Uncovering new substrates for Aurora A kinase
Here a new strategy to identify Aurora A substrates and their phosphorylation sites is presented, which generates a list of 90 potential Aurora A targets. Specific phosphorylation of Nuclear and Spindle-Associated Protein (NuSAP) by Aurora A is validated in vivo.
Keywords: Aurora substrates, pattern discovery, phosphorylation motifs
Abstract
Aurora A is a serine/threonine kinase that is essential for a wide variety of cell-cycle-related events, but only a small number of its substrates are known. We present and validate a strategy by which to identify Aurora A substrates and their phosphorylation sites. We developed a computational approach integrating various types of biological information to generate a list of 90 potential Aurora substrates, with a prediction accuracy of about 80%. We also demonstrated the specific phosphorylation of NUSAP (nucleolar and spindle-associated protein) by Aurora A in vivo. Our results provide a means by which to develop an understanding of Aurora A function and suggest unexpected roles for this kinase.
Introduction
The aurora kinases are a family of closely related serine/threonine protein kinases, with three members in metazoans (A, B and C) that are functionally related to different cancers (Mahadevan & Beeck, 2007). In humans, the gene which codes for Aurora A (AurA) is often amplified in primary tumours and cancer cell lines (Bischoff et al, 1998), and AurA and AurB overexpression is associated with poor prognosis in several tumour types (Naruganahalli et al, 2006). The Aurora kinases are important in cell division and the maintenance of genome stability. The functions of each family member are linked to their specific localization at critical cell-cycle times (Vader & Lens, 2008). AurA is a centrosomal kinase that participates in cell-cycle progression and has a well-characterized role in centrosome maturation during late G2 and prophase, as well as in spindle assembly (Sardon et al, 2008). AurB localizes to the nucleus in interphase, to the kinetochores and the spindle midzone in anaphase and is required for chromosome bi-orientation and cytokinesis (Vader & Lens, 2008). AurC acts during meiosis, and its function is closely related to that of AurB in cytokinesis (Slattery et al, 2009). The three Aurora kinases have a similar protein domain organization, with divergent amino-terminal and carboxy-terminal domains and a conserved catalytic domain. AurA and AurB show specificity for certain substrates, but also share some of them in vitro, including HH3 and MBP. A consensus motif for these kinases was first determined for Ipl1, the only Aurora kinase in the budding yeast Saccharomyces cerevisiae (Cheeseman et al, 2002).
Although the participation of Aurora kinases in the cell cycle is well established, their mechanism of action is poorly understood and the relatively small number of their substrates that are known do not account for their many functions. Thus, the identification of Aurora substrates is important for a global understanding of their functions.
Here, we present and validate a new strategy for identifying new substrates for AurA kinase. By analysing the available data on AurA substrates and their phosphorylation sites, we developed a computational approach that uses distinct types of biological information to generate a ranked list of potential Aurora substrates. We then validated our predictions in experiments with a group of candidates by using in vitro kinase assays and mass spectrometry analyses. Finally, we also demonstrated the specific phosphorylation of NUSAP (nucleolar and spindle-associated protein) by AurA in vivo.
Results And Discussion
Identification of potential substrates for AurA
The consensus motif of the Aurora kinase was first characterized in S. cerevisiae (Cheeseman et al, 2002) as [KR].[ST][ILV]. Two different patterns have been proposed for the human AurA kinase: [KNR]R.[ST][AFILMV] (Ferrari et al, 2005) and R.[ST][ILV] (Ohashi et al, 2006), the latter being a more restricted form of the yeast motif. However, these motifs only match 8, 5 and 7, respectively, of the 19 known phosphorylation sites of AurA. To broaden the coverage, we aligned the site-containing regions from the list of known AurA phosphorylation sites and defined a more lenient motif—[KR].[ST][^P]—in the light of a report by Ferrari et al (2005) showing that proline at position +1 abolishes phosphorylation by AurA. This ‘notP motif' matches 13 of the 19 sites. It also matches most of the known substrates of AurB (15 of 18 known AurB phosphorylation sites). This is not surprising, as the motif is an extension of the yeast pattern and Ipl1—the only yeast Aurora kinase—shares functions and substrates with both AurA and AurB. In fact, recent data suggest that in vertebrates, the substrate specificity of these two kinases is primarily a consequence of their different subcellular locations and/or interactions with specific cofactors (Fu et al, 2009; Hans et al, 2009).
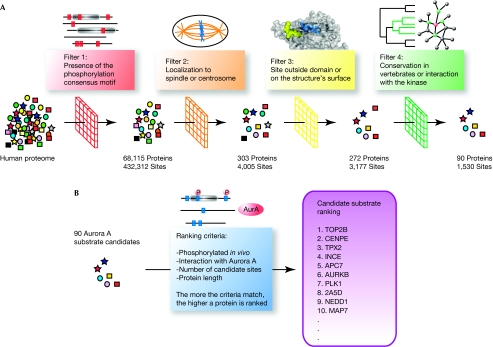
The notP motif matched 432,312 sites in 68,115 of 77,683 non-redundant protein sequences in the human proteome, many of which are probably false positives. We therefore developed a filtering strategy with which to increase the specificity of our predictions by integrating different types of contextual data including subcellular localization, in vivo phosphorylation, interactions with the kinase, site accessibility and motif conservation across vertebrate species (Fig 1).
Figure 1.
Schematic representation of the bioinformatics approach. (A) Candidate substrate selection. Aurora substrate candidates were selected on the basis of a series of filters applied to the whole human proteome: presence of an Aurora phosphorylation motif in the sequence, localization to the centrosome or the spindle, accessibility of the consensus motif and conservation of the potential phosphorylation site among vertebrates. (B) Ranking of candidate Aurora substrates (see main text for details).
As AurA specifically localizes to the centrosomes, spindle poles and spindle microtubules (Carmena & Earnshaw, 2003), we focused our analysis on the subset of proteins associated with these structures, as identified by large-scale proteomic analyses of human cells (Andersen et al, 2003; Sauer et al, 2005; Nousiainen et al, 2006). We also included proteins that were annotated as localizing to the spindle in the GeneOntology database (Ashburner et al, 2000; Fig 1), some of which localize to the kinetochores and could therefore be substrates of AurB. By reducing the number of potential candidates from 77,683 human proteins to 308, we restricted our analysis to the specific biological context of the Aurora kinases, thereby reducing the possibility of false-positives, but also risking discarding some substrates.
As phosphorylation motifs need to be accessible to their corresponding kinase, they are often located in hinge and loop regions outside globular domains (Neduva et al, 2005; Gnad et al, 2007). We therefore used information from high-resolution three-dimensional structures—when available—to calculate the accessibility of each site and retained only those on the protein surface. If no reliable structural information was available we excluded the sites inside domains, although this means that we potentially discarded a few real ones. Phosphorylation sites that were predicted to be outside globular domains were always considered accessible (Fig 1).
Functionally relevant phosphorylation motifs are expected to be better conserved during evolution than those that appear randomly (Malik et al, 2008). We therefore included a further filtering step, to eliminate biologically irrelevant hits. To assess the conservation of candidate sites, we created sets of orthologues for all proteins in our spindle/centrosome set from the Ensembl database (Flicek et al, 2008). We limited the conservation analysis to include only vertebrates, because short motifs usually evolve quickly and can therefore only be detected in closely related species (Neduva et al, 2005). As phosphorylation sites often occur outside domains (Gnad et al, 2007) and are fast evolving (Neduva et al, 2005), typical multiple sequence alignment strategies would not provide sensible results (Chica et al, 2008; Perrodou et al, 2008). We therefore devised two alternative ways of assessing motif conservation.
Our first strategy was to examine whether the motif was found at a similar position in the human sequence and the corresponding orthologue, allowing a deviation of 1% in the position. As the relative positions of sites might not be preserved across domain insertions and deletions or gene fusion events, this method could only be applied to orthologues of similar length.
Our second strategy used the protein Basic Local Alignment Search Tool (BLAST; Altschul et al, 1997) to search for locally matching sequence stretches in orthologues containing the phosphorylation motif. As protein BLAST produces local alignments of the residues surrounding the given phosphorylation sites, this approach is also applicable in cases of non-uniform changes in sequence length in some species, and thus did not require any restriction for selecting the set of orthologues. Similar approaches have been used by Malik et al (2008) to identify functionally relevant phosphorylation sites.
We only considered sites in the top five ranks for each substrate candidate that were conserved in at least 90% of the orthologues by one of our methods. Among the known AurA substrates, 12 of 19 phosphorylation sites were ranked in the top five according to the notP motif conservation. The complete filtering procedure reduced the number of candidate substrates to 90, with 347 potential phosphorylation sites conserved (Fig 1).
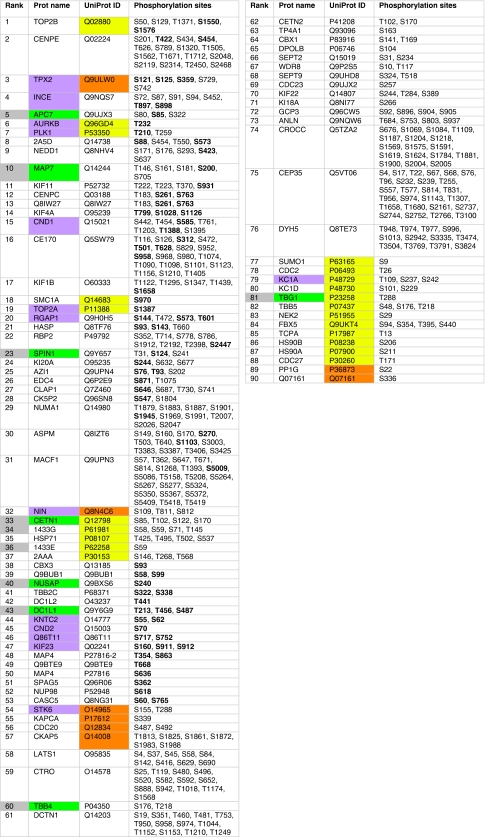
Finally, we ranked the candidates by using a scheme that awards one point to proteins (i) interacting directly or indirectly with AurA, (ii) with conserved potential phosphorylation sites according to the presence-based method or (iii) with conserved potential phosphorylation sites according to the BLAST-based method. Two extra points were attributed to proteins with a notP motif coinciding with a known in vivo phosphorylated site (Nousiainen et al, 2006; Dephoure et al, 2008), which strongly indicates it is a real substrate. In substrate candidates with the same number of points, we considered the number of sites phosphorylated in vivo and conserved according to both the presence- and BLAST-based method (‘threefold overlap'), as well as the number of sites fulfilling at least two of these three criteria (‘twofold overlap'). The final ranked list of candidate substrates for Aurora kinases and their predicted phosphorylation sites are shown in Fig 2.
Figure 2.
Ranking of the 90 substrate candidates for Aurora kinases. Known substrates of AurA or B are shown in purple. Experimentally tested candidates are shown in grey, with new confirmed substrates shown in green. Known direct or indirect AurA interactors are shown in orange and yellow, respectively. All accessible and conserved matches to the notP motif ([KR].[ST][^P]) are listed and known in vivo phosphorylation sites are marked in bold.
Experimental validation of the predictions
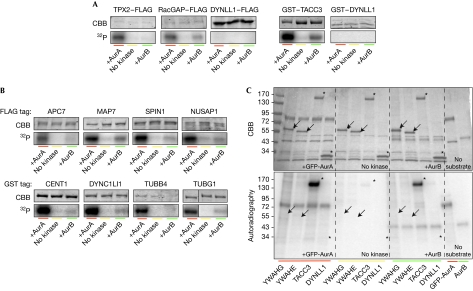
To test the predictions of our analyses we performed AurA in vitro phosphorylation assays on a set of ten candidates selected from throughout the ranked list on the basis of the availability of full-length complementary DNA clones in appropriate vectors for expression in mammalian cells (Fig 2). Kinase assay conditions were set on the known AurA substrates TPX2 (Kufer et al, 2002) and TACC3 (Kinoshita et al, 2005), each with a different tag (TPX2–FLAG and GST–TACC3). As a negative control, we used DYNLL1—a protein that is present in the spindle/centrosome proteome but lacks the consensus motif for AurA phosphorylation (Fig 3A).
Figure 3.
Validation of candidate substrates by in vitro phosphorylation assays. (A) Specificity of the in vitro Aurora kinase assay tested on known AurA substrates and on a protein not containing the phosphorylation motif (DYNLL1, negative control). Substrates were expressed in human cells, and pulled down by their FLAG or GST tag. Precipitated proteins were mixed with AurA, AurB or only buffer in the presence of 32P-ATP, then separated by SDS–PAGE and stained with Coomassie brilliant blue. Incorporated 32P was visualized by autoradiography. The gel fragments shown belong to representative experiments repeated at least three times. Exposure time of the autoradiographs was adapted for each substrate to allow the visualization of the 32P incorporation. (B) AurA phosphorylation assays were performed as in A on potential substrates from the candidate list. (C) Autoradiography (32P) and Coomassie staining of an in vitro kinase assay gel, where YWHAG (14-3-3γ) and YWHAE (14-3-3ɛ) were tested in comparison with a positive (TACC3) and a negative control (DYNLL1). Arrows and asterisks, positions of candidate proteins. APC7, anaphase-promoting complex 7; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
The kinase assays showed that 8 of the 10 candidate substrates incorporated 32P on incubation with AurA (Fig 3B), whereas 2—YWHAG (14-3-3γ) and YWHAE (14-3-3ɛ)—did not, suggesting that they are not AurA substrates in vitro (Fig 3C). In order to quantify the results and compare ATP incorporation between substrates, we normalized the autoradiography band signals to exclude differences due to the time of film exposure and the amount of protein (supplementary Fig S1A online), concluding that CENT1, DYNC1LI1, TUBB4, TUBG1, MAP7, NUSAP and SPIN are phosphorylated by AurA in vitro. However, we found no correlation between the number of predicted phosphorylation sites of a protein and the amount of ATP that it incorporated, suggesting that not all the sites are phosphorylated and/or that they show different affinities for the kinase. As the level of anaphase-promoting complex 7 (APC7) phosphorylation was much lower than that for the other candidates, we confirmed phosphorylation at the predicted site Ser 85 in the AurA-treated sample by using mass spectrometry (supplementary Fig S1B,C online).
As the notP motif also matches the AurB recognition sites, we tested whether the selected substrates were also phosphorylated by AurB in vitro, with RacGAP1 as a positive control (Minoshima et al, 2003). We found that AurB weakly phosphorylated all confirmed AurA substrates (Fig 3B) and did not phosphorylate YWHAG and YWHAE (Fig 3C). However, AurA and AurB phosphorylation concentrations should not be directly compared, as it has been shown that AurB activity in vitro is lower than that of AurA (Eyers et al, 2005). Overall, our experimental validation assays showed that 8 of 10 candidate proteins are Aurora substrates in vitro—a prediction success rate of about 80%. The accuracy with respect to individual phosphorylation sites remains to be assessed.
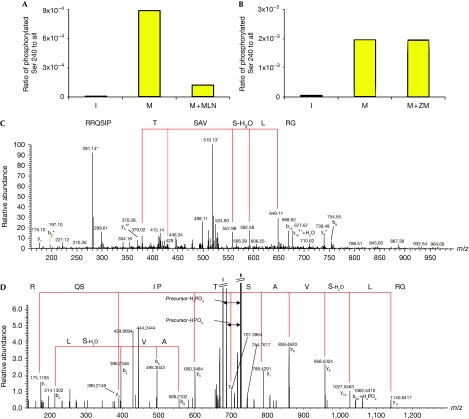
To determine whether these candidates are AurA substrates in vivo, we immunoprecipitated recombinant NUSAP from transfected interphase cells or mitotic monastrol-arrested cells, with or without the selective AurA inhibitor MLN8237 (Fig 4). The mitotic or interphase state of the cells in the different conditions was confirmed by fluorescence-activated cell sorting analysis (supplementary Fig S2 online). Mass spectrometry analyses identified a phosphorylated peptide—containing the predicted site Ser 240—in the sample obtained from the mitotic cells (Fig 4C). This phosphopeptide was absent in interphase cells and strongly reduced in mitotic cells treated with the inhibitor (Fig 4A). To test whether AurB could also phosphorylate NUSAP, we performed an MS/MS analysis on recombinant NUSAP incubated with the AurB inhibitor ZM447439 (Fig 4D). We did not find any difference between the amounts of phosphorylated peptide observed in mitotic cells with or without inhibitor (Fig 4B), suggesting that AurB does not phosphorylate NUSAP Ser 240. These data suggest that NUSAP is an AurA substrate in vivo.
Figure 4.
NUSAP mitotic phosphorylation at Ser 240 correlates with Aurora A activity. Protein samples of FLAG–NUSAP immunoprecipitated from I, M and M+MLN or with M+ZM were analysed using LC-MS/MS, focusing on the predicted phosphorylated residue Ser 240. The histograms (A, B) show the calculated ratios based on peptides carrying the phosphorylated Ser 240 compared with all matched peptides containing this residue. (C, D) MS/MS spectra of two peptides matched the triply charged phosphorylated peptide GRLSphosVASTPISQRR and the doubly charged phosphorylated peptide GRLSphosVASTPISQR, respectively. In both spectra, y-ion series are marked with red lines indicating matched amino-acid sequences (C: doubly charged y-ion series and D: singly charged y-ion series) together with some b ions. The conversion of phosphorylated Ser 240 into dehydroalanine is marked in the spectra. I, interphase cells; LC-MS/MS, liquid chromatography-mass spectrometry; M, mitotic cells arrested with monastrol; M+MLN, mitotic cells co-treated with monastrol and MLN8237; M+ZM, monastrol and ZM447439.
One of our other predictions—that CENP-E Thr 422 is phosphorylated by AurA and AurB—has been validated in vivo (Kim et al, 2010) since we performed our analyses, providing further support for our strategy.
Uncovering new functions for Aurora kinases
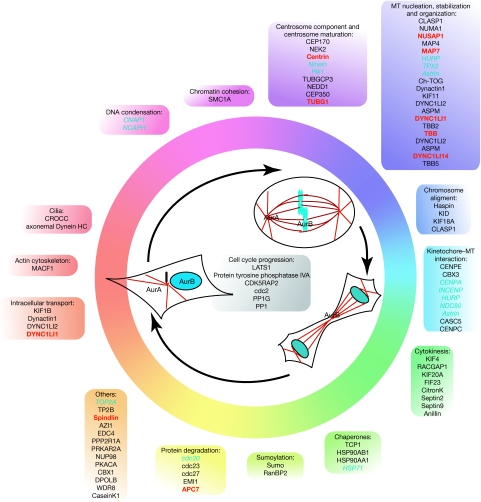
To explore the different roles of the putative Aurora substrates, we grouped them according to their cellular functions using the information in the UniprotKB database (Fig 5). This analysis showed that several of the new candidates clustered into groups, defining specific functions previously shown to be regulated by the Aurora kinases—for example, centrosome maturation and chromosome alignment. The clustering of potential substrates suggests that there might be a coordinated regulation of different proteins for any given function. Our analysis also indicates that the Aurora kinases might regulate yet more processes, such as sumoylation and protein degradation. Indeed, we found five different proteins of the APC-dependent degradation machinery in our candidate list, including APC7, which we have validated in vitro. Another interesting functional cluster contains proteins related to cilia, and a recent work has shown that AurA is essential for cilia reabsorption before the entry of quiescent cells into mitosis (Pugacheva et al, 2007). The putative phosphorylation of two cilia proteins—CROCC and DNAHC5—by AurA suggests a wider role for this kinase in the regulation of cilia function. The identification of new putative substrates for the Aurora kinases will enable not only the further investigation of some of their known functions, but also the examination of their potential regulatory role in other cellular processes.
Figure 5.
Functional distribution of the predicted Aurora substrates. Known and candidate substrates classified according to their functions throughout the cell cycle. In the cell schemes, microtubules appear in red and DNA in blue. In the text boxes, blue, red and black indicate known, validated and proposed Aurora substrates, respectively.
CONCLUSIONS
We describe a new approach to identify candidate substrates for the Aurora kinases. Our in vitro results and the in vivo validation of NUSAP and CENP-E suggest that our method has predictive potential. As a consequence of our stringent filtering criteria—designed to maximize specificity—we might have excluded some Aurora substrates, the total number could therefore be more than 100. Although we have only applied our method to Aurora, it could be adapted to identify substrates of other kinases. To estimate its general applicability, we investigated how many of the 57 known mitotic kinases (Schmit & Ahmad, 2007) have enough phosphorylation data (at least ten different substrates) available from which to derive reliable phosphorylation patterns. We found that it would be possible to apply our strategy to 14 of them (25%). Considering all 113 human kinases implicated in mitosis—including the pleiotropic kinase families protein kinase A and mitogen-activated protein kinase—we can automatically use our strategy on 32 of them, increasing the potential applicability of the method to about 30%. Overall, we anticipate that the general approach presented here and the strategies to align functional motifs will help to decipher the many cellular functions that are regulated by kinases, including some that are yet to be discovered.
Methods
All the methodological details referring to the biological data, computational analyses, protein expression and purification, in vitro kinase assays, immunoprecipitation of NUSAP from interphase and mitotic cells, and mass spectrometry analyses are provided in the figure legends and the supplementary information (online) accompanying the paper.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Núria Mallol and Leonor Ávila (Centre for Genomic Regulation) for technical support. We acknowledge financial support from the Spanish Ministry of Education and Science through the grants BIO2007-62426 and PSE-010000-2009 (to P.A.), BFU2006-04694, BFU2005-24990-E and CSD2006-00023 (to I.V.). An FPU fellowship to R.A.P. MLN8237 was kindly provided by Selleck Chemicals.
Footnotes
The authors declare that they have no conflict of interest.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Ashburner M et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JR et al. (1998) A homologue of Drosophila Aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J 17: 3052–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Earnshaw WC (2003) The cellular geography of Aurora kinases. Nat Rev Mol Cell Biol 4: 842–854 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G (2002) Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Chica C, Labarga A, Gould CM, Lopez R, Gibson TJ (2008) A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinform 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP (2008) A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers PA, Churchill ME, Maller JL (2005) The Aurora A and Aurora B protein kinases: a single amino acid difference controls intrinsic activity and activation by TPX2. Cell Cycle 4: 784–789 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Marin O, Pagano MA, Meggio F, Hess D, El-Shemerly M, Krystyniak A, Pinna LA (2005) Aurora-A site specificity: a study with synthetic peptide substrates. Biochem J 390: 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P et al. (2008) Ensembl 2008. Nucleic Acids Res 36: D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Bian M, Liu J, Jiang Q, Zhang C (2009) A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci USA 106: 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M (2007) PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol 8: R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F, Skoufias DA, Dimitrov S, Margolis RL (2009) Molecular distinctions between Aurora A and B: a single residue change transforms Aurora A into correctly localized and functional Aurora B. Mol Biol Cell 20: 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland A, Weijie L, Cleveland D (2010) Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142: 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA (2005) Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol 170: 1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA (2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol 158: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D, Beeck S (2007) Aurora kinase targeted therapeutics in oncology: past, present and future. Expert Opin Drug Discov 2: 15. [DOI] [PubMed] [Google Scholar]
- Malik R, Nigg EA, Korner R (2008) Comparative conservation analysis of the human mitotic phosphoproteome. Bioinformatics 24: 1426–1432 [DOI] [PubMed] [Google Scholar]
- Minoshima Y et al. (2003) Phosphorylation by Aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell 4: 549–560 [DOI] [PubMed] [Google Scholar]
- Naruganahalli KS, Lakshmanan M, Dastidar SG, Ray A (2006) Therapeutic potential of Aurora kinase inhibitors in cancer. Curr Opin Investig Drugs 7: 1044–1051 [PubMed] [Google Scholar]
- Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB (2005) Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol 3: e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R (2006) Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA 103: 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Sakashita G, Ban R, Nagasawa M, Matsuzaki H, Murata Y, Taniguchi H, Shima H, Furukawa K, Urano T (2006) Phospho-regulation of human protein kinase Aurora-A: analysis using anti-phospho-Thr288 monoclonal antibodies. Oncogene 25: 7691–7702 [DOI] [PubMed] [Google Scholar]
- Perrodou E, Chica C, Poch O, Gibson TJ, Thompson JD (2008) A new protein linear motif benchmark for multiple sequence alignment software. BMC Bioinform 9: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA (2007) HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T, Peset I, Petrova B, Vernos I (2008) Dissecting the role of Aurora A during spindle assembly. EMBO J 27: 2567–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH (2005) Proteome analysis of the human mitotic spindle. Mol Cell Proteomics 4: 35–43 [DOI] [PubMed] [Google Scholar]
- Schmit TL, Ahmad N (2007) Regulation of mitosis via mitotic kinases: new opportunities for cancer management. Mol Cancer Ther 6: 1920–1931 [DOI] [PubMed] [Google Scholar]
- Slattery SD, Mancini MA, Brinkley BR, Hall RM (2009) Aurora-C kinase supports mitotic progression in the absence of Aurora-B. Cell Cycle 8: 2984–2994 [PubMed] [Google Scholar]
- Vader G, Lens SM (2008) The Aurora kinase family in cell division and cancer. Biochim Biophys Acta 1786: 60–72 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.