Traf7, a MyoD1 transcriptional target, regulates nuclear factor-κB activity during myogenesis
Here, a novel mechanism is presented of how the E3 ligaseTraf7 couples MyoD1 function to NF-kB activity, regulating the balance between cell cycle progression and differentiation during myogenesis.
Keywords: myogenesis, MyoD1, Traf7
Abstract
We have identified the E3 ligase Traf7 as a direct MyoD1 target and show that cell cycle exit—an early event in muscle differentiation—is linked to decreased Traf7 expression. Depletion of Traf7 accelerates myogenesis, in part through downregulation of nuclear factor-κB (NF-κB) activity. We used a proteomic screen to identify NEMO, the NF-κB essential modulator, as a Traf7-interacting protein. Finally, we show that ubiquitylation of NF-κB essential modulator is regulated exclusively by Traf7 activity in myoblasts. Our results suggest a new mechanism by which MyoD1 function is coupled to NF-κB activity through Traf7, regulating the balance between cell cycle progression and differentiation during myogenesis.
Introduction
Myogenesis is governed by myogenic regulatory factors (MRFs), a group of transcription factors including MyoD1, Myf5, myogenin and Mrf4. Ectopic expression of a single MRF (such as MyoD1) is sufficient to force nonmuscle cells to complete the myogenic programme, demonstrating its central function in myogenesis (Tapscott et al, 1988). It is known that the cyclin-dependent kinase inhibitor p21 is induced after enforced MyoD1 expression (Halevy et al, 1995), but all the connections between MyoD1 activity and cell cycle exit—an obligatory step before differentiation—have not been elucidated. Furthermore, although MyoD1 is abundantly expressed in cycling myoblasts, its specific function in cell proliferation has not been thoroughly explored. Nuclear factor-κB (NF-κB) transcription factor has a function in myogenesis but, this function is enigmatic, as the protein has been shown to both promote and antagonize differentiation (Guttridge et al, 1999; Wang et al, 2007).
In this study, we show that MyoD1 activates expression of Traf7—a relatively unexplored member of the tumour necrosis factor receptor-associated factor family—which in turn enhances ubiquitylation of NF-κB essential modulator (NEMO; inhibitor of κB kinase (IKKγ)), a component of the IKK signalling complex important for NF-κB pathway activation. We show for the first time that NEMO can be ubiquitylated in a Traf7-dependent manner in growing myoblasts. This promotes the activation of the NF-κB pathway, enhancing cyclin D expression and cell proliferation. The ablation of Traf7 abrogates NEMO ubiquitylation and provokes premature cell cycle exit and myogenic differentiation. Thus, we have identified a new mechanism by which MyoD1 enhances expression of Traf7, leading to the ubiquitylation of a key component of the NF-κB pathway.
Results And Discussion
MyoD1 targets involved in ubiquitin-associated processes
Cellular commitment to myogenesis begins with the decision to exit the cell cycle and is followed by the activation of a temporally regulated transcriptional programme directed by MRFs. However, the mechanism by which specific genes bound by factors such as MyoD1, transcriptionally regulate the cell cycle during myogenesis remains largely unknown. Indeed, while MyoD1 has been classically linked to myogenic differentiation, MyoD1 protein expression is robust in myoblasts (Mal & Harter, 2003). To address the role of MyoD1 in growing cells, we conducted genome-wide MyoD1 factor-binding analyses in C2C12 myoblasts to identify new targets with potential functions in proliferating cells.
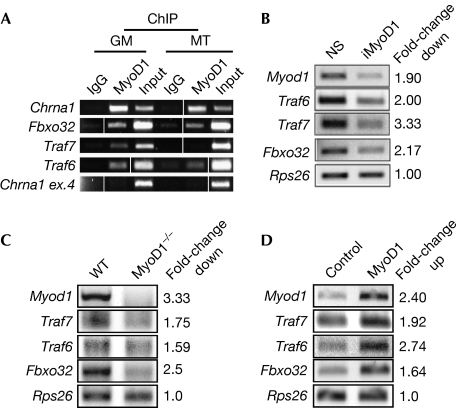
We performed chromatin immunoprecipitation (ChIP)-on-chip analyses using proximal promoter microarrays, thus extending our previous analysis (Blais et al, 2005). Several previously reported targets were identified as well as novel targets, including a cohort of E3 ubiquitin-protein ligases (supplementary Table S1 online). Two of these genes, Fbxo32 and Trim16, were shown to be MyoD1 transcriptional targets in mouse embryonic fibroblasts. The recruitment of MyoD1 to the promoters of selected targets is shown in supplementary Fig S1 online. Furthermore, our data show that Fbxo32, Traf6 and Traf7, which are all E3 ligases, are direct targets of MyoD1 in growing myoblasts (Fig 1A). Thus, our data suggest for the first time, to our knowledge, a direct transcriptional link between the crucial myogenic regulatory protein MyoD1 and a cohort of ubiquitylation-associated genes.
Figure 1.
MyoD1 regulates ubiquitin-related genes. (A) ChIP validation for MyoD1 binding to selected targets in C2C12 growing myoblasts (GM) and myotubes (MT). Input is 0.5% of total starting material. Chrna1, positive control; Chrna1 exon 4, negative control. Vertical lines indicate that intervening lanes were removed. (B) RT–PCR analysis after MyoD1 RNAi (iMyoD1). NS, nonspecific control. (C) RT–PCR analysis in wild-type (WT) and MyoD1−/− primary myoblasts. (D) RT–PCR analysis after MyoD1 overexpression in growing C2C12 cells. Rps26 is a nonspecific input and loading control. Fold-change in mRNA levels (B–D) determined by densitometric quantification using Image J software. ChIP, chromatin immunoprecipitation; mRNA, messenger RNA; NS, nonspecific; RNAi, RNA interference; RT–PCR, reverse transcriptase PCR; WT, wild type.
We examined the functional impact of MyoD1 binding by analysing changes in the expression of three E3 ligases identified in our screen—Traf6, Traf7 and Fbxo32—after RNA interference-mediated depletion of MyoD1 in growing C2C12 myoblasts. The expression of each of these genes decreased by at least twofold after MyoD1 depletion (Fig 1B). To examine whether MyoD1 regulates these genes in primary cells, we performed analogous studies using myoblasts isolated from wild-type and MyoD1−/− mice and observed that expression of each of these targets was similarly reduced (Fig 1C). We also measured transcription of these targets after ectopic expression of MyoD1 in C2C12 myoblasts and observed that MyoD1 significantly enhanced their expression (Fig 1D). Together, our studies demonstrate that MyoD1 activates the transcription of several E3 ligases in growing myoblasts.
A distinct function for Traf7 in differentiation
To determine how transcription of these target genes correlated with MyoD1 binding and to understand their functions in myogenesis, we examined their steady-state expression levels during differentiation (Fig 2A). We compared these with those of Fbxo21 and Trim63—these are also E3 ligases that are expressed in C2C12 cells, but their proximal promoters are not bound by MyoD1. As expected, expression of Traf6 and Fbxo32 increased during differentiation (Zhao et al, 2005). By contrast, expression of Traf7 steadily declined from the mid-point of the differentiation programme at t=48 (T48). Furthermore, MyoD1 recruitment to the Traf7 promoter was not observed in myotubes (Fig 1A), and additional ChIP analyses indicated robust loading of RNA polymerase II and acetylation of histone H3 lysine 9—markers of active transcription—in growing cells and almost complete absence of these markers in differentiated cells (data not shown).
Figure 2.
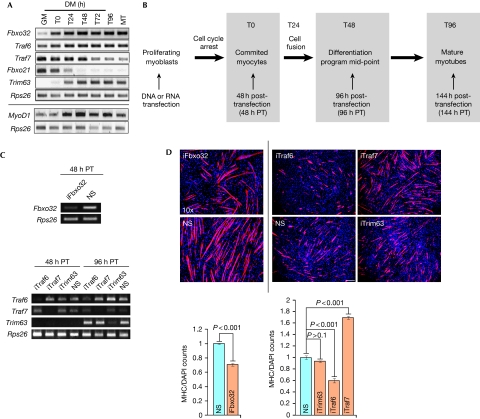
A distinct function for Traf7 in differentiation. (A) RT–PCR analysis of indicated E3 ligases (top) and MyoD1 (bottom). Rps26, input and loading control. GM and MT indicate growing myoblasts and myotubes, respectively. Cells are confluent at T0. T24–T96 indicate time (hours) in DM. (B) Diagram of experimental scheme. Transfections were performed in growing myoblasts at <50–60% confluence. (C) RT–PCR analysis of Fbxo32 (T0) and Traf6, Traf7 and Trim63 depletion at T0 (48 h PT) or T48 (96 h post-transfection). (D) Top panels show immunofluorescence on myoblasts depleted of Fbxo32 (left) or Traf6, Traf7 or Trim63 (right, independent experiment) and differentiated for 2 days. Scale bar, 40 μm, magnification × 10. Trim63, control; RNAi performed with pool of four siRNAs. Cells were stained with MHC antibodies (red) and DAPI (blue). Quantitation of data in top panel is also shown; MHC/DAP1 signal for each siRNA was normalized against NS. Error bars indicate s.e.m. values; P-values shown for n=5 independent experiments. DAPI, 4′,6-diamidino-2-phenylindole; DM, differentiation medium; MHC, myosin heavy chain; NS, nonspecific; PT, post-transfection; RNAi, RNA interference; RT–PCR, reverse transcriptase PCR.
To gain insight into the functions of these MyoD1 target genes during myogenesis, we examined the impact of acute gene silencing on myotube formation (Fig 2B). Suppression was robust 48 h after transfection (T0) and persisted through the mid-point of the differentiation programme (T48; Fig 2C). Cell densities were comparable for all small interfering RNA (siRNA)-transfected populations, ruling out growth defects and toxicity (Fig 2D). Myosin heavy chain (MHC) is principally expressed late in myogenesis and not in cycling myoblasts. We quantified entry into the myogenic programme by determining the percentage of MHC-positive cells at 48 h after induction of differentiation (T48) and by examining cellular fusion and formation of multinucleated myotubes. Depletion of Fbxo32 resulted in a reduction in the number of myotubes compared with controls (Figs 2D). Similarly, Traf6 depletion largely abrogated differentiation (Fig 2D). By this study, we observed 50% fewer myotubes that were, on average, 35% shorter than controls, suggesting that Traf6 is essential during the execution of the myogenic programme. By contrast, myogenesis was greatly accelerated after acute depletion of Traf7 (Fig 2D). Furthermore, we observed a significant increase in the number of longer, multinucleated myotubes compared with control cells when we transfected individual siRNAs targeting Traf7 or siRNA pools (Fig 2D; supplementary Fig S2A online). Depletion of other E3 ligases (Trim63 or Fbxo21) had no effect on differentiation (Fig 2D; data not shown), suggesting that Traf7 has a specific function in muscle differentiation. To our knowledge, our data provide the first evidence that Traf6 and Traf7 operate through distinct rather than collaborative pathways during myogenesis (Ha et al, 2009); Traf6 promotes differentiation, whereas Traf7 opposes it.
Traf7 depletion promotes precocious differentiation
The characterization of Traf7 has been limited to non-muscle cells (Ha et al, 2009). Therefore, we focused on the function of Traf7 in muscle differentiation. First, we quantified the number of myotubes after Traf7 depletion during differentiation and observed 33% more myotubes at early differentiation (T24), compared with controls, and this effect was sustained through the mid-point of the myogenic programme (T48). Furthermore, the average myotube length under the conditions of Traf7 depletion was almost three times that observed in the control (supplementary Fig S3A online). However, by the end of the differentiation time course (T96; supplementary Fig S3A online), there was no difference between treated and untreated cells, suggesting that Traf7 could function within a narrow time window of myogenesis.
Next, we tested the function of Traf7 during cell cycle exit and differentiation. Traf7-depleted myoblasts were kept in growth medium for 24 h beyond confluence. Myoblasts cultured under these conditions can commit to the myogenic programme. This approach enabled us to separate the effect of Traf7 deficiency from that of mitogen withdrawal, as we observed significantly more MHC-positive myotubes than controls (T24-growth medium; supplementary Fig S3A online), strongly suggesting that Traf7 depletion alone can trigger premature differentiation. Finally, to eliminate the contribution of density-dependent cell cycle exit, we depleted Traf7 from a sub-confluent population of cells and subsequently removed mitogens. Remarkably, there was a sixfold increase in the number of cells expressing MHC when Traf7 was depleted, indicating that cells had exited the cell cycle and commenced differentiation (supplementary Fig S3B online).
Conversely, ectopic expression of Traf7 substantially delayed differentiation (assessed at the differentiation mid-point, T48), as shown by shorter and fewer myotubes, and cells expressing the highest levels of Traf7 exhibited no MHC staining (supplementary Fig S3C online). We note that no apoptosis was observed on Traf7 overexpression (supplementary Fig S4 online). By contrast, expression of a Traf7 mutant lacking the RING, Really Interesting New Gene, domain had no effect on differentiation, strongly suggesting that ubiquitylation of a Traf7 target blocks differentiation (supplementary Fig S5A online). Together, these data support a new function for Traf7 whereby a reduction in Traf7 levels below a crucial threshold can trigger differentiation.
Traf7 depletion reduces cyclin D1 expression
The enhanced differentiation phenotype observed on depleting Traf7 could stem from premature cell cycle exit or myoblast fusion. We observed that several genes required for myoblast fusion (Adam12, Boc and Cdon) were not upregulated in Traf7-depleted cells compared with control cells. By contrast, expression of several cell-cycle-associated genes in confluent myoblasts was substantially reduced when Traf7 was depleted (supplementary Fig S6A online), suggesting that this E3 ligase directly or indirectly controls cell cycle exit. Reduced expression of cell cycle genes could represent a secondary effect of cell cycle deregulation resulting from a combination of Traf7 depletion and confluence arrest. To determine whether there was a direct effect on expression, we repeated our experiments using growing myoblasts. Interestingly, Traf7 depletion significantly affected the expression of cyclin D1 (Ccnd1), although it had no impact on the expression of additional cell cycle genes (supplementary Fig S6B online). Cells lacking Traf7 also showed reduced levels of 5-bromo-2-deoxyuridine incorporation, further suggesting that premature cell cycle exit had occurred (supplementary Fig S6C online).
Traf7 regulates cell cycle exit through the NF-κB pathway
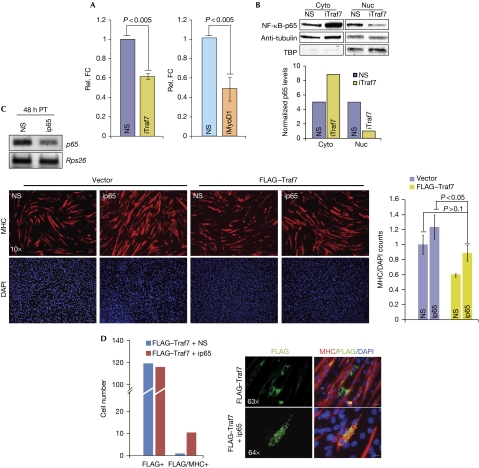
Next, we investigated how cyclin D1 expression might relate to the premature differentiation provoked by Traf7 depletion. Downregulation of NF-κB transcriptional activity in C2C12 cells precedes the induction of myogenin and the initiation of myoblast fusion (Guttridge et al, 1999). Furthermore, NF-κB binding to the Ccnd1 promoter is reduced during myogenesis, explaining in part how NF-κB (p50/p65) functions to promote myoblast proliferation and how its loss could accelerate differentiation (Guttridge et al, 1999; Bakkar et al, 2008). As our data strongly suggested that Traf7 regulates Ccnd1 transcription, we reasoned that Traf7 depletion might lead to reduced NF-κB activity, thereby diminishing transcription from the Ccnd1 locus. Indeed, luciferase reporter assays revealed a significant reduction in NF-κB transcriptional activity in growing myoblasts after Traf7 depletion (Fig 3A). Depletion of MyoD1—which led to a strong reduction in Traf7 expression—had a similar effect (Fig 3A). Interestingly, the majority of p65 remained in the cytoplasm after Traf7 depletion (Fig 3B), suggesting that Traf7 promotes NF-κB activity through regulation of the p65 subunit of NF-κB. Indeed, p65-deficient and RelA/p65 null myoblasts undergo accelerated differentiation (Bakkar et al, 2008). Accordingly, when we ablated p65 in growing myoblasts, we observed a premature differentiation phenotype similar to that of Traf7 depletion (Fig 3C). This result led us to test whether p65 depletion could antagonize the suppression of differentiation induced by ectopic Traf7 expression. We observed that p65-depletion rescued the phenotype associated with Traf7 overexpression, suggesting that Traf7 and p65 participate in a common pathway (Fig 3C). Furthermore, although cells that ectopically express Traf7 show little MHC staining, ablation of p65 forced a subset of these cells to initiate the myogenic programme (Fig 3D).
Figure 3.
A pathway connecting Traf7 and p65. (A) NF-κB luciferase reporter assay in growing myoblasts depleted of Traf7 (left) or MyoD1 (right). Luciferase counts were normalized against Renilla to show relative fold changes (FC). (B) Top panel: Immunoblot (IB) analysis of p65 in cytoplasmic (Cyto) and nuclear (Nuc) fractions from lysates of confluent myoblasts. TATA-binding protein (TBP) and α-tubulin represent loading controls for nuclear and cytoplasmic extracts, respectively. Bottom panel: normalized densitometric quantification of top panel. (C) Top panel: RT–PCR analysis of p65 mRNA in myoblasts 48 h after siRNA transfection. Bottom, left: IF of MHC expression (red) in myoblasts transfected with vector or FLAG–Traf7 and depleted of p65 (ip65) then differentiated for 48 h (scale bar, 40 μm; magnification × 10). Bottom, right: Normalized MHC expression for ip65-treated cells compared with NS controls. (D) Quantitation of MHC- and FLAG/MHC-positive cells in the ip65 rescue experiment. Panels show IF of FLAG–Traf7 (green) and MHC (red) in myoblasts transfected with or without p65 siRNA and FLAG–Traf7 and differentiated for 48 h. Scale bars, 5 μm for magnification × 63 and 4.92 μm for magnification × 64. Error bars indicate s.e.m. values; P-values shown for three independent experiments. DAPI, 4′,6-diamidino-2-pheylindole; FC, fold-change; IB, immunoblot; IF, immunofluorescence; MHC, myosin heavy chain; mRNA, messenger RNA; NF, nuclear factor; NS, nonspecific; TBP, TATA-binding protein.
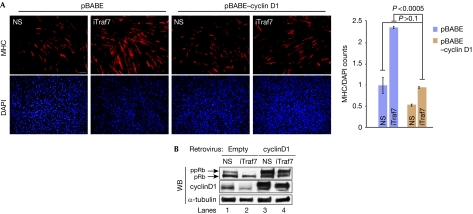
Next, we asked whether expression of cyclin D1 could rescue the phenotype observed on Traf7 depletion in growing cells. Expression of cyclin D1 alone resulted in increased proliferation, as expected, and in a substantial reduction (approximately 50%) of normalized MHC staining in cells induced to differentiate (Fig 4A). By contrast, stable expression of cyclin D1 in Traf7-depleted myoblasts restored MHC staining to control levels (Fig 4A). Thus, cyclin D1 overexpression rescued the Traf7 depletion phenotype. We also observed increased hypo-phosphorylation of the retinoblastoma tumour suppressor protein, a signature of cell cycle exit, on Traf7 RNAi treatment (Fig 4B). Overexpression of cyclin D1 in Traf7-ablated cells restored the retinoblastoma tumour suppressor protein phosphorylation to levels near those of controls, confirming that depletion of Traf7 causes cell cycle exit and overexpression of cyclin D1 rescues it. Collectively, these results suggest a pathway in which Traf7 depletion leads to reduced p65 nuclear translocation, downregulation of NF-κB activity, resulting in diminished cyclin D1 transcription and, ultimately, premature myogenic differentiation. These data are important because they identify a new link between two crucial regulators of myogenesis—MyoD1 and NF-κB—and suggest that Traf7, a direct transcriptional target of MyoD1, is a likely candidate for this link.
Figure 4.
Traf7 expression positively correlates with pRB phosphorylation state. (A) Immunofluorescence of MHC expression in myoblasts ectopically expressing cyclin D1 and depleted of Traf7 (iTraf7) and differentiated for 48 h. Scale bars, 40 μm; magnification × 10. Histogram shows normalized MHC signal in iTraf7 cultures compared with that of control. Error bars indicate s.e.m. values; P-values for three independent experiments. (B) pRB and cyclin D1 immunoblots on 100 μg of lysate. DAPI, 4′,6-diamidino-2-phenylindole; MHC, myosin heavy chain; NS, nonspecific; pRb, hypo-phosphorylated pRb; ppRb, hyper-phosphorylated pRb.
Traf7 interacts with IKKα/β through NEMO
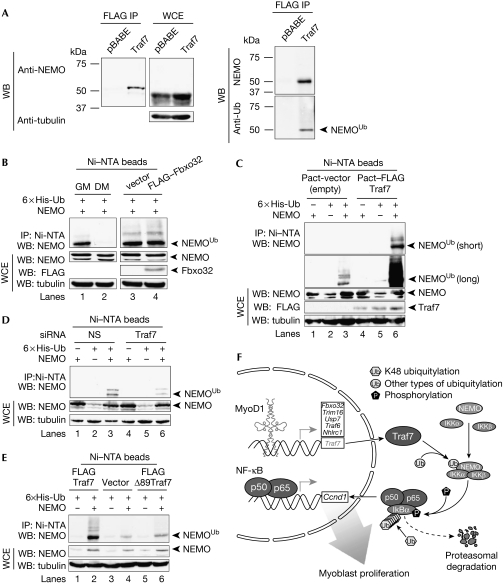
To explore whether Traf7 participates in NF-κB regulation, we performed proteomic analyses to identify Traf7-associated proteins in confluent C2C12 cells (T0). We stably expressed FLAG–Traf7, and purified associated proteins using a combination of immuno-affinity and ion exchange (MonoQ) chromatography. The resulting polypeptides were identified by tandem mass spectrometry (supplementary Fig S7A online). From these analyses we identified NEMO (IKKγ) as a potential interacting protein. Importantly, NEMO was not identified in parallel proteomic screens after anti-FLAG immuno-affinity purification of lysates derived from a control (FLAG-only) or a FLAG-tagged Fbxo32 stable cell line (data not shown). As antibodies that detect native Traf7 are not available, we performed experiments with C2C12 cells engineered to stably express low levels of FLAG–Traf7 to confirm the Traf7–NEMO interaction. The mRNA levels of FLAG–Traf7 in this cell line are comparable with physiological levels of Traf7 in control cells based on reverse transcriptase PCR analyses (data not shown). We confirmed the association between endogenous NEMO and FLAG–Traf7 (Fig 5A) with two different NEMO antibodies using these cells. We observed two major species of NEMO, differing by around 10–15 kDa, and Traf7 preferentially interacted with the larger species of endogenous NEMO (Fig 5A). Immunoblotting with a ubiquitin antibody revealed that this species represents a ubiquitylated form of NEMO (Fig 5A, right panel). The shift in molecular weight observed after expression of Traf7 is consistent with ubiquitylation of NEMO, and it seems that this species is preferentially retained in complexes with Traf7.
Figure 5.
Traf7 specifically enhances ubiquitylation of NEMO in myoblasts. (A) IB analysis of lysates from confluent stable FLAG–Traf7 C2C12 cells. (Left) IB of FLAG IP and whole-cell extract (WCE) probed with NEMO (top; from MBL) and tubulin (bottom) antibodies. (Right) IB was probed with NEMO (top; from NCI) and ubiquitin antibodies (bottom). (B) Lanes 1 and 2, IB of lysates from confluent myoblasts (GM) or myotubes differentiated for 48 h (DM) transiently transfected with NEMO and His6–Ub expression constructs. Lanes 3 and 4, IB of lysates from GM transiently transfected with FLAG–Fbxo32 or vector alone along with NEMO and His6–Ub expression constructs. The latter were included in transfections in (C–E). Purification of lysates using Ni–NTA beads under denaturing conditions and input WCE are indicated. (Top) IB with NEMO antibodies. (Bottom) IB of WCE with NEMO, FLAG, and α-tubulin (loading control) antibodies. (C) IB of lysates from GM transfected with FLAG–Traf7 or vector alone. (Top) IB with NEMO antibodies. Two exposures shown. (Bottom) IB as in (C). (D) IB of lysates from GM transfected with Traf7 or control (NS) siRNAs. IB as in (C). Unmodified and modified NEMO are indicated. (E) IB of lysates from GM transiently transfected with vector alone, Traf7, or a Traf7 mutant (Δ89 Traf7), together with NEMO and His6–Ub expression constructs. IB as in (C). (F) Model of process whereby Traf7, a MyoDI transcriptional target, enhances NEMO ubiquitylation in myoblasts. DM, differentiation medium; IB, immunoblot; IP, immunoprecipitation; MBL, MBL International; NCI, National Cancer Institute; NEMO, nuclear factor-κB essential modulator; NS, nonspecific; NTA, nitrilotriacetic acid; siRNA, small interfering RNA; Ub, ubiquitin; WB, western blot.
Ubiquitylation of NEMO that is not associated with the proteasome is thought to be required for the activation of the NF-κB pathway (Shifera, 2010). This type of ubiquitylation can be detected in the absence of proteasome inhibitors. Termination of this type of signal can be achieved by deubiquitylation or through proteasomal targeting. Thus, we determined the status of NEMO ubiquitylation at T0, when the Traf7–NEMO interaction is robust, in the absence and presence of MG132. Importantly, we observed robust ubiquitylation of NEMO in the absence of MG132 (Fig 5), and the total cellular pool of ubiquitylated NEMO was modestly increased in the presence of MG132 (supplementary Fig S7B online). These data suggest that this form of NEMO is relatively stable and that some fraction of the total pool of ubiquitylated NEMO has a function unrelated to its degradation.
We next asked whether ubiquitylation of NEMO is Traf7-dependent. Histidine-tagged ubiquitin and untagged NEMO were co-expressed in myoblasts and myotubes in the absence of MG132, and the tagged ubiquitin conjugates were then purified under denaturing conditions, followed by immunoblotting to detect NEMO (supplementary Fig S7D online). The major species detected in growing myoblasts was mono-ubiquitylated NEMO (as shown by its migration), whereas ubiquitylated NEMO was undetectable in differentiated cells (Fig 5B).
Importantly, expression of Traf7 significantly enhanced levels of modified NEMO, suggesting that NEMO can be ubiquitylated in vivo in response to Traf7 expression, whereas co-expression of Fbxo32, an E3 ligase not linked to NF-κB signalling, had no effect on NEMO ubiquitylation (Fig 5B,C). To investigate whether endogenous Traf7 is important for NEMO ubiquitylation, we performed analogous experiments after Traf7 depletion. The depletion of Traf7 resulted in less modified NEMO, particularly the mono-ubiquitylated form (Fig 5D). Furthermore, Traf7 upregulation altered ubiquitylation of endogenous NEMO (Fig 5C). The E3 ligase activity of Traf7 was required for NEMO modification, as expression of a Traf7 mutant lacking the RING domain was able to bind to NEMO but failed to enhance NEMO modification (Fig 5E; supplementary Fig S7D online) and block differentiation.
In summary, our observations show that in vivo, the E3 ligase activity of Traf7 has an important physiological function in NEMO ubiquitylation, providing a plausible mechanism for the activation of the NF-κB pathway in proliferating muscle cells. Together, these data support a model (Fig 5F) in which Traf7, through its control of NEMO ubiquitylation, mediates MyoD1 regulation of the NF-κB pathway and cell cycle progression in myoblasts.
Methods
Cell culture, transfections and luciferase assays. C2C12 mouse myoblasts were differentiated as described previously (Blais et al, 2005). Wild-type and primary MyoD1−/− mouse myoblasts (supplementary Fig S1A online) were a gift from M. Rudnicki, University of Ottawa. C2C12 cells were used in all other experiments. For quantification of myotube formation, we used MetaMorph software to calculate the ratio of MHC to 4',6-diamidino-2-phenylindole fluorescence.
Plasmids encoding His6-tagged ubiquitin (gift of R. Baer, Columbia University), FLAG–Traf7 (gift of S. Ishii, RIKEN, Japan) and FLAG-Δ89–Traf7, were transfected into myoblasts using Lipofectamine 2000 (Invitrogen). pcDEF3–FLAG–Δ89–Traf7 complementary DNA lacks the amino-terminal first 88 amino acids. For luciferase assays, Traf7 siRNA silencing in growing cells was performed 24 h before transfecting with 4 μg of 3 × κB-Luc and 1 μg of cytomegalovirus–Renilla reporter plasmids. Luciferase assays were performed at T48.
Purification of Traf7-associated proteins and mass spectrometry. Confluent (T0) C2C12 cells stably expressing FLAG–HA–Traf7 were lysed in 10 mM HEPES (pH 8), 250 mM KCl, 0.1% NP-40, 0.1 mM EDTA and 10% glycerol. A total of 40 mg of protein was purified with FLAG–M2 agarose (Sigma). Bound material was eluted with FLAG peptide and purified on a MonoQ column eluted with a linear gradient from 50 mM to 750 mM KCl in 20 mM Tris (pH 7.6), 0.2 mM EDTA, 10 mM β-mercaptoethanol, 10% glycerol and protease inhibitors. FLAG–HA–Traf7 eluted in two peaks between 350 and 570 mM KCl, which were processed separately. Co-eluting proteins from each peak were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis, excised, trypsinized and subjected to microcapillary liquid chromatography coupled to tandem mass spectrometry.
Detailed protocols for retroviral transduction, RNAi, cellular fractionation, ChIP and purification of Traf7-associated proteins are provided in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank M. Rudnicki, R. Baer, I. Aifantis and S. Ishii for reagents; P. Asp and V. Vethantham for insightful discussions; R. Dasgupta and C.Yun (New York University RNA interference facility) for assistance with RNA interference screens. B.D.D. was supported by National Institutes of Health (1 R21 CA125734-01 and 2R01 GM067132) and the Muscular Dystrophy Association. I.S. was supported by funds from National Institutes of Health (K22 CA115998-03).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC (2008) IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol 180: 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS (1999) NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol 19: 5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H, Han D, Choi Y (2009) Traf-mediated TNFR-family signaling. Curr Protoc Immunol Ch. 11, Unit 11.9D [DOI] [PubMed] [Google Scholar]
- Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB (1995) Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267: 1018–1021 [DOI] [PubMed] [Google Scholar]
- Mal A, Harter ML (2003) MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci USA 100: 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifera AS (2010) Proteins that bind to NEMO and downregulate the activation of NF-κB. Biochem Biophys Res Commun 36: 585–589 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB (1988) MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405–411 [DOI] [PubMed] [Google Scholar]
- Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC (2007) NF-κB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol 27: 4374–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Wu Y, Zhao J, Guo S, Bauman WA, Cardozo CP (2005) Structure and function of the upstream promoter of the human Mafbx gene. J Cell Biochem 96: 209–219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.