed in Fig. 1, GEP mRNA was expressed in the osteoblast (Ob) cell but not in tooth cells at E17.5 (1A). After birth, GEP was shown in odontoblasts (Od, B-D), ameloblasts (Am, B-D), and osteoblast cells (B-D). Immunohistochemistry data confirmed high expression of GEP protein in ameloblasts and enamel matrix (E). This information suggests that the expression profile of GEP is linked to osteogenesis, odontogenesis and amelogenesis during postnatal development.
Fig 1.
GEP expression in tooth during development. Panels A-D show in situ hybridization for GEP expression in teeth at different stages (red signal). At E17.5 (A), GEP was detected in the osteoblast (Ob) cells but not in the odontoblast (Ob) cells. At day one (B), the expression of GEP mRNA was observed in both odontoblasts (Ob) and ameloblasts (Am). At day 10 (C), the expression of GEP mRNA in odontoblasts was high; At day 21 (D), GEP mRNA was detected in ameloblasts, odontoblasts, periodontal ligament (PDL) and osteoblast cells. Panel E showed that GEP protein was particularly high in newly formed enamel.
GEP KD Mice Display Porous, Hypomineralized, and Immature Alveolar Bone
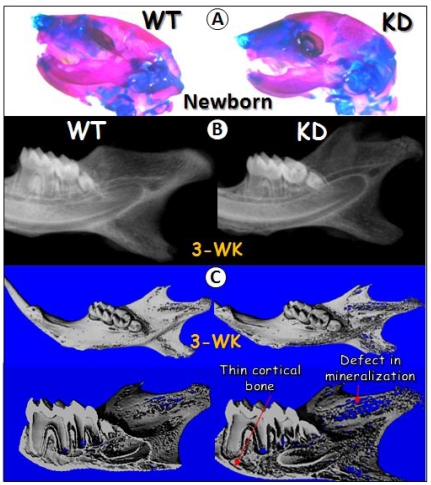
The in vivo knockdown techniques used in this study was originally developed by C. X. Deng's laboratory at NIH/NIDDK, which has been successfully used to knock down FGF receptor2 in cartilage 32. In this study, we crossed U6-ploxPneo-GEP-RNAi knockdown (KD) transgenic mice 7 to Sox2-Cre transgenic mice.. The GEP KD newborn head, stained with Alizarin Red/Alcian Blue, displayed no apparent difference from the age-matched control head (Fig 2A), suggesting that GEP may not be essential for early development. However, at age of 3-week, radiograph and μ-CT images obtained from the GEP KD mandible showed a defect in mineralization as revealed by a thin porous mandible (Fig 2B-C). Overall, the tooth was slightly small with thin dentin in GEP KD mice (Fig. 2C).
Fig 2.
Effects of GEP KD on craniofacial development. A) Alizarin red/Alcian blue staining of the wild-type (WT, left panel) and the knockdown (KD, right panel) newborns heads were shown. B) Radiographs of the 3-week-old WT and GEP KD mandibles were shown. C) Micro-CT images of the 3-week-old WT (left panels) and GEP KD (right panels) mandibles were shown. The whole mount scanning images were shown on upper panels and the sagittal sections were shown on lower panels.
GEP KD Mice Display Reduced Dentin Mass
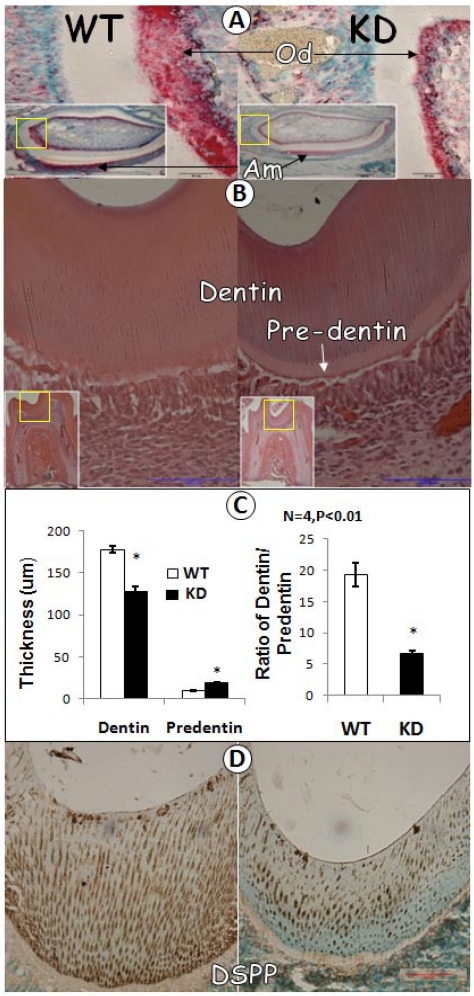
To further characterize GEP KD tooth, we carried out in situ hybridization for comparison of GEP levels in GEP KD and the control tooth. As shown in Fig. 3A, there was a remarkably reduction in GEP mRNA expression in the GEP KD odontoblast cells. H&E staining revealed that the thickness of dentin was decreased in the GEP KD mice, while the predentin was increased, compared to the age matched control (Fig. 3A, B and D). Quantitative data showed that the above changes are statistically significant (Fig 3C, left panel), and the ratio of dentin/predetin was significantly decreased in the GEP KD mice compared to that of the WT mice (Fig. 3C, right panel). Furthermore, the expression level of DSPP protein (dentin sialophosphoprotein, a marker for dentin formation) in the GEP KD mouse was dramatically reduced (Fig. 3D).
Fig 3.
Dentin defect in 3-week-old GEP knockdown (KD) Mice. A) In situ hybridization displayed a reduction of GEP mRNA in the GEP knockdown odontoblast layer (right panel; Od; signal in red) compared to the age matched wild type control (WT, left panel). B) HE staining of the first molars showed a reduction of dentin thickness and an increase in pre-dentin thickness in the GEP knockdown mice (right panel) compared to the control (left panel). C) Quantitative data showed that a significant changes in the thickness of the GEP KD dentin and predentin compared to those in the control (left panel), and that the ratio of dentin / predetin was significantly decreased in GEP KD mice compared to the WT control mice (right panel). All the values were presented as mean ± SE. n=4, *p = 0.05. D) The immunostaining of DSPP (brown staining with green nuclear counterstain) showed a lower level in GEP KD mice (right panel) compared to that of the WT control mice (left panel).
GEP Accelerates Odontoblast Cell Proliferation and Differentiation
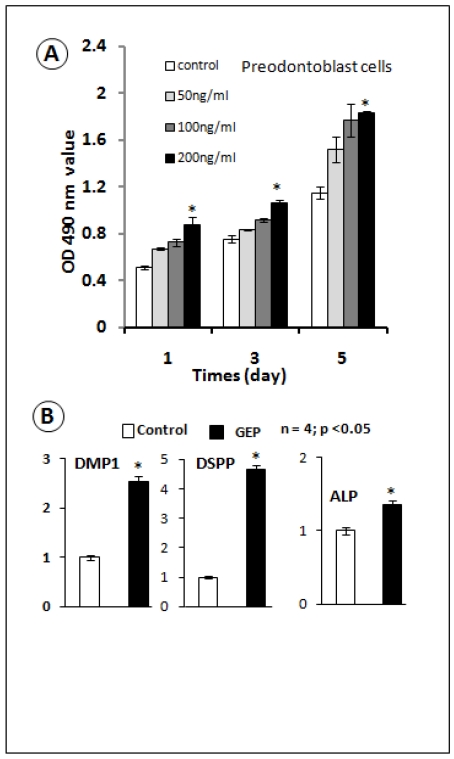
The GEP knockdown dentin phenotype prompted us to determine whether recombinant GEP was able to change odontoblasts proliferation and differentiation. We previously created a GEP stable cell line using 293 EBNA cells and generated recombinant GEP 7. In this study, we tested the effect of GEP on a preodontoblast cell line 31, 33. Our data showed that recombinant GEP stimulated proliferation of the preodontoblast cell line at all tested concentrations, although the significant difference was only observed at the 200 ng/ml concentration (Fig. 4A). Next, we tested the effects of GEP (200ng/ml) on cell differentiation using an odontoblast cells line. Our real-time RT-PCR data showed that GEP increased expressions of DMP1, DSPP and ALP (markers for odontogenesis, Fig. 4B). These in vitro data support a positive role of GEP in odontogenesis.
Fig 4.
Recombinant GEP protein stimulates cell proliferation and accelerates cell differentiation in the odontoblast cell line. A) MTT assay data showed an increase in cell proliferation in all 3 concentrations of recombinant GEP protein, although there was a significant difference only at the concentration of 200 ng (Data are mean ± SEM, n=4, p <0.05). B) Recombinant GEP protein also increased levels of cell markers for odontoblast cell differentiation: DMP1, DSPP and ALP. These values were normalized by GAPDH. (Data are mean ± SEM, n=4, p <0.05).
GEP KD Mice Developed an Enamel Phenotype
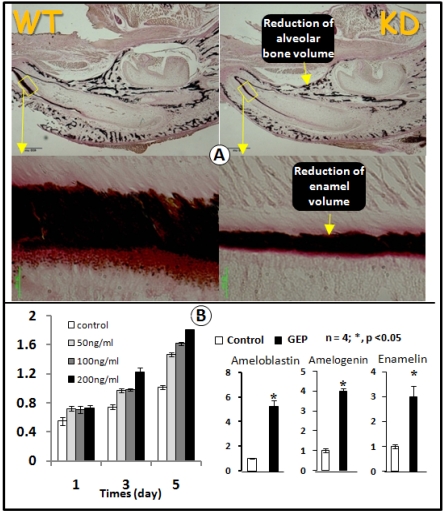
Because GEP was expressed in ameloblast cells (Figs 1 and 3) and the GEP expression level was reduced in the GEP KD ameloblast cells (Fig. 3A), we next asked whether GEP played a role in amelogenesis. The von Kossa staining showed a sharp reduction of the total enamel volume (less than 50% of the normal enamel volume) in the GEP KD lower jaw (Fig. 4A). To further address whether GEP controls cell proliferation and differentiation in the ameloblast cells, we next added recombinant GEP to an ameloblast cell line (LS8) for testing if GEP had effects on cell proliferation and cell differentiation. As shown in Fig. 5B, recombinant GEP stimulated ameloblast proliferation at day3 and day5 (left panel). GEP also increased expressions of ameloblastin, amelogenin, and enamelin (markers for amelogenesis, Fig. 5B, right panel). Taken together, both the in vivo and in vitro data support a positive role of GEP in amelogenesis.
Fig 5.
GEP KD mice display a reduction in enamel mineralization. A) Von Kossa staining showed a reduction of mineralization in the GEP knockdown mice (KD, right panel) compared to the age matched wild type (WT, left panel). B) MTT assay data showed an increase in cell proliferation in the ameloblast cell line with all 3 concentrations of recombinant GEP protein, although there was a significant difference only at the concentration of 200 ng. C) Recombinant GEP protein increased levels of cell markers for ameloblast cell differentiation: ameloblastin, amelogenin, and enamelin. These values were normalized by GAPDH. (Data are mean ± SEM, n=4, p <0.05).
DISCUSSIONS
GEP, as a local growth factor, is closely associated with development, tissue regeneration, tumorigenesis, and inflammation 13, 34-36. Through a functional genetic screen, we previously isolated GEP as a binding partner of cartilage oligomeric matrix protein (COMP, a noncollagenous matrix protein whose mutations lead to pseudoachondroplasia, multiple epiphyseal dysplasia, and short-limb dwarfism) 37. Our recent studies showed that GEP-mediated chondrocyte proliferation was regulated by GEP/COMP interaction, suggesting its importance in skeletogenesis 7. Although the GEP receptor is largely unknown, the signaling pathway activated by GEP has been revealed based on the following two pieces of evidence. First, GEP was reported to activate Erk1/2 signaling in SW-13 adrenal carcinomas 38. Second, our recent finding showed that GEP greatly accelerates chondrogenesis whose activity was abolished for more than 60% when Erk1/2 blocker was applied 7.
In this study we used GEP-knockdown transgenic mouse model and cell culture to investigate the role of GEP during odontogenesis and enamelogenesis. Our key findings are 1) GEP is expressed in the ameloblast (the cell for enamel formation), and the odontoblast (the cell for dentin formation), as well as the osteoblast (the cell for bone formation); 2) recombinant GEP accelerates cell proliferation and differentiation in both ameloblast and odontoblast cell lines in vitro; and 3) knockdown of GEP in the above cells leads to mineralization defects in enamel, dentin and jawbone. The above data supports a role of GEP in tooth formation during postnatal development.
To our best knowledge, there was no report on GEP expression and function in the craniofacial region. In this study, our result showed that GEP was expressed in all these tissues, although knockdown of this gene led to no apparent embryonic phenotype (Figs 2-3), suggesting that this gene may not be critical in early bone and tooth development. However, the μ-CT data showed that the size and bone density of GEP KD mouse mandibles were markedly reduced, supporting a role of GEP in osteogenesis during postnatal development.
Our quantitative analysis showed a significant difference in the thickness of dentin and predentin between the WT and the GEP KD mice. The ratio of dentin to predentin also decreased sharply from the GEP KD mice to WT mice. Because DSPP, a gene critical for dentin formation, has been shown to be a good phenotypic marker for secretory odontoblasts 39-42, we compared its protein expression profile in the knockdown and the control dentin. As expected, the expression of DSPP protein in GEP KD mice was indeed much lower than that in the WT mice. Furthermore, using the cell culture system, we clearly demonstrated that recombinant GEP protein increased preodontoblast cell proliferation, and induced the odontoblast differentiation as shown by an increase in mRNA expressions of DMP1, ALP and DSPP (Fig.3B). All these data suggest that GEP is likely critical for odontogenesis.
In this research, we also studied GEP function in amelogenesis. The enamel thickness was sharply decreased in the GEP KD mice (Fig. 4A). Von Kossa staining revealed that the mineral was considerably reduced in GEP KD mice (Fig. 4A). It is known that three 'structural' enamel proteins--amelogenin, enamelin, and ameloblastin are likely important for enamel formation 16-17, 43-45. Therefore, we tested the effects of the recombinant GEP on their expression in vitro. Our result showed that GEP stimulated ameloblast proliferation in a dosage-dependent manner (Fig. 4B). In addition, GEP increased mRNA expressions of ameloblastin, amelogenin and enamelin (Fig. 4B). Based on this finding, we believe that GEP is likely involved in the amelogenesis.
Cre-LoxP-based RNA interference is a newly developed approach, which combines RNA interference-mediated gene knockdown using a plasmid-based structure and Cre-LoxP system. It was initially used in cell culture system 27 and late in animals for studies of FGF receptor2 in cartilage 32. Comparison with the traditional knockout approach, this method provides a much fast (approximately 6 months or less), yet efficient way to knockdown gene functions in vivo in a tissue-specific manner. For example, Coumoul et al. reported over 95% reduction of FGF receptor 2 after removing the neomycin gene in the U6 promoter by crossing with transgenic Cre mice in the mouse germline. As a result, knockdown of Fgfr2, a critical growth factor for many organ developments, led to embryonic lethality. Because of the Cre-loxP nature, this system can be very useful for the tissue specific knockdown purpose. With this Cre-loxP plasmid we have also successful knocked down GEP in cartilage tissue 7. The GEP knockdown mice developed dwarfism and striking defects in the skeletal system, including delayed endochondral bone growth and reduced bone length and volume. However, this technique has certain limitations. For example, we found that the knockdown efficiency was not consistent. Consequently, the phenotype varies.
In summary, this study provides novel insights into the role of GEP in regulation of odontogenesis and amelogenesis. Our work supports a hypothesis that GEP, as a local growth factor, controls expression levels of DSPP, ALP, DMP1, AMELX, ENAM and AMBN during postnatal tooth development.
Acknowledgments
This work was partially supported by NIH grants: DE015209 (JQF).
References
- 1.Zhou J, Gao G, Crabb JW, Serrero G. Purification of an autocrine growth factor homologous with mouse epithelin precursor from a highly tumorigenic cell line. J Biol Chem. 1993 May;268(15):10863–9. [PubMed] [Google Scholar]
- 2.Anakwe OO, Gerton GL. Acrosome biogenesis begins during meiosis: evidence from the synthesis and distribution of an acrosomal glycoprotein, acrogranin, during guinea pig spermatogenesis. Biol Reprod. 1990 Feb;42(2):317–28. doi: 10.1095/biolreprod42.2.317. [DOI] [PubMed] [Google Scholar]
- 3.Baba T, Hoff HB3rd, Nemoto H, Lee H, Orth J, Arai Y, Gerton GL. Acrogranin, an acrosomal cysteine-rich glycoprotein, is the precursor of the growth-modulating peptides, granulins, and epithelins, and is expressed in somatic as well as male germ cells. Mol Reprod Dev. 1993 Mar;34(3):233–43. doi: 10.1002/mrd.1080340302. [DOI] [PubMed] [Google Scholar]
- 4.Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000 Jul;48(7):999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- 5.Zanocco-Marani T, Bateman A, Romano G, Valentinis B, He ZH, Baserga R. Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res. 1999 Oct 15;59(20):5331–40. [PubMed] [Google Scholar]
- 6.Lu R, Serrero G. Inhibition of PC cell-derived growth factor (PCDGF, epithelin/granulin precursor) expression by antisense PCDGF cDNA transfection inhibits tumorigenicity of the human breast carcinoma cell line MDA-MB-468. Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):3993–8. doi: 10.1073/pnas.97.8.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng JQ, Guo FJ, Jiang BC, Zhang Y, Frenkel S, Wang DW, Tang W, Xie Y, Liu CJ. Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates Erk1/2 signaling and JunB transcription factor in chondrogenesis. FASEB J. 2010 Jun;24(6):1879–92. doi: 10.1096/fj.09-144659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sell C, Dumenil G, Deveaud C, Miura M, Coppola D, DeAngelis T, Rubin R, Efstratiadis A, Baserga R. Effect of a null mutation of the insulin-like growth factor I receptor gene on growth and transformation of mouse embryo fibroblasts. Mol Cell Biol. 1994 Jun;14(6):3604–12. doi: 10.1128/mcb.14.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Gulyas M, Hjerpe A. Mesothelial differentiation as reflected by differential gene expression. Am J Respir Cell Mol Biol. 2004 Apr;30(4):510–8. doi: 10.1165/rcmb.2003-0266OC. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki M, Nishiahara M. Granulin precursor gene: a sex steroid-inducible gene involved in sexual differentiation of the rat brain. Mol Genet Metab. 2002 Jan;75(1):31–7. doi: 10.1006/mgme.2001.3274. [DOI] [PubMed] [Google Scholar]
- 11.Barreda DR, Hanington PC, Walsh CK, Wong P, Belosevic M. Differentially expressed genes that encode potential markers of goldfish macrophage development in vitro. Dev Comp Immunol. 2004 Jun;28(7-8):727–46. doi: 10.1016/j.dci.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Justen HP, Grunewald E, Totzke G, Gouni-Berthold I, Sachinidis A, Wessinghage D, Vetter H, Schulze-Osthoff K, Ko Y. Differential gene expression in synovium of rheumatoid arthritis and osteoarthritis. Mol Cell Biol Res Commun. 2000 Mar;3(3):165–72. doi: 10.1006/mcbr.2000.0211. [DOI] [PubMed] [Google Scholar]
- 13.He Z, Ong CH, Halper J, Bateman A. Progranulin is a mediator of the wound response. Nat Med. 2003 Feb;9(2):225–9. doi: 10.1038/nm816. [DOI] [PubMed] [Google Scholar]
- 14.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004 Dec;22(4):233–41. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 15.Xu K, Zhang Y, Ilalov K, Carlson CS, Feng JQ, Di Cesare PE, Liu CJ. Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J Biol Chem. 2007 Apr 13;282(15):11347–55. doi: 10.1074/jbc.M608744200. [DOI] [PubMed] [Google Scholar]
- 16.Lacruz RS, Nanci A, Kurtz I, Wright JT, Paine ML. Regulation of pH During Amelogenesis. Calcif Tissue Int. 2010 Feb;86(2):91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CE, Wazen R, Hu Y, Zalzal SF, Nanci A, Simmer JP, Hu JC. Consequences for enamel development and mineralization resulting from loss of function of ameloblastin or enamelin. Eur J Oral Sci. 2009 Oct;117(5):485–97. doi: 10.1111/j.1600-0722.2009.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caton J, Luder HU, Zoupa M, Bradman M, Bluteau G, Tucker AS, Klein O, Mitsiadis TA. Enamel-free teeth: Tbx1 deletion affects amelogenesis in rodent incisors. Dev Biol. 2009 Apr 15;328(2):493–505. doi: 10.1016/j.ydbio.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takamori K, Hosokawa R, Xu X, Deng X, Bringas PJr, Chai Y. Epithelial fibroblast growth factor receptor 1 regulates enamel formation. J Dent Res. 2008 Mar;87(3):238–43. doi: 10.1177/154405910808700307. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi K, Nagata H, Horiguchi T, Abe K, Arie Wahyudi I, Baba Y, Harada H, Noma T. BMP2-induced gene profiling in dental epithelial cell line. J Med Invest. 2008 Aug;55(3-4):216–26. doi: 10.2152/jmi.55.216. [DOI] [PubMed] [Google Scholar]
- 21.Gibson CW, Yuan ZA, Li Y, Daly B, Suggs C, Aragon MA, Alawi F, Kulkarni AB, Wright JT. Transgenic mice that express normal and mutated amelogenins. J Dent Res. 2007 Apr;86(4):331–5. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- 22.Pispa J, Mustonen T, Mikkola ML, Kangas AT, Koppinen P, Lukinmaa PL, Jernvall J, Thesleff I. Tooth patterning and enamel formation can be manipulated by misexpression of TNF receptor Edar. Dev Dyn. 2004 Oct;231(2):432–40. doi: 10.1002/dvdy.20138. [DOI] [PubMed] [Google Scholar]
- 23.Paine ML, Zhu DH, Luo W, Snead ML. Overexpression of TRAP in the enamel matrix does not alter the enamel structural hierarchy. Cells Tissues Organs. 2004;176(1-3):7–16. doi: 10.1159/000075023. [DOI] [PubMed] [Google Scholar]
- 24.Tabata MJ, Fujii T, Liu JG, Ohmori T, Abe M, Wakisaka S, Iwamoto M, Kurisu K. Bone morphogenetic protein 4 is involved in cusp formation in molar tooth germ of mice. Eur J Oral Sci. 2002 Apr;110(2):114–20. doi: 10.1034/j.1600-0722.2002.11194.x. [DOI] [PubMed] [Google Scholar]
- 25.Simmer JP, Hu JC. Dental enamel formation and its impact on clinical dentistry. J Dent Educ. 2001 Sep;65(9):896–905. [PubMed] [Google Scholar]
- 26.Sui W, Xiao MZ, Hong YL. Expression of amelogenins in developing embryonic and neonatal rat teeth. Chin J Dent Res. 2000 May;3(1):51–4. [PubMed] [Google Scholar]
- 27.Coumoul X, Li W, Wang RH, Deng C. Inducible suppression of Fgfr2 and Survivin in ES cells using a combination of the RNA interference (RNAi) and the Cre-LoxP system. Nucleic Acids Res. 2004;32(10):e85. doi: 10.1093/nar/gnh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980 Dec;22(3):299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 29.Feng JQ, Huang H, Lu Y, Ye L, Xie Y, Tsutsui TW, Kunieda T, Castranio T, Scott G, Bonewald LB, Mishina Y. The Dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. J Dent Res. 2003 Oct;82(10):776–80. doi: 10.1177/154405910308201003. [DOI] [PubMed] [Google Scholar]
- 30.Feng JQ, Zhang J, Dallas SL, Lu Y, Chen S, Tan X, Owen M, Harris SE, MacDougall M. Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. J Bone Miner Res. 2002 Oct;17(10):1822–31. doi: 10.1359/jbmr.2002.17.10.1822. [DOI] [PubMed] [Google Scholar]
- 31.Priam F, Ronco V, Locker M, Bourd K, Bonnefoix M, Duchene T, Bitard J, Wurtz T, Kellermann O, Goldberg M, Poliard A. New cellular models for tracking the odontoblast phenotype. Arch Oral Biol. 2005 Feb;50(2):271–7. doi: 10.1016/j.archoralbio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Coumoul X, Shukla V, Li C, Wang RH, Deng CX. Conditional knockdown of Fgfr2 in mice using Cre-LoxP induced RNA interference. Nucleic Acids Res. 2005;33(11):e102. doi: 10.1093/nar/gni100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang B, Cao Z, Lu Y, Janik C, Lauziere S, Xie Y, Poliard A, Qin C, Ward LM, Feng JQ. DMP1 C-terminal mutant mice recapture the human ARHR tooth phenotype. J Bone Miner Res. 2010 Oct;25(10):2155–64. doi: 10.1002/jbmr.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med. 2003 Oct;81(10):600–12. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 35.Kessenbrock K, Frohlich L, Sixt M, Lammermann T, Pfister H, Bateman A, Belaaouaj A, Ring J, Ollert M, Fassler R, Jenne DE. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J Clin Invest. 2008 Jul;118(7):2438–47. doi: 10.1172/JCI34694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM, Lacomis L, Erdjument-Bromage H, Tempst P, Wright CD, Ding A. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002 Dec 13;111(6):867–78. doi: 10.1016/s0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 37.Liu CJ, Kong W, Ilalov K, Yu S, Xu K, Prazak L, Fajardo M, Sehgal B, Di Cesare PE. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 2006 May;20(7):988–90. doi: 10.1096/fj.05-3877fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Z, Ismail A, Kriazhev L, Sadvakassova G, Bateman A. Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res. 2002 Oct 1;62(19):5590–6. [PubMed] [Google Scholar]
- 39.Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, Yuan GH, Dong J, Gay I, MacDougall M. Runx2, osx, and dspp in tooth development. J Dent Res. 2009 Oct;88(10):904–9. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu JC, Simmer JP. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007 May;10(2):45–52. doi: 10.1111/j.1601-6343.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 41.MacDougall M. Refined mapping of the human dentin sialophosphoprotein (DSPP) gene within the critical dentinogenesis imperfecta type II and dentin dysplasia type II loci. Eur J Oral Sci. 1998 Jan;106(Suppl 1):227–33. doi: 10.1111/j.1600-0722.1998.tb02180.x. [DOI] [PubMed] [Google Scholar]
- 42.McKnight DA, Simmer JP, Hart PS, Hart TC, Fisher LW. Overlapping DSPP mutations cause dentin dysplasia and dentinogenesis imperfecta. J Dent Res. 2008 Dec;87(12):1108–11. doi: 10.1177/154405910808701217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michon F, Tummers M, Kyyronen M, Frilander MJ, Thesleff I. Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev Biol. 2010 Apr 15;340(2):355–68. doi: 10.1016/j.ydbio.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Sehic A, Peterkova R, Lesot H, Risnes S. Distribution and structure of the initial dental enamel formed in incisors of young wild-type and Tabby mice. Eur J Oral Sci. 2009 Dec;117(6):644–54. doi: 10.1111/j.1600-0722.2009.00676.x. [DOI] [PubMed] [Google Scholar]
- 45.Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. Proteomics and genetics of dental enamel. Cells Tissues Organs. 2005;181(3-4):219–31. doi: 10.1159/000091383. [DOI] [PubMed] [Google Scholar]