Abstract
This study focused on concatemer formation and integration pattern of transgenes in zebrafish embryos. A reporter plasmid based on enhanced green fluorescent protein (eGFP) driven by Cytomegalovirus (CMV) promoter, pCMV-pax6in-eGFP, was constructed to reflect transgene behavior in the host environment. After removal of the insertion fragment by double digestion with various combinations of restriction enzymes, linearized pCMV-pax6in-eGFP vectors were generated with different combinations of 5′-protruding, 3′-protruding, and blunt ends that were microinjected into zebrafish embryos. Repair of double-strand breaks (DSBs) was monitored by GFP expression following religation of the reporter gene. One-hundred-and-ninety-seven DNA fragments were amplified from GFP-positive embryos and sequenced to analyze the repair characteristics of different DSB end combinations. DSBs involving blunt and asymmetric protruding ends were repaired efficiently by direct ligation of blunt ends, ligation after blunting and fill-in, or removed by cutting. Repair of DSBs with symmetric 3′-3′ protrusions was less efficient and utilized template-directed repair. The results suggest that non-homologous end joining (NHEJ) was the principal mechanism of exogenous gene concatemer formation and integration of transgenes into the genome of transgenic zebrafish.
Keywords: Transgene, Concatemer, DSB (double strand breaks), NHEJ (non-homologous end joining), Zebrafish
Introduction
The mechanism by which exogenous genes are integrated into the host genome has been a major concern of transgene research. In an effort to develop effective methods for site-directed integration and enhance the efficiency of stable transgene integration, researchers have analyzed transgene flanking sequences to identify potential integration hotspots 1,2. However, studies on transgene integration in mammals have shown that integration appears to be a random process and, although sequences in integration sites have some common structural features, no so-called integration hotspots exist. Furthermore, transgenes are prone to forming concatemers prior to integration 3,4,5,6. The integrated foreign DNA appears mainly as random end-to-end concatemers 4,7,8,9,10,11,12. There are 3 types of concatemers: head-to-tail, head-to-head, and tail-to-tail. Head-to-tail concatemers can be stably integrated into the chromosomes 9. Head-to-head and tail-to-tail concatemers are observed when the concentration of linear foreign molecules in the host nucleus is high; these types of concatemers are unstable and can change into head-to-tail concatemers 9. In transgenic mammals, repair mechanism associated with double strand breaks (DSBs) may affect transgene behavior, as well 3,5. Studies of transgenic fish have also shown that exogenous genes had consistently integrated as head-to-tail concatemers into the host fish genome and the integration process exhibits characteristics of non-homologous recombination 1,2,12,13. Taken together, those studies suggest that DSB repair is related to the molecular mechanism of concatemer formation during exogenous gene integration in transgenic fish.
Cellular genomes are usually sensitive to ionizing radiation (IR), DNA mutagens, and other physical, chemical and biological factors that result in DNA damage in the form of DSBs 14,15. For example, V(D)J recombination during lymphocyte maturation in the immune system of higher organisms generates a large number of endogenous DSBs 16,17,18. As DSBs are considered to be the most severe form of DNA damage, they must be repaired immediately to prevent cell death 19,20,21.
Cells repair DSBs by two mechanisms: homologous recombination (HR) and non-homologous end joining (NHEJ) 22,23,24. HR is the principle mechanism for DSB repair in bacteria and lower eukaryotes, such as yeast 25,26, while NHEJ is the main mechanism used in higher eukaryotes 27. Although NHEJ is the predominant mechanism for DSB repair in mammals 28,29, it is an error-prone repair process 30,31,32,33,34. Since the NHEJ machinery directly ligates the ends of DSBs, and there is little or no homologous sequence, it frequently leads to the loss or insertion of base pairs 22,26.
The majority of research on NHEJ in zebrafish embryos has concentrated on gene expression patterns and functions of the NHEJ repair complex core components. It has been shown that injection of the Ku70 morpholino does not affect zebrafish embryogenesis, but exposure of Ku70 morpholino-injected embryos to low doses of ionizing radiation results in marked cell death throughout the developing brain, spinal cord and tail 35. In addition, Li et al. found that V(D)J recombination in the lymphocytes of zebrafish was different from that in mammals in that the signal ends were lost and the coding ends did not form circle-hairpin structures 36. Zhong found that early immunoglobulin gene rearrangement occurred during the maturation process of zebrafish eggs; however, there was no ligation of immunoglobulin gene coding ends, presumably because of the instability of NHEJ-associated components in zebrafish lymphocytes 37. These findings suggest that zebrafish have an NHEJ repair complex that is similar to that in mammals. The studies described above, however, did not systematically investigate the characteristics of NHEJ or the molecular mechanism of concatemer formation during exogenous gene integration.
In this report, we characterized the end ligation process and DSB repair via NHEJ in zebrafish embryos. To monitor DSB repair, we constructed a reporter plasmid with a CMV promoter and an enhanced green fluorescent protein (eGFP) coding sequence, having a linker sequence between them. The linker contained pax6in and SV40poly(A) to prevent the CMV promoter from driving eGFP expression. The reporter plasmid was digested with different pairs of restriction endonucleases to remove the linker and generate linearized vectors with non-homologous ends. Linearized vectors were then microinjected into zebrafish embryos, and the ability to ligate and repair different fragment ends was monitored by restoration of GFP expression. In addition, the sequences of 197 junction regions were determined in order to analyze their ability to repair different fragment ends. The results indicated that the NHEJ repair mechanism in zebrafish is similar to that in mammals in that repair occurs through direct ligation of DNA ends, rather than depending on multiple homologous sequences. Based on these results, we propose a two-step mechanism for the integration of exogenous genes via NHEJ repair in zebrafish. Specifically, after microinjection the NHEJ complex would first process DSBs between exogenous gene molecules and form concatemers. As the concentration of exogenous gene DSBs decreases, the NHEJ repair mechanism would begin to skew towards endogenous DSBs generated in the host genome during successive generations of cellular division. Since endogenous DSBs are distributed on various sites in the chromosomes, ligation between transgene DSBs and endogenous DSBs would result in random integration of exogenous genes. We presume that ligases, polymerases and endonucleases related to DNA repair are involved in this process.
Materials and Methods
Zebrafish and Microinjection
Wild type zebrafish (AB) were maintained according to the established guide for zebrafish care 38 in a glass aquarium with a circulating water system maintained at a constant temperature of 28℃. The ratio of illumination time to darkness time was 12hrs:12hrs. Fish handling and embryo generation were performed in accordance with IACUC regulations. Fifteen minutes after artificial fertilization, zebrafish embryos were microinjected with DNA at a concentration of 100 ng/μl.
Construction of pCMV-pax6in-eGFP plasmid
With consideration to the study of NHEJ repair mechanism in mammalian cells 28, our zebrafish NHEJ reporter plasmid, pCMV-pax6in-eGFP, was derived from the pEGFP-N1 vector (Clontech, Mountain View, CA, USA). The pEGFP-N1 vector backbone was PCR amplified using reverse primer P1 (5′-GAGCTCGAGAATTCACTA AACCAGCTCT-3′) with SacI, XhoI and EcoRI restriction sites and forward primer P2 (5′-AAGCTTGGTACCCACGTGGGATCC ACCGGT CGCCACC-3′) with HindIII, KpnI, PmaCI and BamHI restriction sites (Figure 1). Underlined text and strikethroughs correspond to the various restriction endonuclease target sequences. The amplified vector backbone was digested with SacI and HindIII (Fermentas, MBI, Shenzhen, China). The PCR amplified 1.2 kb pax6 fragment was digested with SacI and EcoRI, and an amplified SV40 poly(A) fragment was digested with EcoRI and HindIII. The three digested amplification products, vector, pax6 fragment, and SV40 poly(A) fragment were ligated and transformed into DH5α competent bacteria to obtain the NHEJ reporter plasmid pCMV-pax6in-eGFP (Figure 1).
Figure 1.
Features of the plasmid pCMV-pax6in-eGFP. The plasmid contains a CMV promoter and the enhanced green fluorescent protein (eGFP) coding sequence, separated by a linker sequence. The linker contains pax6in and SV40poly(A).
NHEJ reporter assay
The pCMV-pax6in-eGFP reporter plasmid was digested with seven different pairs of restriction endonucleases to produce linearized vectors with different DNA ends. The corresponding enzyme combinations were used to generate DSBs: 5′-5′(XhoI+HindIII and HindIII+EcoRI), 3′-3′(KpnI+SacI), 5′-3′(EcoRI+KpnI), 5′-B (HindIII + EcoRV), 3′-B(KpnI+EcoRV) and B-B(PmaCI+EcoRV), where 5′, 3′, and B denote digestions that result in 5′-protruding, 3′-protruding and blunt ends, respectively. After removal of the insertion fragment by restriction endonuclease digestion, linearized vectors were recovered and injected into artificially fertilized zebrafish embryos, and the fluorescence was monitored 10 hours after sample injection. Ligation of the linearized fragment ends between the CMV promoter and eGFP coding sequence was expected to result in GFP expression and a readout of a functional NHEJ repair mechanism (Figure 2).
Figure 2.
Schema of the NHEJ reporter assay. A: The pEGFP-N1 vector backbone was PCR amplified using reverse primer P1 and forward primer P2. B: pCMV-pax6in-eGFP plasmid in which the pax6 fragment and SV40poly(A) was ligated with the pEGFP-N1 vector backbone, in order to prevent the CMV promoter from driving eGFP expression. C: The insertion fragment was removed from the pCMV-pax6in-eGFP plasmid through double digestion, and the linearized plasmid was injected into zebrafish embryos or eggs. D: The linearized vector was rejoined by NHEJ in zebrafish embryos or eggs.
Each linearized vector was injected into zebrafish embryos separately; 10 hours later, twenty GFP-emitting embryos were collected for DNA extraction and sequencing. Each group of embryos were ground separately with a plastic rod, and 500 μL of DNA extraction solution [10 mM Tris·HCl (pH 8.0), 300 mM NaCl, 10 mM EDTA, 2.0% (w/v) SDS] was added. Samples were incubated for 3 hours at 65ºC to extract the genomic DNA. The fragment between the religated CMV promoter and eGFP was amplified by using the forward primer P3 (5′-AGAGCTGGTTTA GTGAA-3′) targeting the CMV promoter and the reverse primer P4 (5′-TGCCGTTCTTCTGCTTGTC-3′) targeting the eGFP coding sequence; the predicted amplified fragment length was 531bp. The amplified PCR fragment was ligated into pMD18-T vector (Takara, Shiga, Japan), and 30 clones from each group were selected for sequencing (Invitrogen, Carlsbad, CA, USA).
The mechanism by which exogenous transgenes are processed in zebrafish embryos may require expression of the zygotic gene that processes DSBs; alternatively, residual maternal substances in eggs may be utilized to repair exogenously introduced DSBs. To analyze the mechanism for processing exogenous genes in zebrafish embryos, unfertilized eggs were microinjected with linearized fragments digested with 5′-5′(XhoI+HindIII), 3′-3′(KpnI+SacI) and 5′-5′(HindIII+EcoRI). Samples were collected 15 minutes after microinjection, and DNA extraction solution was added. DNA extracted from 20 embryos was amplified with primers P3 and P4 to detect NHEJ reaction products. In addition, 20 unfertilized zebrafish eggs were placed into 1×PBS and triturated with a plastic grind rod. Homogenized samples were incubated with 100 ng of linearized vector digested with 5′-5′(XhoI+ HindIII), 3′-3′(KpnI+SacI) or 5′-5′(HindIII+EcoRI). After incubation for 15 minutes, DNA extraction solution was added to stop the reaction and extract DNA. PCR amplification with primers P3 and P4 was used to determine if ligation occurred between the CMV promoter and eGFP.
Results
NHEJ reporter plasmid
To monitor NHEJ DSB repair in zebrafish embryos, we constructed pCMV-pax6in-eGFP, in which pax6 cDNA and SV40 poly(A) terminator fragments were inserted between the CMV promoter and eGFP coding sequence to block eGFP expression. The use of SV40 poly(A) terminator was necessary for eliminating background fluorescence from the plasmid; use of a fragment without the SV40 poly(A) terminator did not completely eliminate the green fluorescence expression (data not shown). The pax6 fragment contained a number of restriction sites on each end to facilitate generation of restriction fragment ends with various combinations of 5′-protruding, 3′-protruding or blunt ends. After removal of the insertion fragment through double digestion, ligation of non-homologous ends via NHEJ DSB repair restored GFP expression from the reporter plasmid.
Microinjection of NHEJ reporter plasmid
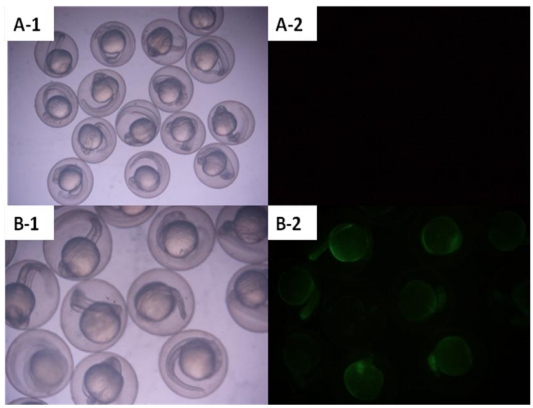
Zebrafish embryos that were microinjected with the intact pCMV-pax6in-eGFP plasmid, including pax6 fragment and SV40 poly(A) terminator, did not express GFP. However, embryos injected with pCMV-pax6in-eGFP after removal of the insertion fragment exhibited increased GFP fluorescence (Figure 3). DNA extracted from GFP expressing embryos was amplified with primers P3 and P4 and yielded the expected ~531 bp fragment which resulted from religation of the CMV promoter and eGFP (Figure 4). This result suggested that direct ligation occurred between DSBs in linearized fragments after injection into the zebrafish embryo. In addition, the amplified fragment length demonstrated that the ligation direction between transgenes occurred in a head-to-tail manner.
Figure 3.
Zebrafish embryos were microinjected with circular type plasmid pCMV-pax6in-eGFP and linearized plasmid which had the insertion fragment removed from between the CMV promoter and the eGFP reporter gene. A: Zebrafish embryos microinjected with circular type plasmid pCMV-pax6in-eGFP (no fluorescence was observed). B: Zebrafish embryos microinjected with linearized plasmid which had the insertion fragment removed from between the CMV promoter and the eGFP reporter gene in the pCMV-pax6in-eGFP plasmid. Re-expression of eGFP was observed as the embryo emitted fluorescence.
Figure 4.
Gel electrophoresis of the PCR products amplified from the genomes of six fluorescent zebrafish embryos which were microinjected with linearized pCMV-pax6in-eGFP plasmids. A band of 531 bp length was amplified from all the fluorescent zebrafish embryo genomes, which represented the expected product from a religation event between the CMV promoter and eGFP. Lanes 1-6: PCR products of the amplified beta-actin gene, used for genome quality control. Lanes 1'-6': PCR products of the junction sites between the CMV promoter and eGFP. -: negative control in which the template was the wild type zebrafish genome DNA. +: positive control in which the template was circular type pCMV-pax6in-eGFP plasmid. M: DNA molecular weight marker.
Characteristics of DSB repair in zebrafish embryos
The pCMV-pax6in-eGFP reporter plasmid was digested with seven different pairs of restriction enzymes to generate seven groups of linearized vectors with different forms of DSBs. Following microinjection into zebrafish embryos, DNA was extracted and sequenced, yielding 197 sequences that crossed the junction site of the CMV promoter and eGFP. We found that the majority of linearized vectors were directly ligated and formed head-to-tail junctions; only a minority of the ends resulted in recombination through as few as one or even no base pair homology.
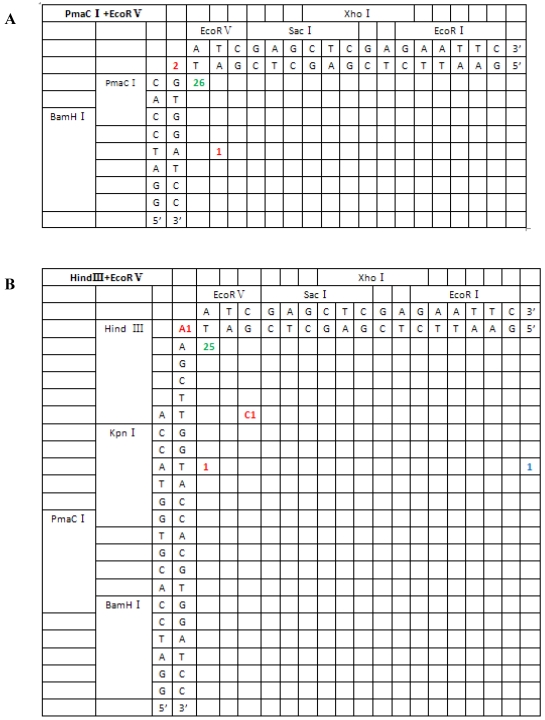
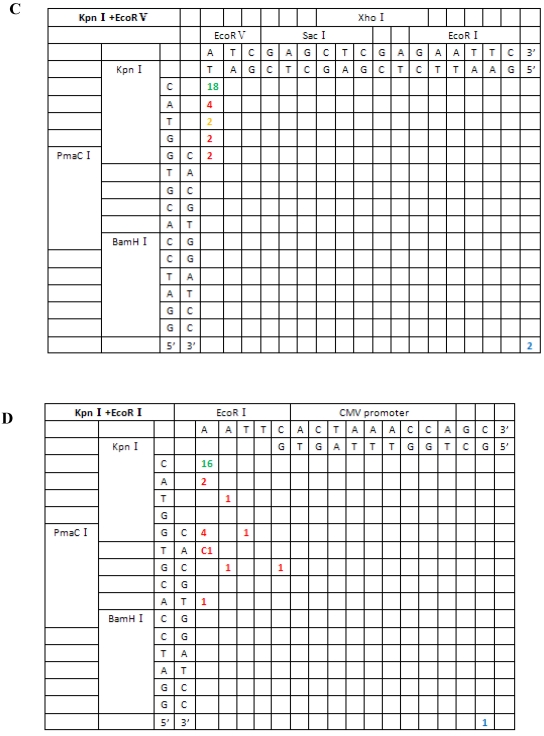
From the 197 PCR products, we were able to deduce the repair mechanism for different pairs of blunt and/or protruding ends. The characteristics of junction region formation were typical of the NHEJ repair mechanism. According to the ends formed, linearized vectors could be divided into groups with blunt ends, asymmetrical protruding ends and symmetrical protruding ends (Figures 5 and 6). For linearized vectors with blunt ends and asymmetrical ends, the primary repair mechanisms were direct blunt-end ligation, ligation after blunting of protruding ends or fill-in and removal post-ligation (shown with green numbers) (Figure 5). In most cases, ligation was homology-independent and the terminal sequences were kept intact, i.e. no additional bases were added or deleted on the basis of terminal sequences. For DSBs with two blunt ends, B-B(PmaCI+EcoRV), 89.5% of the ends were found to have been directly ligated, while linearized vectors with one blunt and one protruding 5′ end also exhibited 86% direct ligation. The amount of direct ligation for 3′-B(KpnI+EcoRV) and 5′-3′(EcoRI+KpnI) combinations was lower for both (60% and 53.3%), and in this two linearized vectors contained KpnI 3′-protruding ends; the proportions followed incorrect pairing between corresponding base pairs (shown with red numbers) were quite high, reaching 30% and 43.3%, respectively.
Figure 5.
Sequence analysis of the junction site between CMV and eGFP isolated from transgenic zebrafish embryos microinjected with blunt-end (PmaCI+EcoRV) and asymmetrical end (HindIII+EcoRV, KpnI+EcoRV, KpnI+EcoRI) linearized vectors. Nucleotide sequences of the ends of the indicated substrates are shown along the axes. DNA strands are illustrated in the 5′-3′ direction (from bottom to top and from left to right). Arabic numerals indicate the position of the junction, occurrence frequency of specific ligation types. Ligation types are shown with different colours:direct ligation of blunt-end and ligation after blunting of protruding ends, or fill-in and removal by excision post-ligation are shown with green numbers; correct base-pairings following repair are shown with orange numbers; incorrect base-pairings following repair attempt are shown with red numbers; blue numbers represent the repair of bases where more than 15 bps were lost following base repair. The nucleotide sequence of these junctions starts with the last protruding or extension nucleotide of the left enzyme and the first protruding or extension nucleotide of the upper enzyme. For example, the green number 18 shown in boldface in KpnI+EcoRV indicates the type of this junction site is direct ligation, and it appeared 18 times in a total of 30 sequenced DNA ends; the sequence of this junction site is 5′-GTACATG-3′. This denotation scheme is preserved in Figure 6.
Figure 6.
Sequence analysis of the junction site between CMV and eGFP isolated from transgenic zebrafish embryos microinjected with symmetrical end (HindIII+XhoI, HindIII+EcoRI, KpnI+SacI) linearized vectors.
For linearized vectors generated with symmetrical protruding ends, 5′-5′(XhoI+HindIII) and 5′-5′(EcoRI+HindIII) (Figure 6), 47.8% (11/23) and 30% (9/30) of the ends were repaired, respectively, by direct ligation of blunt ends following fill-in. In addition, repair after ligation involved base deletion in 17.4% (4/23) and 43.3% (13/30) of the cases, respectively. On the contrary, ligation after end blunting and fill-in or removal by excision repair post-ligation was not observed for linearized vectors with symmetrical 3′-protruding ends, 3′-3′(KpnI+SacI). The majority of ligation types for this type of vector was template-directed repair and depended on microhomology of terminal bases. Following pairing of complementary bases, extra bases were excised, and the ends were finally religated; 84.6% (22/26) were ligated with the correct base pairing (shown with orange numbers) compared to 15.4% (4/26) with incorrect base pairing. The above results indicated that 3′-protruding ends were quite active in ligation reactions and the repair proportion of correct base pairing in this process was high, suggesting that the invasion of free radical -OH at the 3′ end required highly homologous base pairs.
Based on the junction sites of head-to-tail-ligated junctions formed by the repair of seven different kinds of DSBs, we divided repair types into two classes: (I) terminal base homology-independent repair, including direct ligation of blunt ends, ligation after blunting of protruding ends and fill-in or removal by excision post-ligation of protruding ends; and (II) terminal base homology-dependent repair, which included ligation following template-directed repair. The first repair type acted on all DSB combinations except for the 3′-3′ type, which were primarily repaired by the second mechanism. In addition, template-directed repair could be further divided into two groups, depending on whether bases were correctly or incorrectly paired. The repair mechanism for each type of DSB combination is summarized in Table 1.
Table 1.
Ligation types of the seven groups of different DNA end joining
| DSBs type | NHEJ repair type and proportion | ||
|---|---|---|---|
| DSBs types generated by pairs of restriction endonucleases | Direct ligation of blunt ends, ligation after blunting of protruding ends and fill-in or cut-off after ligation | Template-directed repair (correct base pairing) | Others (incorrect base pairing and base loss of more than 15 bp) |
| XhoI+HindIII(5′-5′) | 11/23 (ligation after fill-in) +4/23 (excision after ligation){65.0%} | 4/23 (17.5%) | 4/23 (17.5%) |
| EcoRI+HindIII(5′-5′) | 9/30 (ligation after fill-in)+(6/30+7/30)(excision after ligation) {73.3%} | 2/30 (6.7%) | 6/30 (20.0%) |
| HindIII+EcoRV(5′-B′) | 25/29 (ligation after fill-in) {86.0%} | 0 | 4/29 (14.0%) |
| EcoRV+PmaC I(B′-B′) | 26/29 (direct ligation) {89.5%} | 0 | 3/29 (10.5%) |
| EcoRV+KpnI (B′-3′) | 18/30 (fill-in after ligation) {60.0%} | 2/30 (6.7%) | 10/30 (33.3%) |
| KpnI+SacI(3′-3′) | 0 | 22/26 (84.6%) | 4/26 (15.4%) |
| KpnI+EcoRI(3′-5′) | 16/30 (fill-in after ligation) {53.3%} | 0 | 14/30 (46.7%) |
NHEJ complex pre-exist in zebrafish unfertilized eggs
To test if the machinery for NHEJ was present in unfertilized eggs, zebrafish eggs were injected with linearized vectors, or homogenates of unfertilized eggs were incubated with linearized vectors. The DNA from both cases was extracted and amplified with primers P3 and P4. In both cases, the amplified PCR product corresponded to the religation between the CMV promoter and eGFP (Figure 7). This result suggested that the NHEJ machinery for repairing DSBs was present in the mature eggs and was not a result of embryogenesis.
Figure 7.
NHEJ effects in unfertilized zebrafish eggs. 1, 4, 7 junction site PCR product amplified from the genomes of fluorescent embryos microinjected with linearized DNA. 2, 5, 8 junction site PCR product amplified from homogenates of unfertilized eggs incubated with linearized DNA. 3, 6, 9 linearized vector used as the PCR template. 1-3: XhoI+HindIII linearized DNA; 4-6: KpnI +SacI linearized DNA; 7-9: HindIII+EcoRI linearized DNA. M: DNA molecular weight marker. DNA, 10: template was the wild type zebrafish genome. +: template was the pCMV-pax6in-eGFP plasmid. A band of 531 bp length was amplified from all the fluorescent zebrafish embryo genomes, which was the expected product for the event of religation between the CMV promoter and eGFP.
Discussion
In our previously published studies on the F4 hGH-transgenic common carp, the integration pattern and host sequences adjacent to the inserted pMThGH-transgene were extensively studied by using methods of plasmid rescue and Southern analysis. We found that the transgenes had consistently integrated into the host genome with concatemers 1-2. Many other researchers have previously reported that linearized exogenous genes form concatemers prior to integration into the host genome when used in microinjection 3-8. Many studies have explained the mechanism of transgene integration, but the mechanisms of concatemer formation have not yet been clearly elucidated. Studies of transgenic fish have shown that the integration process of exogenous genes shows characteristics of non-homologous recombination 1,2,12,13.
This study analyzed the sequence characteristics of 197 PCR products isolated from transgenic zebrafish embryos, all of the sequence in the junction region examined was the product of direct ligation, a typical characteristic of NHEJ repair. This indicated that NHEJ plays an important role in concatemer formation of DSB repair in zebrafish. In contrast to repair mechanisms in mammals, the efficiency of single-strand ligation was quite high. These results suggest that although the NHEJ repair mechanism in zebrafish appears to be similar to that in higher eukaryotes, it also retains some distinct characteristics.
Linearized vectors with double blunt ends showed the highest frequency of ligation without base pair insertions or deletions, and appeared to be the optimal substrate for NHEJ repair.
The repair ratio of substrates containing 3′-protruding ends, especially those with 3′-3′, was sufficiently less than the corresponding ligation of B-B and 5′-B. The results suggested that the presence of a 3′ end may reduce the efficiency of the direct ligation reaction.
It is technically challenging to treat 3′-protruding ends with polymerase, but 5′-protruding ends are receptive to polymerase activity to fill-in overhangs. This may be the reason why 3′- and 5′-protruding ends have different effects on the DSB repair efficiency 39. On the other hand, the free 5′-PO4 group from the protruding end base appeared to be more prone to attack and subsequent direct ligation with the vicinal 3′-OH group; but, when the 5′-PO4 group was absent, the 3′-OH group preferentially served as the primer for polymerase. Since direct ligation of DSBs is the main target of the NHEJ repair mechanism, our results support the hypothesis that NHEJ is the principal mechanism for exogenous gene concatemer formation and integration of transgenes into host genome in zebrafish.
The correction rate for NHEJ repair of asymmetric protruding ends in zebrafish is quite high, and the proportions of base loss and insertion are quite small. This feature is consistent with the results for NHEJ repair in Kluyveromyces lactis, as reported by Kegel et al. 22. However, the repair efficiencies without nucleotide deletion and insertion in the three combinations we examined, 5′-5′(HindIII+EcoRI and XhoI+HindIII) and 3′-3′(KpnI+SacI), was significantly less when compared with that of asymmetric protruding end and blunt-end combinations, suggesting that when polarity of protruding ends is the same, the ligation reaction may be hindered and result in base loss or insertion through other NHEJ processes.
Although 5′-5′ protruding ends displayed no ligation effects after end fill-in or cleavage in mammalian cells 3, our study found that the efficiencies of ligation after protruding end cleavage were higher than 30%. In addition, the proportion of fill-in and cleavage events following joining of 5′-5′ protruding ends was significantly smaller in zebrafish, as compared to that in mammals.
Researchers have hypothesized that significant differences exist between the NHEJ complexes of zebrafish and mammals 35,36,37. Our results indicated that there was, at least, a small divergence among the NHEJ repair pathways of zebrafish and mammals.
The proper repair events we observed for linearized vectors injected into unfertilized eggs or incubated with homogenates of unfertilized eggs indicated that the NHEJ complex is present and functional in zebrafish eggs, and it can mediate the ligation of exogenous DSB ends. In addition, we predict that this NHEJ complex also plays a principal role in repairing the large quantity of endogenous DSBs generated by the rapid cleavage rate that occurs in the early stage of zebrafish embryogenesis.
Similar conclusions have been obtained from concatemer type and transgene integration patterns in three-year-old P0 common carp tissues expressing an antisense sGnRH transgene 40. In those studies, we found that concatemers were the primary form of transgenes in the genome, and the ligation type mediating concatemer formation was similar to what we found in zebrafish here (data not shown).
Taken together, the above data demonstrated that fish utilize a DSB repair mechanism that is similar to that used by mammals. Since DSB-directed ligation appears to be the main form of NHEJ repair mechanism, our results indicate that NHEJ is the principal mechanism for exogenous gene concatemer formation and integration of transgenes into the host genome of zebrafish. We propose the following two-step model to explain these mechanisms.
The NHEJ repair components functioning in zebrafish embryos could repair DSBs with high efficiency, even in vitro. The extremely high concentration of exogenous genes microinjected into zebrafish embryos would result in large accumulations of DSBs. Following microinjection, the NHEJ complex would first process DSBs between exogenous gene molecules and form concatemers. As the concentration of exogenous gene DSBs decreases, the NHEJ repair mechanism would begin to skew towards endogenous DSBs generated in the host genome during successive generations of cellular division. Since endogenous DSBs are distributed on various sites in the chromosomes, ligation between transgene DSBs and endogenous DSBs would result in random integration of exogenous genes.
Foreign genes introduced by microinjection inevitably form concatemers and randomly integrate into the host chromosomes. Many studies have shown that the transgenes are efficiently expressed, only with fewer integrated copy numbers of the transgene and allowing for their stable transmission in the germ-line of fish. Our results suggest that it is necessary to modify the end of a transgene before microinjection, in order to prevent concatemer formation and increase the integration efficiency and expression level of the transgene.
Acknowledgments
We thank Mrs. Shangpin Chen for assistant in microinjection during the study. This work was supported by the Development Plan of the state Key Fundamental Research of China (grant number 2007CB109206), National Natural Science Foundation (grant numbers 30930069, 30430540) and the '863' High Technology Project of China (grant number 2006AA10Z141).
References
- 1.Zeng ZQ, Zhu ZY. The molecular polymorphism of transgenes in F4 generation red carp transfected with pMThGH gene. Chinese Sci Bulletin. 2000;45:1957–1962. [Google Scholar]
- 2.Wu B, Sun YH, Wang Y W, Wang YP, Zhu ZY. Characterization of transgene integration pattern in F4 hGH-transgenic common carp (cyprinus carpio) Cell Res. 2005;15:447–454. doi: 10.1038/sj.cr.7290313. [DOI] [PubMed] [Google Scholar]
- 3.Roth DB, Wilson JH. Non-homologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada T, Sasaki H, Seki R, Sakaki Y.Mechanism of chromosomal integration of transgenes in microinjected mouse eggs. sequence analysis of genome-transgene and transgene-transgene junctions at two loci. Gene. 1993;128:197–202. doi: 10.1016/0378-1119(93)90563-i. [DOI] [PubMed] [Google Scholar]
- 5.Chan AWS, Homan EJ, Ballou LU, Burns JC, Bremel R. Transgenic cattle produced by reverse-transcribed gene transfer in oocyte. Proc Nat Acad Sci. 1998;95:14028–14033. doi: 10.1073/pnas.95.24.14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Nat Acad Sci. 2004;101:11135–11140. doi: 10.1073/pnas.0403925101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houdebine LM, Chourrout D. Transgenesis in fish. Experientia. 1991;47(9):891–897. doi: 10.1007/BF01929879. [DOI] [PubMed] [Google Scholar]
- 8.Bishop JO, Smith P. Mechanism of chromosomal integration of microinjected DNA. Mol Biol Med. 1989;6(4):283–298. [PubMed] [Google Scholar]
- 9.Folger KR, Wong EA, Wahl G, Capecchi MR. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Molecular and cellular biology. 1982;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellaire G, Yan J, Little KC, Drouin R, Chartrand P. Evidence that extrachromosomal double-strand break repair can be coupled to the repair of chromosomal double-strand breaks in mammalian cells. Chromosoma. 2002;111(5):304–312. doi: 10.1007/s00412-002-0212-6. [DOI] [PubMed] [Google Scholar]
- 11.Roth DB, Wilson JH. Relative rates of homologous and nonhomologous recombination in transfected DNA. PNAS. 1985;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penman DJ, Iyengar A, Beeching AJ, Rahman A, Sulaiman Z, Maclean N. Patterns of transgene inheritance in rainbow trout (oncorhynchus mykiss) Mol Reprod and Dev. 1991;30:201–206. doi: 10.1002/mrd.1080300306. [DOI] [PubMed] [Google Scholar]
- 13.Hsiao CD, Hsieh FJ, Tsai HJ. Enhanced expression and stable transmission of transgenes flanked by inverted terminal repeats from adeno-associated virus in zebrafish. Dev Dyn. 2001;220:323–36. doi: 10.1002/dvdy.1113. [DOI] [PubMed] [Google Scholar]
- 14.Jillian LY, O'neil NJ, Rose AM. Homologous recombination is required for genome stability in the absence of DOG-1in Caenorhabditis elegans. Genetics. 2006;173:697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siim P, Burkert JS, Martin J. et al. Alternative Induction of Meiotic Recombination From Single-Base Lesions of DNA deaminases. Genetics. 2009;182:41–54. doi: 10.1534/genetics.109.101683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber RM. Immunoglobulin diversity: rearranging by cutting and repairing. Curr Biol. 1996;6:134–136. doi: 10.1016/s0960-9822(02)00443-8. [DOI] [PubMed] [Google Scholar]
- 17.Vangent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 18.Ramsden DA, Van Gent DC, Gellert M. Specificity in V(D)J recombination: new lessons from biochemistry and genetics. Curr Opin Immunol. 1997;8:114–120. doi: 10.1016/s0952-7915(97)80167-7. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 20.Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a Dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623–1634. doi: 10.1105/tpc.11.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenichiro M, Shinohara A, Shinohara M. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes non-homologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics. 2008;179:213–25. doi: 10.1534/genetics.107.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kegel A, Martinez P, Carter SD, Åström SU. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 2006;34:1633–1645. doi: 10.1093/nar/gkl064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin L, Wen TJ, Schnable PS. Role of RAD51 in the repair of MuDR-induced double-strand breaks in Maize (zea mays L.) Genetics. 2007;178:57–66. doi: 10.1534/genetics.107.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansour WY, Schumacher S, Rosskopf R. et al. Hierarchy of non-homologous end joining, single-strand annealing and gene converse at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastwa E, Błasiak J. Non-homologous DNA end joining. Acta Biochimica Polonica. 2003;50:891–908. [PubMed] [Google Scholar]
- 26.Xin Y, Abram G. Ku-dependent and Ku-independent end joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanaar R, Hoeijmakers J, Van Gent DC. Molecular mechanisms of DNA double-strand break repair. Trends in Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 28.Liang F, Han M, Romanienko MP, Jasin M. Homology-directed repair is major double-strand break repair pathway in mammalian cells. Proc Nat Acad Sci. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mia RL, Meier B, Lee TW, Hall J, Ahmed S. End joining at Caenorhabditis elegans telomeres. Genetics. 2008;180:741–754. doi: 10.1534/genetics.108.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees-Miller SP, Godbout R, Chan D. et al. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;5201:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 31.Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Nat Acad Sci. 1996;93:13949–54. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 33.Jason AS, Waldman BC, Waldman AS. A Role for DNA mismatch repair protein Msh2 in error-prone double-strand-break repair in mammalian chromosomes. Genetics. 2005;170:355–363. doi: 10.1534/genetics.104.039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev. of Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 35.Catherine L, Bladen WKL, Dynan WS, Kozlowski DJ. DNA damage response and Ku80 function in the vertebrate embryo. Nucleic Acids Res. 2005;9:3002–3010. doi: 10.1093/nar/gki613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Chang Y. V(D)J recombination in zebrafish: Normal joining products with accumulation of unresolved coding ends and deleted signal ends. Mol Immunol. 2007;44:1793–1802. doi: 10.1016/j.molimm.2006.07.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong H, Li Z, Lin S, Chang Y. Initiation of V(D)J recombination in zebrafish (danio rerio) ovaries. Mol Immunol. 2007;44:1784–1792. doi: 10.1016/j.molimm.2006.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westerfield M. The zebrafish book:a guide for the laboratory use of zebrafish danio (brachydanio) rerio. Eugene: University of Oregon Press; 2000. [Google Scholar]
- 39.Kornberg A. DNA replication. San Francisco: WH Freeman & Co; 1980. [Google Scholar]
- 40.Hu W, Li SF, Tang B, Wang YP, Lin HR, Liu XC, Zou J, Zhu ZY. Antisense for gonadotropin-releasing hormone reduces gonadotropin synthesis and gonadal development in transgenic common carp (cyprinus carpio) Aquaculture. 2007;271:498–506. [Google Scholar]