RNA channelling by the eukaryotic exosome
Cryo-EM structures of the yeast RNA exosome in the apo and the RNA-bound states provide direct structural evidence for a substrate threading mechanism.
Keywords: exosome, RNA degradation, cryo-electron microscope, macromolecular complex
Abstract
The eukaryotic exosome is a key nuclease for the degradation, processing and quality control of a wide variety of RNAs. Here, we report electron microscopic reconstructions and pseudo-atomic models of the ten-subunit Saccharomyces cerevisiae exosome in the unbound and RNA-bound states. In the RNA-bound structures, extra density that is visible at the entry and exit sites of the exosome channel indicates that a substrate-threading mechanism is used by the eukaryotic exosome. This channelling mechanism seems to be conserved in exosome-like complexes from all domains of life, and might have been present in the most recent common ancestor.
Introduction
The RNA exosome is a multiprotein complex that participates in several RNA degradation and processing reactions. Exosome functions range from processing of stable RNAs—such as ribosomal RNA, small nuclear and nucleolar RNA—to the complete turnover of mRNA (Ibrahim et al, 2008). In addition, the exosome is involved in several surveillance pathways that target pre-mRNA with maturation defects, hypomethylated tRNA and incorrectly assembled pre-ribosomes (Hilleren et al, 2001; Dez et al, 2006; Kadaba et al, 2006). Consistent with its role in RNA metabolism, the exosome is conserved in a range of eukaryotic organisms including yeast, trypanosomes, flies, plants and humans, and it is also present in archaea (Schilders et al, 2006; Lorentzen et al, 2008a).
All exosomes share a structural core of nine subunits (Exo9), as found in the crystal structures of archaeal and human exosomes (Buttner et al, 2005; Liu et al, 2006; Lorentzen et al, 2007). Six subunits (Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3 in eukaryotes) adopt the RNase PH fold that is present in phosphorolytic RNases and form a hexameric ring structure around a central channel (Fig 1). One side of this hexamer associates with the three additional core subunits Rrp4, Rrp40 and Csl4, which contain S1 and KH putative nucleic-acid binding domains. This structural arrangement creates a continuous channel spanning Exo9 from the S1/KH trimeric cap to the hexameric RNase PH ring. All of the exosome RNase PH subunits in both the yeast and the human complexes have accumulated mutations in the phosphorolytic active sites, which have rendered the Exo9 complex inactive as a nuclease (Liu et al, 2006; Dziembowski et al, 2007). Instead, the RNase activity is mediated by a tenth subunit—Rrp44—which, in yeast, stably associates with Exo9 to form the active ten-subunit complex (Exo10).
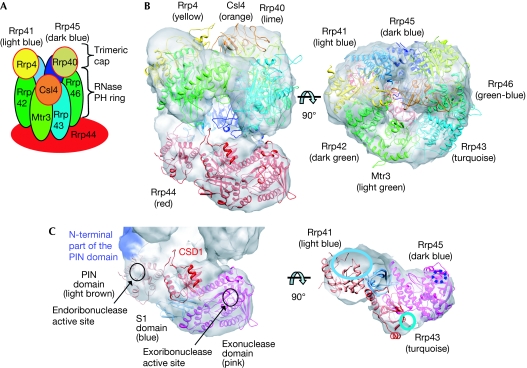
Figure 1.
Cryo-electron microscopic structure of the Saccharomyces cerevisiae exosome. (A) Schematic representation of the eukaryotic exosome architecture. The ribonuclease Rrp44 is positioned at the bottom of Exo9. (B) Side and top views of the apo exosome electron microscopic map are shown as a white surface, with the fitted atomic coordinates of the exosome subunits displayed as ribbons. (C) Zoom-in views of the domains of the Rrp44 subunit. The PIN, CSD1, exoribonuclease and S1 domains are shown as ribbons and the position of the amino-terminal of the PIN domain is coloured purple. The right-hand panel shows an orthogonal slice of the electron microscopic map. Interactions of Rrp44 domains with RNase PH subunits of the exosome are indicated. PIN, PilT N-terminal.
Rrp44 is a multidomain protein composed of a PilT amino-terminal (PIN) domain that has endonucleolytic activity and an RNase II/R homology region with hydrolytic exoribonuclease activity (Cheng & Deutscher, 2002; Lebreton et al, 2008; Schaeffer et al, 2009; Schneider et al, 2009). Consistent with negative-stain electron microscopy data (Wang et al, 2007), a recently published crystallographic structure has shown the way in which yeast Rrp44 binds directly to the Rrp41 and Rrp45 subunits at the surface opposite to the binding interface for the S1/KH subunits (Bonneau et al, 2009). These structural data are in agreement with data from the biochemical mapping of a 31–33-nucleotide (nt)-long path, suggesting that RNA substrates are threaded through the central channel of Exo9 to the Rrp44 exoribonuclease site (Bonneau et al, 2009). To investigate this threading mechanism, we have determined the structures of apo and RNA-bound exosome complexes from Saccharomyces cerevisiae by electron microscopy.
Results And Discussion
Organization of the ten-subunit core
The Saccharomyces cerevisiae Exo10 complex was reconstituted from individually purified proteins and subcomplexes according to previously published protocols (supplementary Fig S1A online; Liu et al, 2006; Bonneau et al, 2009). From this sample, negative-stain and cryo-electron microscopy data were collected and three-dimensional maps were reconstructed, with resolutions of 16 and 14 Å, respectively (supplementary Figs S1B,C & S2A,B online). The two maps show similar ring-shaped structures with a central channel that is characteristic of exosomes (Fig 1).
To obtain a pseudo-atomic model of the entire yeast exosome complex, crystal structures and homology models were docked into the cryo-electron microscopic map and refined using a flexible fitting method (Topf et al, 2008). Yeast Rrp40, Rrp41, Rrp45 and Rrp44 crystal structures (Oddone et al, 2006; Lorentzen et al, 2008b; Bonneau et al, 2009) and homology models of yeast Rrp4, Rrp42, Mtr3, Rrp43 and Rrp46 based on the structure of the human exosome (Liu et al, 2006) were used in the fitting procedures. For the RNase PH ring, pseudo threefold symmetry representing the three dimers Rrp41–Rrp45, Rrp43–Rrp46 and Rrp42–Mtr3, is clearly visible in the cryo-electron microscopic map (supplementary Fig S3A online). The three cap proteins Rrp4, Rrp40 and Csl4 associate at the top of the RNase PH ring in an asymmetrical manner (Fig 1). The smaller N-terminal domains of subunits Rrp40 and Csl4 had no clear density and were not included in the model. The elongated piece of density at the bottom of the RNase PH ring corresponds to the Rrp44 nuclease. Whereas positions of the PIN, exonucleolytic and S1 domains of Rrp44 seem to be conserved when compared with available crystal structures (Lorentzen et al, 2008b; Bonneau et al, 2009), CSD1 undergoes a small movement and CSD2 does not show well-ordered density, suggesting that this domain is flexible in the apo Exo10 (supplementary Fig S3B online). When all subunits are positioned in the map, the only remaining unfilled density is located close to the PIN domain and probably corresponds to the 36 N-terminal residues that are not modelled in the crystal structure used for fitting (Fig 1C, shown in purple). The interactions observed between Rrp44PIN and Rrp41/45 are consistent with the previously published crystal structure of the Rrp44/41/45 complex (Bonneau et al, 2009). In addition, our cryo-electron microscopic map shows that Rrp43 (residues 97–121, 176–208 and 245–276) and Rrp44CSD1 (residues 260–271 and 394–397) are in close proximity to each other.
RNA protection by exosomes lacking cap subunits
Comparison between the pseudo-atomic model of the S. cerevisiae exosome presented here and previous models of human and yeast exosomes (Liu et al, 2006; Wang et al, 2007) show that the hexameric RNase PH ring structure is well conserved, with a larger degree of flexibility for the putative RNA-binding subunits Csl4, Rrp4 and Rrp40 that cap the RNase PH ring (supplementary Fig S3C online). The electron microscopy maps suggest smaller movements of Rrp4 and larger movement of Csl4 and Rrp40 compared with the crystal structure of the human exosome. In accordance with these observations, comparisons of high-resolution crystal structures of archaeal and eukaryotic exosomes have shown increased flexibility of the KH/S1 proteins (Lorentzen et al, 2007; Lu et al, 2010).
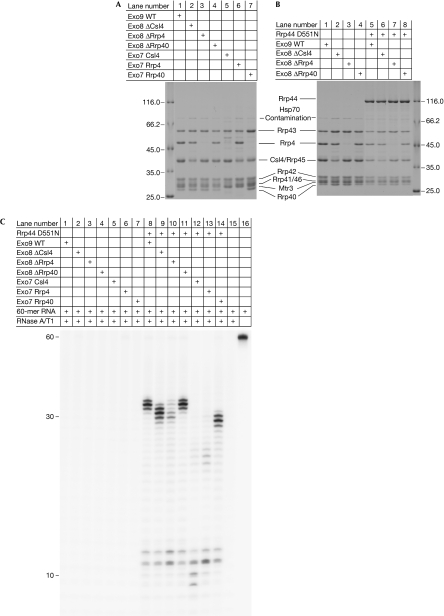
We explored the role of the individual cap proteins in exosome assembly and RNA binding. It has been shown that the six RNase PH subunits of the eukaryotic exosome do not form a stable complex in the absence of the three cap proteins (Liu et al, 2006). Work with truncation mutants has shown that yeast Rrp4 and Rrp40 have an important role in stabilizing the exosome structure, whereas Csl4 does not (Schaeffer et al, 2009). We reconstituted and purified exosome complexes lacking either one or two cap subunits (Fig 2A). Exosomes lacking one cap subunit (Exo ΔCsl4, Exo ΔRrp4 and Exo ΔRrp40) eluted as stable complexes in size-exclusion chromatography (data not shown) and were all able to bind to Rrp44 in pull-down experiments (Fig 2B). This suggests that none of the cap proteins has an essential function in forming a stable exosome. We then tested the RNA-binding abilities of Exo8 and the Exo8–Rrp44 complex in RNA protection assays similar to those that have been reported for Exo10 (Bonneau et al, 2009). In this assay, a 60-nt-long unstructured RNA was incubated with exosome complexes, and the unprotected parts of the RNA were degraded with RNase A/T1. We used an Rrp44(D551N) mutant that lacks catalytic activity but binds to RNA for this (Lorentzen et al, 2008b). Whereas wild-type Exo9 in the absence of Rrp44(D551N) does not protect RNA, Exo9+Rrp44(D551N) does, yielding fragments of 31–33 nt in length (Bonneau et al, 2009). In agreement with this, none of the reconstituted exosome complexes lacking one cap subunit were able to protect RNA in the absence of Rrp44(D551N) (Fig 2C, lanes 2–4). With Rrp44(D551N), exosome complexes lacking one cap subunit still protect RNA (Fig 2C, lanes 9–11). In the presence of Rrp44 (D551N), Exo ΔRrp40 behaved similarly to Exo10, protecting 31–33 nt RNA fragments (Fig 2C, lanes 8 and 11), whereas Exo ΔCsl4 protected RNA fragments that were 2 nt shorter (Fig 2C, lane 9). This relatively small change in the protection pattern might be due to differences in the RNA-binding properties of Rrp4 and Csl4, or could reflect that the different sizes of the cap proteins allow the RNase A/T1 to approach the RNA to different extents. The Exo ΔRrp4 complex (Fig 2C, lane 10) shows a weaker protection pattern with a greater accumulation of degradation products, suggesting a critical role for Rrp4 in forming an exosome–RNA complex.
Figure 2.
Role of the cap subunits in RNA recruitment. (A) Sodium dodecyl sulphate gel showing the reconstituted Exo9 (lane 1), Exo8 (lanes 2–4) and Exo7 (lanes 5–7) after size-exclusion chromatography. (B) Purified Exo9 (lane 1) and exosome complexes containing any two of the three cap subunits (Exo ΔCsl4, Exo ΔRrp4 or Exo ΔRrp40; lanes 2–4) form stable complexes that bind to Rrp44 (D551N RNase inactive mutant) in pull-down experiments (lanes 5–8). (C) RNA protection assay. The purified exosome complexes were incubated with a 60-nucleotide-long RNA molecule and unprotected RNA was degraded with RNase A/T1. RNA protection by exosomes without Rrp44 is shown in lanes 1–7 and the protection by Rrp44(D551N)-containing exosomes in lanes 8–14. WT, wild type.
The finding that exosome complexes can still form and protect RNA in the absence of any one of the individual cap subunits prompted us to investigate the RNA protection patterns of exosomes lacking two cap subunits. Exosome samples containing only one cap subunit (Exo7 Csl4, Exo7 Rrp4 and Exo7 Rrp40) in the presence and absence of Rrp44(D551N) were tested in RNA protection experiments (Fig 2C). Similar to the wild-type Exo9 complex, in the absence of Rrp44(D551N), Exo7 complexes were unable to protect RNA (Fig 2C, lanes 5–7). In the presence of Rrp44(D551N), the Csl4-containing complex only protected short 9–12 nt fragments, indicating the absence of a stable exosome complex (Fig 2C, lane 12). The 9–12 nt fragments correspond to the protection pattern observed for Rrp44 (D551N) alone (Bonneau et al, 2009). RNA protection of Exo7 Rrp4 in the presence of Rrp44(D551N) also displays bands corresponding to smaller 11–12 nt fragments, as well as weak bands corresponding to the 21–23 nt fragments (Fig 2C, lane 13), suggesting that Rrp4 alone is not sufficient to form a fuctional exosome. In the presence of Rrp40 (Exo7 Rrp40+Rrp44(D551N); Fig 2C, lane 14), however, strong bands corresponding to fragments that are 27–29 nt in length are observed in addition to the 11–12 nt fragment. This suggests that of the cap proteins, Rrp40 alone is sufficient to form a stable exosome that is functional in RNA protection in vitro. Judging from the increased intensity of the Rrp40 band in the Exo7 Rrp40 sample (Fig 2A, lane 7), this subunit might be present in multiple copies, probably through binding to the Rrp4 and/or Csl4 binding sites. This is similar to the archaeal exosome, in which Rrp4 and Csl4 have three identical binding sites (Buttner et al, 2005). This observed plasticity in conformation and in RNA binding by the cap proteins probably reflects the large number of RNA substrates that are recruited and metabolized by the exosome.
EM reconstruction of an RNA-bound exosome
To gain structural insight into RNA recruitment and degradation by the eukaryotic exosome, we collected negative-stain and cryo-electron microscopic data from RNA-bound exosome samples. To verify that RNA binds under the conditions of electron microscopic sample preparation, we first collected negative-stain data on exosome particles bound to 5-nm gold-labelled RNA. The large gold particles allow for visualization of single RNA molecules (Fig 3A). The micrographs showed that every gold-labelled RNA molecule is associated with an exosome complex. The gold particles were visible mostly at the centre of the exosome complexes where the central channel is located.
Figure 3.
Structural evidence of RNA binding at the channel entrance. (A) Negative-stain micrograph of exosomes bound to gold-labelled RNA. The 5′ biotinylated RNA is coupled to streptavidin covalently linked to 5-nm gold particles (black dots). (B) Top view of cryo-electron microscopy maps of apo (left) and RNA-bound exosome (right) with fitted exosome subunits. The extra density located at the entrance of the central channel in the RNA-bound map is shown in red. One loop of Rrp4 (shown in black) is in close proximity to this density and contains conserved residues. (C) Top view of negative-stain electron microscopy maps of apo (left) and RNA-bound exosome (right), with extra density present in the RNA-bound map shown in red. (D) Sequence alignment of the Rrp4 loop in close proximity to the extra density observed in the RNA-bound exosome (loop coloured black in B). Saccharomyces cerevisiae, Schizosaccharomyces pombe, Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans and Sulfolobus solfataricus sequences are aligned. Conserved basic residues are shown in blue and conserved acidic residues are shown in red. (E) Growth phenotypes resulting from the expression of plasmid-borne wild type (WT) and R149E, R150D, K151D mutant Rrp4 in a yeast strain, in which the chromosomal Rrp4 copy is under the control of a doxycycline-repressible promoter. Alleles were assessed in the presence or absence of doxycycline (DOX) after incubation for 72 h at 30°C. −LEU, without leucine.
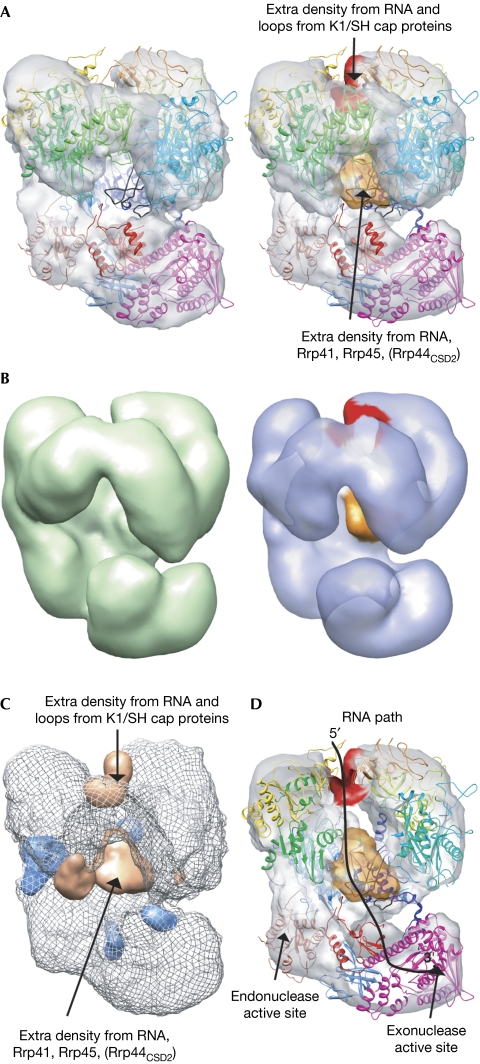
Next, negative-stain and cryo-electron microscopic data were collected from RNA-bound exosomes and three-dimensional maps were reconstructed at 17 and 12 Å resolution, respectively (supplementary Figs S2C,D online). The RNA molecules have 30–40-nt-long single-stranded RNA extensions at the 3′-end, but contain bulky 5′-ends that are predicted to stall at the exosome channel entrance. A pseudo-atomic model of the RNA-bound exosome was obtained for the cryo-electron microscopic map using fitting procedures, as described for the apo exosome. Both the negative-stain and cryo-electron microscopic three-dimensional reconstructions of the substrate-bound exosome show extra density at the channel entrance compared with the reconstructions of the apo exosome (Fig 3B,C). Class averages of negative-stain apo and RNA-bound exosomes also show this extra density (supplementary Fig S4 online) that probably corresponds to bound RNA as well as the RNA-binding loops of Rrp4, Rrp40 and Csl4. Interestingly, the RNA density is located in close proximity to a loop of Rrp4 (residues 149–158) that contains well-conserved basic residues that could interact with RNA (Arg 149, Arg 150 and Lys 151; Fig 3D). To investigate the physiological importance of these basic residues, we introduced a wild-type or a mutant (R149E, R150D and K151D) version of Rrp4 into centromeric plasmids in a yeast strain in which the chromosomal Rrp4 copy was controlled by a doxycycline-repressible promoter and tested the growth phenotypes in the presence or absence of doxycycline (Fig 3E). The mutant Rrp4, in contrast to the wild-type allele, did not support the growth, indicating that charged residues in the Rrp4 loop are essential for exosome structure and/or function.
In addition to the density at the entrance channel, extra density is observed inside the central channel of the RNA-bound structures (Fig 4). The extra density is located close to the secondary structure elements of Rrp41 and Rrp45 that contain well-conserved arginines (Arg 95rrp41, Arg 96Rrp41, Arg 106Rrp45 and Arg 113Rrp45) that are shown to be directly involved in RNA binding by the archaeal exosome (Lorentzen & Conti, 2005). In vitro biochemical experiments have also shown that these conserved arginines of Rrp41 are involved in RNA binding in the yeast exosome (Bonneau et al, 2009). As the density inside the channel seems too much to be accounted for by single-stranded RNA alone, it seems likely that it is partly due to loops of Rrp41 and Rrp45 that become ordered on substrate binding. In particular, loops 63–82, 125–138 and 167–175 of Rrp45—which interact with the CSD2 domain in the crystal structure of Rrp41/44/45—are ideally positioned to account for this density. The extra density could therefore, at least partly correspond to CSD2 of Rrp44. Such a rearrangement of CSD2 on RNA binding in Exo10 could explain previous data showing a protection pattern of 11–12 nt for Exo10, while isolated Rrp44 gives a protection pattern of 9–12 nt (Bonneau et al, 2009).
Figure 4.
RNA threading through the exosome channel. (A) Side views of cryo-electron microscopy maps of apo (left) and RNA-bound exosomes (right). The extra densities at the entrance and exit of the channel are coloured in red and orange, respectively. Loops of Rrp45 that become visible in the electron microscopy map on RNA binding are shown in black. (B) Side views of negative-stain electron microscopy maps of apo (left) and RNA-bound exosomes (right). Extra densities present in the RNA-bound map are shown as in A. (C) Difference maps calculated between the RNA-bound and the apo cryo-electron microscopy maps. Positive (density appearing on RNA binding) and negative difference maps are shown in orange and blue, respectively. (D) Slice through the RNA-bound exosome showing the central channel of the complex. Extra densities present in the RNA-bound map are shown in red and orange. The proposed RNA recruitment path through the central channel is shown as a black line.
Taken together, the data implicate an RNA-threading mechanism by the eukaryotic Exosome, mediated by conserved sequence motifs at the entrance to and inside the central channel (Fig 4D). The Exo9 channel could serve as a selection filter that ensures preferential degradation of RNAs with unfolded 3′-ends over that of properly folded RNAs or RNAs that are bound to protein partners. Such a substrate-recruitment model provides an explanation for why the Exo9, despite being catalytically inactive, is conserved. RNA threading appears to be an evolutionarily conserved mechanism used by all exosome-like complexes and might well have been present in an RNA-degrading complex found in an ancestor common to all three domains of life.
Methods
Exosome sample preparation. S. cerevisiae exosome preparation, pull-down experiments and RNA protection assays were carried out as previously described (Liu et al, 2006; Bonneau et al, 2009). The seven-, eight- and nine-subunit exosomes were prepared by incubating purified subunits and subcomplexes, and were further purified by size-exclusion chromatography. In the case of Exo7 Rrp4, Exo7 Rrp40 and Exo8, peak fractions at a similar exclusion volume to that of the Exo9 were combined. For the Exo7 Csl4 complex, analysis of the peak fractions suggested that this complex is not stably associated during size-exclusion chromatography. In the protection assay, the exosome samples were incubated with Rrp44(D551N) in a 1.5:1 molar ratio before labelled RNA was added. For cryo-electron microscopy data collection, the S. cerevisiae exosome sample was diluted to a final concentration of 0.3 mg/ml, and 3.5 μl was put on glow-discharged C-flat grids (CF-2/2-4C-100; Protochips). After 30 s, excess solution was blotted, and the grids were flash frozen in liquid ethane. To obtain the RNA-bound complex, a concentration of 0.3 mg/ml of exosome was incubated with RNA in a 1:1.2 molar ratio (synthesized by Dharmacon, sequence: 5′-CCCCCGAAAGGGGG(A)30-3′) for 45 min at 22°C. The complex was then vitrified as described above.
Electron microscopic data collection and processing. Cryo-electron microscopy data were collected on a Polara cryo-electron microscope operated at 300 kV (FEI) using a 4k × 4k charge-coupled device camera (Gatan). Particles were manually picked in Ximdisp (Crowther et al, 1996), corrected for the effect of the contrast transfer function by phase flipping, filtered between 150 and 8 Å, and normalized in SPIDER (Frank et al, 1996). Image processing and three-dimensional reconstructions were done using SPIDER and IMAGIC-5 (van Heel et al, 1996), and homology models were made with MODELLER-9v6 (Sali & Blundell, 1993). Fitting of atomic structures and models to the electron microscopy maps was done using UCSF Chimera (Goddard et al, 2007) and Flex-EM (Topf et al, 2008). A more comprehensive description of data processing and fitting procedures can be found in the supplementary information online. The cryo-electron microscopy map has been deposited in EMDB with accession numbers EMD_1708 and EMD_1709 for apo and RNA-bound data sets, respectively.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by the 3D Repertoire European Union grant to H.R.S., E.C. and A.D. and the EUxosome European Science Foundation grant to H.R.S. and A.D. Funding by an EMBO installation grant, Polish State grants (N N301 160335 and N N301 251336), as well as a TEAM grant from the Foundation for Polish Science to A.D., by the Wellcome Trust to H.R.S., and by the Max Planck Gesellschaft, the SFB646 and the Leibniz Programme of the German Research Foundation to E.C. is acknowledged. E.L. was supported by the Federation of European Biochemical Societies and Human Frontier Science Program postdoctoral fellowships. M.T. is funded by a Medical Research Council Career Development Award.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E (2009) The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559 [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP (2005) Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP (2002) Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem 277: 21624–21629 [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM (1996) MRC image processing programs. J Struct Biol 116: 9–16 [DOI] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D (2006) Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157: 281–287 [DOI] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Ibrahim H, Wilusz J, Wilusz CJ (2008) RNA recognition by 3′-to-5′ exonucleases: the substrate perspective. Biochim Biophys Acta 1779: 256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT (2006) Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B (2008) Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E (2005) Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell 20: 473–481 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E (2007) RNA channelling by the archaeal exosome. EMBO Rep 8: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Conti E (2008a) Structural organization of the RNA-degrading exosome. Curr Opin Struct Biol 18: 709–713 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E (2008b) Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell 29: 717–728 [DOI] [PubMed] [Google Scholar]
- Lu C, Ding F, Ke A (2010) Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring. PLoS ONE 5: e8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M (2006) Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep 8: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A (2009) The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Raijmakers R, Pruijn GJ (2006) Cell and molecular biology of the exosome: how to make or break an RNA. Int Rev Cytol 251: 159–208 [DOI] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D (2009) The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A (2008) Protein structure fitting and refinement guided by cryo-EM density. Structure 16: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A (2007) Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci USA 104: 16844–16849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.