DNA end resection by CtIP and exonuclease 1 prevents genomic instability
This study reports on the collaborative action of human CtIP and EXO1 in DNA end resection during DNA double-strand break repair to prevent the formation of abnormal chromosome structures.
Keywords: DNA double-strand break repair, CtIP, exonuclease 1, camptothecin
Abstract
End resection of DNA—which is essential for the repair of DNA double-strand breaks (DSBs) by homologous recombination—relies first on the partnership between MRE11–RAD50–NBS1 (MRN) and CtIP, followed by a processive step involving helicases and exonucleases such as exonuclease 1 (EXO1). In this study, we show that the localization of EXO1 to DSBs depends on both CtIP and MRN. We also establish that CtIP interacts with EXO1 and restrains its exonucleolytic activity in vitro. Finally, we show that on exposure to camptothecin, depletion of EXO1 in CtIP-deficient cells increases the frequency of DNA–PK-dependent radial chromosome formation. Thus, our study identifies new functions of CtIP and EXO1 in DNA end resection and provides new information on the regulation of DSB repair pathways, which is a key factor in the maintenance of genome integrity.
Introduction
DNA double-strand breaks (DSBs) are the most cytotoxic lesions that can be generated by ionizing radiation, certain chemotherapeutic drugs, collapse of stalled DNA replication forks or during physiological processes such as meiotic recombination (Bassing et al, 2002; Whitby, 2005). DSBs that are not properly repaired can cause gross chromosomal aberrations that trigger carcinogenesis through activation of oncogenes or inactivation of tumour suppressor genes. Cells use two main mechanisms to repair DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR; Wyman & Kanaar, 2006; Misteli & Soutoglou, 2009). NHEJ repair takes place throughout the cell cycle, whereas HR is restricted to S- and G2-phases. HR is initiated by 5′–3′ resection of DSBs to produce single-strand DNA (ssDNA) tails that function as a signal for ATR-mediated DNA damage checkpoint activation before the recruitment of recombination proteins (Pardo et al, 2009).
Several studies have investigated the molecular mechanisms of DNA end resection in genetically amenable organisms. These have implicated the MRE11–RAD50–NBS1/Xrs2 (MRN/MRX) complex and CtIP/Sae2 in the early stages of DSB processing, followed by the redundant action of BLM/Sgs1 helicase and exonuclease 1 (EXO1) in the generation of long stretches of ssDNA (Mimitou & Symington, 2009). Accordingly, only the simultaneous depletion of BLM and EXO1 resulted in accumulation of partly resected intermediates and hypersensitivity to DSB-inducing agents (Gravel et al, 2008; Mimitou & Symington, 2008; Zhu et al, 2008). These studies led to a two-step model according to which in human cells the initial ‘end-trimming' is carried out by MRN and CtIP, followed by resection by two mechanisms depending on either EXO1 or BLM (Niu et al, 2009).
In this study, we show that EXO1 is recruited to laser-induced DSBs in a CtIP-dependent manner and that CtIP interacts with EXO1, thereby retarding processive degradation of DNA by EXO1 in vitro. Furthermore, we provide evidence that concomitant depletion of CtIP and EXO1 in camptothecin-treated cells leads to chromosomal rearrangements, probably as a result of illegitimate NHEJ-dependent repair of DSBs.
Results And Discussion
EXO1 localization to sites of DNA damage
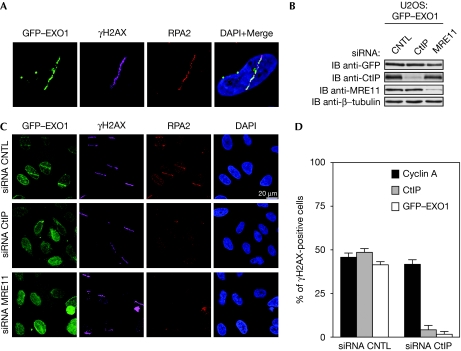
On the basis of the two-step model of DNA end resection, we hypothesized that the recruitment of EXO1 to DSBs was dependent on the initial processing by MRN and CtIP. As endogenous levels of EXO1 are undetectable by direct immunostaining (El-Shemerly et al, 2005), we examined EXO1 localization to laser microirradiation-induced DSBs (Bekker-Jensen et al, 2006) by using U2OS cells stably expressing green fluorescent protein (GFP)-tagged EXO1 (Gravel et al, 2008). Similarly to previous reports, we observed accumulation of EXO1 at sites of DSBs (Bolderson et al, 2010; Fig 1A; supplementary Fig S1A,B online). We therefore asked whether depletion of CtIP or the MRN complex (Fig 1B) would affect EXO1 recruitment to DSBs. Both fixed and live-cell imaging showed that depletion of either CtIP or MRE11 impaired the recruitment of EXO1 and replication protein A (RPA) 2 to DSBs (Fig 1C; supplementary Fig S1B online). Furthermore, in CtIP-depleted cells we did not observe any localization of EXO1 to sites of DSBs, even at late time points (supplementary Fig S1A,B online). In addition, downregulation of BLM did not impair EXO1 recruitment to DSBs (data not shown). Consistent with the S/G2-specific recruitment of CtIP to DSBs (Sartori et al, 2007), accumulation of GFP–EXO1 at DSBs was only observed in cyclin A-positive cells and was strictly CtIP-dependent (Fig 1D). Finally, EXO1, but not CtIP, did not localize to sites of laser-induced DSBs in ataxia telangiectasia-like disorder 1 (ATLD1) cells, which are deficient in DSB resection due to a hypomorphic mutation of the MRE11 gene (Stewart et al, 1999; Carson et al, 2003; supplementary Fig S1C–E online). This defect was rescued on re-expression of wild-type MRE11 (supplementary Fig S1F online). From these observations, we concluded that the recruitment of EXO1 to DSBs depends on the initial DSB-end trimming carried out by MRN together with CtIP.
Figure 1.
Exonuclease 1 localization to sites of DNA damage requires both CtIP and MRE11. (A) U2OS cells stably expressing GFP–EXO1 were microirradiated and 30 min later fixed, immunostained with γH2AX and RPA2 antibodies, and analysed by fluorescence microscopy. Nuclei were visualized with DAPI. (B) GFP–EXO1 cells were transfected with Luciferase- (CNTL), CtIP- or MRE11-specific siRNA oligonucleotides and grown for 72 h. Protein expression was examined by immunoblot (IB) using the indicated antibodies. (C) siRNA-transfected GFP–EXO1 cells from (B) were microirradiated, fixed and analysed as in (A). (D) GFP–EXO1 cells treated as in (A) were co-immunostained with γH2AX, Cyclin A and CtIP antibodies. γH2AX-positive cells were quantified for EXO1, Cyclin A and CtIP staining. A total of 50 cells per sample were counted in two independent experiments. EXO1, exonuclease 1, GFP, green fluorescent protein; RPA2, replication protein A; siRNA, small interfering RNA.
CtIP interacts with EXO1 and restrains its activity
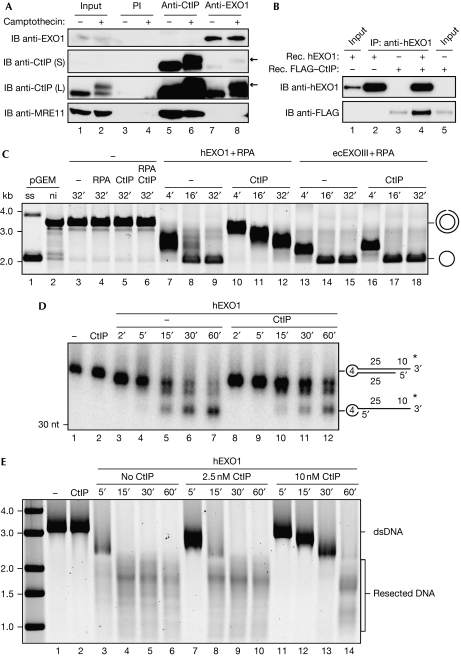
Although they are independently recruited to sites of DSB (supplementary Fig S1E online; Lisby et al, 2004; Chen et al, 2008), CtIP was shown to interact with MRN and stimulate its endonuclease activity in vitro (Sartori et al, 2007), indicating a functional relationship between these factors during DNA end resection. This prompted us to examine whether EXO1 physically associates with the MRN–CtIP complex. To test this, CtIP or EXO1 was immunoprecipitated from HEK293T whole-cell extracts and the recovered complexes were analysed by immunoblotting. Interestingly, CtIP but not MRE11 was present in anti-EXO1-immunocomplexes both in non-stressed cells and in cells treated with camptothecin (Fig 2A)—a chemotherapeutic agent known to induce DSBs exclusively during DNA replication by trapping DNA topoisomerase 1 cleavage complexes (Pommier, 2006). Although we noticed that EXO1 preferentially interacts with the hyperphosphorylated form of CtIP in damaged cells (Fig 2A, lane 8), we did not observe differences in CtIP–EXO1 interaction on phosphatase treatment of the CtIP–EXO1 immunocomplex (supplementary Fig S2A online). We confirmed the previously reported CtIP–MRE11 interaction, but did not detect EXO1 in anti-CtIP immunocomplexes, probably due to low cellular levels of EXO1 (El-Shemerly et al, 2005). Therefore, we immunoprecipitated CtIP from HEK293T cells transiently expressing OMNI-tagged EXO1. Under these conditions, we detected EXO1 in anti-CtIP immunocomplexes, both in the presence and absence of hydroxyurea (supplementary Fig S2B online).
Figure 2.
CtIP interacts with exonuclease 1 and restrains its exonucleolytic activity. (A) WCEs (5 mg) from mock or camptothecin-treated (1 μM, 1 h) HEK293T cells were immunoprecipitated with preimmune serum, CtIP or EXO1 polyclonal antibodies. Proteins were detected by immunoblot (IB) with the indicated antibodies. For CtIP, short (S) and long (L) exposure times are shown. Arrows indicate the hyperphosphorylated form of CtIP. The membrane was stripped and re-probed using a monoclonal EXO1 antibody. Input=1% of the WCEs used for immunoprecipitation. (B) Purified, recombinant (Rec.) EXO1 (200 ng) and FLAG-tagged CtIP (200 ng) were incubated either alone or together before immunoprecipitation with an hEXO1 antibody. Proteins were visualized with the indicated antibodies. Recombinant hEXO1 (50 ng, lane 1) and FLAG–CtIP (20 ng, lane 5) were loaded as inputs. (C) Nicked plasmid (3.75 nM) was incubated with hEXO1 (15 nM) or ecEXOIII (10 U) in the presence or absence of CtIP (15 nM). RPA (300 nM) was present where indicated. Products were resolved as described in the Methods section. The migration patterns of the double-stranded, nicked plasmid and of the single-stranded product are indicated. (D) Radiolabelled oligonucleotide (1 nM, lane 1) was incubated with CtIP alone (1 nM, lane 2), hEXO1 alone (1 nM, lanes 3–7) or both together at equimolar concentrations (lanes 8–12). Reactions were terminated at the indicated time points and the products were analysed as described in the Methods section. (E) Linearized plasmid with 3′-overhangs (2.5, 5 nM DNA ends) was incubated at 37 °C with hEXO1 (15 nM), RPA (300 nM) and the indicated amounts of CtIP. Reactions were terminated at the indicated time points and the products were analysed as described in the Methods section. ecEXOIII, Escherichia coli exonuclease III; hEXO1, human exonuclease 1; PI, preimmune; RPA, replication protein A; WCE, whole-cell extract.
To investigate whether the interaction between CtIP and EXO1 is direct or reliant on bridging factors, we examined CtIP–EXO1 complex formation by using purified, recombinant proteins in an anti-EXO1 immunoprecipitation experiment (Fig 2B). Although a small fraction of CtIP was unspecifically bound to beads, CtIP was enriched when equimolar amounts of EXO1 were present (Fig 2B, lane 4), demonstrating that the two proteins are able to bind directly to each other in vitro.
Saccharomyces cerevisiae Sae2 shows endonuclease activity on defined substrates (Lengsfeld et al, 2007). Although similar activity has not yet been reported for human CtIP, CtIP was shown to enhance the endonucleolytic activity of the MRE11–RAD50 complex (Sartori et al, 2007). On the basis of these observations, we asked whether CtIP might also affect the 5′–3′ exonuclease activity of EXO1 in vitro. First, we examined the activity of purified recombinant EXO1 (supplementary Fig S2C online) using a singly nicked plasmid DNA substrate. Only wild-type EXO1, not a catalytically dead mutant, was able to completely degrade the nicked strand within 30 min of incubation, indicating that there was no contaminant exonuclease activity in our preparation (supplementary Fig S2D online). The addition of equimolar amounts of CtIP decreased exonucleolytic processing, whereas it did not inhibit the activity of Escherichia coli exonuclease III (Fig 2C). Under similar assay conditions, MRE11–RAD50 did not substantially affect EXO1 activity (supplementary Fig S2E online). We observed a similar inhibitory effect of CtIP on EXO1 activity using either a radiolabelled DNA oligonucleotide substrate (Fig 2D) or a linearized plasmid (Fig 2E) both containing 3′-overhangs, which are the preferred substrate for EXO1 in vitro (supplementary Fig S2G online; Lee & Wilson, 1999). Electrophoretic mobility shift assays showed that, in contrast to EXO1, CtIP did not efficiently bind to the oligonucleotide substrate (supplementary Fig S2F online), excluding the possibility of a nonspecific inhibition of EXO1 by CtIP through steric hindrance. As observed with the nicked plasmid, CtIP did not inhibit the activity of E. coli exonuclease III on the linear substrate (data not shown). Furthermore, neither MRE11–RAD50 nor BLM affected EXO1 activity on the plasmid with 3′-overhangs (supplementary Fig S2H online). Interestingly, pre-incubation of CtIP with either blunt-ended or 5′-overhang substrates facilitated processing by EXO1, which did not occur when the proteins were added in the reverse order (supplementary Fig S2I online).
These biochemical data might suggest that CtIP is able to restrain long-range resection by EXO1, thereby generating appropriate recombinogenic ssDNA structures (Mimitou & Symington, 2009; Niu et al, 2009). Inhibition of EXO1 activity was also reported during repair of DNA mismatches. However, whereas in mismatch repair RPA (Genschel & Modrich, 2009) or possibly MutLα (Zhang et al, 2005) are required for terminating EXO1 activity on removal of the mismatch, our data suggest that CtIP might act to fine tune the nucleolytic activity of EXO1.
CtIP and EXO1 promote error-free repair of DSBs
Our observations indicate that the initial end-trimming activity of MRN–CtIP is required for the recruitment of EXO1 to sites of DNA damage and that CtIP might subsequently control EXO1 exonucleolytic activity to facilitate HR. This prompted us to examine whether some of the phenotypes reported for CtIP-deficient cells in response to camptothecin require EXO1 (Sartori et al, 2007). By using siRNA-mediated downregulation of CtIP and EXO1, we monitored various DNA damage phenotypes in response to camptothecin. Single or combined depletion of CtIP and EXO1 did not significantly affect transition through S-phase, as shown by flow cytometry and incorporation of 5-ethynyl-2′-deoxyuridine in DNA (supplementary Fig S3A,B online). As previously shown, CtIP knockdown led to a decrease in CHK1 and RPA2 phosphorylation (supplementary Fig S3C online), indicative of inefficient resection and impaired ATR activation (Sartori et al, 2007). However, EXO1 depletion had no impact on ATR signalling, apart from a modest increase in RPA2 hyperphosphorylation (supplementary Fig S3C online). Furthermore, the pattern of camptothecin-triggered DNA-damage-signalling events in cells simultaneously depleted for CtIP and EXO1 did not vary from that of CtIP singly depleted cells (supplementary Fig S3C online). Taken together, these data support the view that EXO1 acts downstream from CtIP and MRN in DNA end resection and are consistent with studies reporting an alternative, EXO1-independent mode of processive resection (Gravel et al, 2008; Mimitou & Symington, 2008).
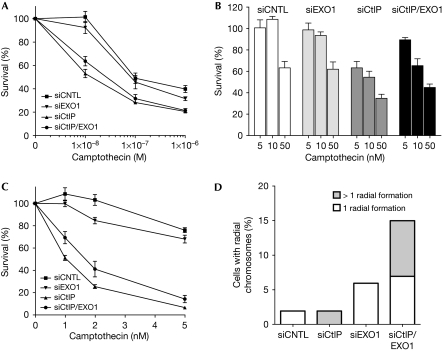
Next, we analysed the sensitivity of these cells to an acute treatment with camptothecin by colony formation assays. Consistent with previous reports, we observed that CtIP downregulation caused hypersensitivity to camptothecin, whereas EXO1 depletion conferred only minor cytotoxicity (Fig 3A; Sartori et al, 2007; Gravel et al, 2008). Interestingly, we observed a partial, but statistically significant rescue of sensitivity at low camptothecin concentrations by simultaneous downregulation of CtIP and EXO1 (Fig 3A,B). Consistent with this, we found a similar increase in survival on chronic treatment with camptothecin (Fig 3C). We then treated cells with Olaparib, an inhibitor of poly(ADP-ribose) polymerase (PARP; Helleday et al, 2008). As PARP is involved in the repair of DNA single-strand breaks (Hoeijmakers, 2001), it was proposed that PARP inhibition results in the accumulation of replication-associated DSBs (Bryant et al, 2005; Farmer et al, 2005), creating lesions similar to those caused by camptothecin (Pommier, 2006). Treating singly or doubly depleted cells with Olaparib resulted in a pattern of hypersensitivity similar to that caused by camptothecin (supplementary Fig S3D online).
Figure 3.
CtIP and exonuclease 1 protect cells from chromosomal damage. (A) At 72 h after transfection with the indicated siRNA oligonucleotides, U2OS cells were treated with either DMSO or camptothecin (1 h, 1 μM; acute treatment) and survival was determined by colony formation. Data represent the mean±s.e.m. of five independent experiments. (B) Cell survival at low doses of camptothecin from the data shown in (A). Data represent the mean±s.e.m. of five independent experiments. (C) Cells transfected as in (A) were treated with either DMSO or camptothecin for 24 h (chronic treatment) and survival was determined by colony formation. Data represent the mean±s.e.m. of three independent experiments. (D) Metaphase spreads from cells transfected and treated as described in (A) were analysed for chromosomal aberrations. A total of 50 metaphase spreads was analysed for each sample. The percentages of metaphase spreads displaying the indicated numbers of radial chromosomes are shown. CNTL, control; DMSO, dimethyl sulphoxide; EXO1, exonuclease 1; siRNA, small interfering RNA.
The above data indicate that CtIP and EXO1 act in the same pathway, but they also point to a potentially new genetic interdependency between these factors during the repair of replication-associated DSBs. To gain further, structure-based insight into the repair of camptothecin-induced lesions, we analysed metaphase spreads from cells lacking CtIP and EXO1. Compared with control cells, EXO1-deficient cells had a slight increase in broken chromatids, whereas depletion of CtIP led to a reduction of this type of chromosomal aberration (supplementary Fig S3E online). However, we noticed a significant increase in the number of radial chromosomes specifically in doubly depleted cells, indicative of illegitimate repair by end joining (Fig 3D; supplementary Fig S3F online).
CtIP and EXO1 cooperate to prevent NHEJ
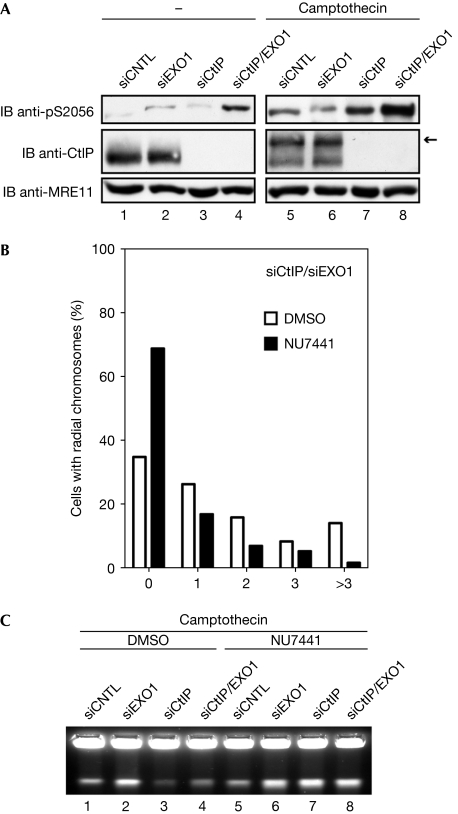
It has been shown that DNA-replication-associated DSBs, such as those induced by camptothecin, activate DNA-PKcs, indicated by autophosphorylation on S2056 (Chen et al, 2005; Sakasai et al, 2010; supplementary Fig S4A online). By analysing S2056 autophosphorylation, we observed increased DNA-PKcs activation particularly in doubly depleted cells, and the signal was further amplified in response to camptothecin (Fig 4A). This prompted us to reexamine camptothecin-induced chromosomal aberrations on inhibition of DNA-PKcs (supplementary Fig S4A online). Under these conditions, we observed an almost threefold reduction in the number of radial structures in EXO1/CtIP-deficient cells, whereas DNA-PKcs inhibition had no major effect in singly depleted cells (Fig 4B; supplementary Fig S4B online; data not shown). Consistent with this and in agreement with an upregulation of NHEJ in the absence of CtIP and EXO1, DNA-PKcs inhibition markedly increased the number of camptothecin-induced breaks measured by PFGE (Fig 4C; supplementary Fig S4C online).
Figure 4.
Inhibition of DNA-PKcs rescues radial chromosomes formation. (A) U2OS cells transfected with siRNA oligonucleotides were treated with DMSO or camptothecin (1 μM, 1 h) and DNA-PKcs autophosphorylation at S2056 was monitored. Arrow indicates the hyperphosphorylated form of CtIP. (B) Metaphase spreads were generated from cells transfected as in (A) and treated with camptothecin (1 μM, 1 h) in the presence or absence of NU7441 (10 μM). In total, more than 100 spreads were analysed for both conditions in two independent experiments. The average number of radial chromsomes per spread was 1.65 (DMSO) and 0.59 (NU7441), equivalent to a 2.8-fold reduction in radial formation. (C) Cells transfected as in (A) were treated with DMSO or camptothecin (2.5 μM, 4 h) in the presence or the absence of NU7441 (10 μM). The amount of broken DNA was assessed by pulsed-field gel electrophoresis. CNTL, control; DMSO, dimethyl sulphoxide; EXO1, exonuclease 1; IB, immunoblot; siRNA, small interfering RNA.
Conclusions
Our data imply that two factors involved in DNA end resection, CtIP and EXO1, probably act together to maintain genomic stability by protecting cells from the deleterious consequences of end-joining-mediated repair of camptothecin-induced DNA lesions. A similar scenario was reported for the repair defects in Fanconi anaemia cells (Adamo et al, 2010; Pace et al, 2010). Therefore, we speculate that hypersensitivity to replication-associated DSBs in resection-compromised cells is probably the result of inappropriate NHEJ rather than HR deficiency alone.
Methods
Live-cell imaging and laser microirradiation. Double-strand breaks were generated in the nuclei of living cells on 18-mm glass cover slips by microirradiation of arbitrarily shaped regions of interest (ROIs) at 355 nm with 15-mW output power of the laser (Genesis 355-80, Coherent; Walter et al, 2003). The ROIs were irradiated 10 times consecutively and identical ROIs were used in all experiments. Subsequently, fluorescence time-lapse imaging was performed for GFP (488 nm excitation, 525–560 nm emission; SP5, Leica, Mannheim, Germany) using an HCX Plan-Apo × 63/NA 1.40 oil immersion objective. Pre-sensitization with 10 μM bromodeoxyuridine was used to avoid artefacts by high local density of DSBs. Non-pre-sensitized control cells showed a lack of EXO1 and CtIP recruitment under the same conditions. Cells were kept in complete growth medium under 5% CO2 at 37 °C during the experiments.
For fixed-cell imaging, DSBs in defined nuclear volumes were generated by microirradiation (MMI CellCut) with a 355-nm UV-A laser adjusted at 50% of the power. Before irradiation, cells were grown for 24 h in the presence of 10 μM bromodeoxyuridine.
See supplementary information online for additional Methods.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank S. Jackson for providing GFP–HA–EXO1 U2OS cells, R. Baer for CtIP antibodies and purified CtIP protein, M. Weitzman for ATLD1-Tert cells, F. Marini for the GFP–EXO1 construct and P. Janscak for purified RPA, BLM and MRE11–RAD50 complex. We also thank M. Stucki and P. Janscak for critical reading of the paper, and U. Ziegler (University of Zurich, Center for Microscopy and Image Analysis) for guidance in the optimization of live-cell imaging. This work was supported by grants of the Swiss National Science Foundation (SNF), the UBS Foundation, the Novartis Foundation for Medical and Biological Research, the University of Zurich Research Priority Program (URPP) and the Désirée and Niels Yde Foundation (to S.F.), the Vontobel-Foundation (to A.A.S.), by an Ambizione fellowship from the Swiss National Science Foundation (SNSF; to A.A.S.) and by the Forschungskredit from the University of Zurich (to M.S.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A (2010) Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell 109: S45–S55 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J (2006) Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK (2010) Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res 38: 1821–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD (2003) The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J 22: 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ (2005) Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem 280: 14709–14715 [DOI] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X (2008) Cell cycle-dependent complex formation of BRCA1–CtIP–MRN is important for DNA double-strand break repair. J Biol Chem 283: 7713–7720 [DOI] [PubMed] [Google Scholar]
- El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S (2005) Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res 65: 3604–3609 [DOI] [PubMed] [Google Scholar]
- Farmer H et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Genschel J, Modrich P (2009) Functions of MutLα, replication protein A (RPA), and HMGB1 in 5′-directed mismatch repair. J Biol Chem 284: 21536–21544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA (2008) DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8: 193–204 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Lee BI, Wilson DM III (1999) The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem 274: 37763–37769 [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11–Rad50–Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Raynard S, Sung P (2009) Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev 23: 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ (2010) Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 329: 219–223 [DOI] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A (2009) DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6: 789–802 [DOI] [PubMed] [Google Scholar]
- Sakasai R, Teraoka H, Tibbetts RS (2010) Proteasome inhibition suppresses DNA-dependent protein kinase activation caused by camptothecin. DNA Repair (Amst) 9: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–587 [DOI] [PubMed] [Google Scholar]
- Walter J, Cremer T, Miyagawa K, Tashiro S (2003) A new system for laser-UVA-microirradiation of living cells. J Microsc 209: 71–75 [DOI] [PubMed] [Google Scholar]
- Whitby MC (2005) Making crossovers during meiosis. Biochem Soc Trans 33: 1451–1455 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R (2006) DNA double-strand break repair: all's well that ends well. Annu Rev Genet 40: 363–383 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122: 693–705 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.