Abstract
In the second article of the ‘Food and Science' series, Paul Chambers and Isak Pretorius explain the central role of yeast in wine making and how biotechnology can contribute to improving the quality of wine.
Wine has been with us since the dawn of civilization and has followed humans and agriculture along diverse migration paths (Fig 1). Serendipity presumably played a part in its genesis more than 7,000 years ago: damaged grapes spontaneously fermented in harvesting vessels; curious farmers tasted the resultant alcoholic beverage; the curious farmers liked what they tasted and enjoyed its effects; said farmers preferred fermented grape juice to the unfermented fruit. The fate of the grape was sealed.
One might argue that the most important test tube in the birth and growth of the modern life sciences is the fermenter…
Figure 1.
A generalized scheme of the spread of Vitis vinifera noble varieties of grapevine and winemaking from their centre of origin in Asia Minor to other parts of the world.
One might argue that the seeds of science and technology, particularly biotechnology, were also sown at this time. Empirical observations of natural events and processes were harnessed in repeat ‘experiments'—which is to say, vintages—and improvements were made by trialling modifications to practices, retaining those that were beneficial and discarding failures, with the results communicated down through the generations. At that time, there was no EMBO reports or alternative means by which to facilitate horizontal dissemination of information, but the principle of development—sans peer review—is clear: experimentation and invention lead to progress—technological and otherwise—and new knowledge is shared and built upon.
Of course, early inventions and innovations in grape and wine production were based on little or no knowledge of the biology of grapevines or the microbes that drive fermentation. In fact, it would be several thousand years before it was even known that microscopic organisms exist: using a primitive microscope, Antonie van Leeuwenhoek observed cells for the first time in 1680 (Fig 2).
Figure 2.
Selected milestones that mark the path of research in microbiology and yeast biology that have affected, directly or indirectly, wine science and winemaking.
Scientific knowledge grows at an exponential rate, and nowhere is this more evident than in the historical milestones of chemistry and biology that have shaped our understanding of the biology of the microorganisms that drive fermentation (Fig 2). This progress has been adorned with some of the most significant names in the chemical and biological sciences, including van Leeuwenhoek, Lavoisier, Gay-Lussac, Pasteur, Buchner and Koch. One might argue that the most important test tube in the birth and growth of the modern life sciences is the fermenter, and the most important model organism has been the yeast Saccharomyces cerevisiae—commonly known as baking, brewing or wine yeast. As readers might know, this is exemplified in the origin of the word enzyme—‘en' meaning within and ‘zyme' meaning leaven. Yeast has been integral to pioneering work in microbiology and biochemistry, particularly in the fields of metabolism and enzymology (Barnett, 1998, 2000; Barnett & Lichtenthaler, 2001).
Throughout the early decades of the twentienth century the place for S. cerevisiae in fundamental research was affirmed, and there are several good reasons for this. Our close relationship with this yeast in food and beverage production over millennia tells us that it is safe to work with; as confirmed by its ‘Generally Recognised as Safe' designation by the US Food and Drug Administration. In addition, it is inexpensive, easy to grow and can be stored for long periods in suspended animation. Perhaps the most important thing is that it has accessible genetics that can be followed through sexual and asexual cycles (Barnett, 2007).
The 1970s set the stage for another explosion of knowledge, sparked by the advent of gene technology and driven by a convergence of genetics, biochemistry, cell biology, microbiology, physical and analytical chemistry, as well as computing brought together under the banner of molecular biology (Fig 3). Yeast molecular biology was established when Gerald Fink's group in the USA demonstrated that yeast could be transformed with foreign DNA (Hinnen et al, 1978). In the same year, Jean Beggs in the UK developed a shuttle vector between Escherichia coli and S. cerevisiae that enabled cloning in yeast (Beggs, 1978). The research community now had a eukaryotic host that was amenable to genetic engineering, benefiting both fundamental research and offering the potential of precise engineering of novel strains for industrial applications. It was the first host cell for industrial-scale production of a recombinant vaccine against hepatitis B and a recombinant food-grade enzyme, chymosin, which is used in cheese processing (Pretorius et al, 2003).
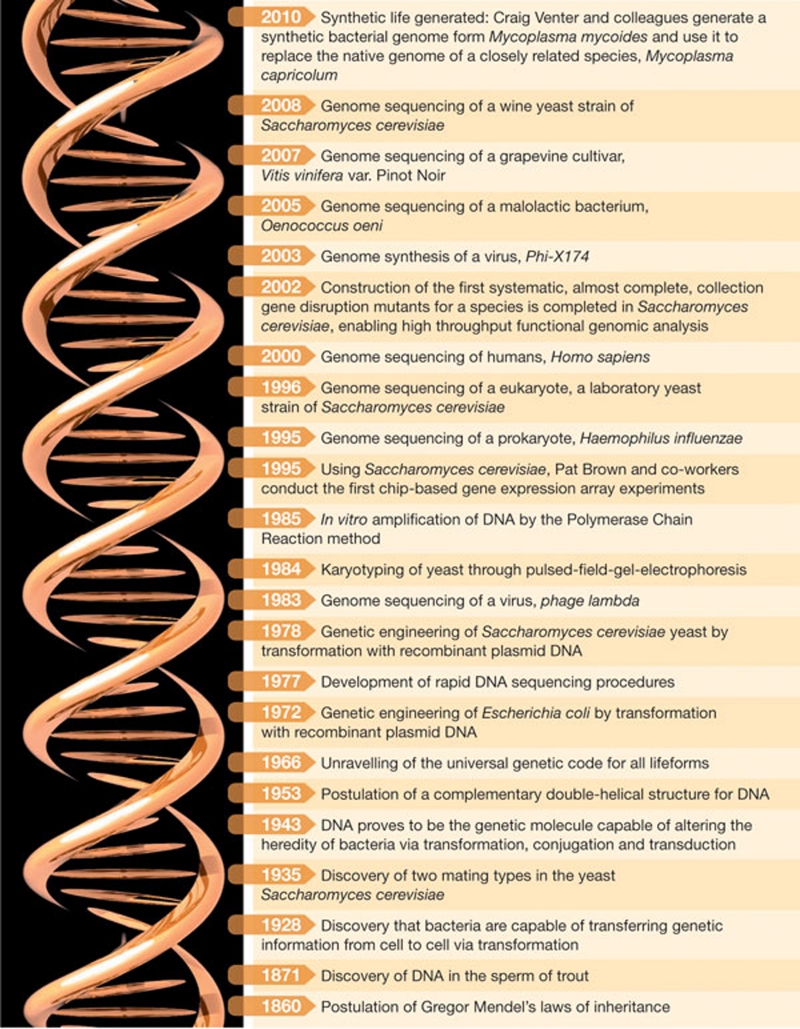
Figure 3.
Selected milestones that mark the path of research in genetics and molecular biology that have affected, directly or indirectly, wine science and winemaking.
Ever since, S. cerevisiae has been one of the most important model organisms in molecular biology and emerging fields; breakthroughs and technological advances in molecular, systems, and now synthetic biology rarely happen without S. cerevisiae figuring somewhere prominently in the story (Fig 3). The international yeast science community has been particularly progressive and proactive in establishing large collaborative projects and building resources that are available to the scientific community. S. cerevisiae was the first eukaryote to have its genome sequenced (Goffeau et al, 1996), a feat that was achieved through an international effort that involved 600 scientists, which paved the way for the first chip-based gene array experiments (Schena et al, 1995). It was the first organism to be used to build a systematic collection of bar-coded gene deletion mutants (Winzeler et al, 1999; Giaever et al, 2002), in which there are deletion strains for most of the open-reading frames in the S. cerevisiae genome. This has enabled high-throughput functional-genomic experiments, and anyone seeking information on just about any aspect of S. cerevisiae biology has access to the amazing community resource: the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/).
All of this is important to wine research; our winemaking workhorse is centre stage in thousands of research projects worldwide, so we know more about this humble eukaryote than any other organism on the planet. It is therefore unsurprising that wine research has benefited enormously from the privileged place that S. cerevisiae occupies in life sciences research. This is particularly evident in the impact that advances in molecular biology and related fields have had on winemaking.
In the hands of molecular biologists, S. cerevisiae is the most tractable of organisms; it is amenable to almost any modification that modern biology can throw at a cell. This makes it an ideal host for generating variants with improved and even exotic phenotypes that will benefit winemaking. The following gives some examples of current research and directions in this field.
In modern winemaking, fermentations are driven largely by single-strain inoculations; pure cultures of selected strains of S. cerevisiae are added to grape must as soon as possible after crushing. This ensures greater control of vinification, leads to more predictable outcomes and decreases the risk of spoilage by other microorganisms. There are many—probably hundreds of—different yeast strains available, and the winemaker's choice can substantially effect the quality of the wine (Lambrechts & Pretorius, 2000; Swiegers et al, 2005).
One of the reasons for the yeast-induced variation in wine quality is that, during fermentation, S. cerevisiae produces an abundance of aroma-active secondary metabolites and releases many aroma compounds from inactive precursors in grape juice, which greatly affect the sensory properties of the wine (Swiegers & Pretorius, 2007). Thus, any genetic variation in wine yeast that affects the production or release of sensorially important molecules will affect wine quality. In this context it has been demonstrated, for example, that different commercial yeast strains generate wines with very different profiles of volatile thiols (Swiegers et al, 2009). These thiols—which are present in grape juice as non-volatile cysteinylated precursors (Tominaga et al, 1998)—are often described as ‘passionfruit', ‘tropical fruits' and ‘citrus' by tasters, flavours that are particularly important in wine varieties such as Sauvignon Blanc (Dubourdieu et al, 2006).
Molecular biology and its tools are crucial to our understanding of the genetic and molecular bases of yeast-driven volatile thiol release from non-volatile precursors in grape juice. Howell et al (2005) have used bioinformatic tools and the SGD to identify candidate S. cerevisiae carbon–sulphur lyase genes that might be involved in the release of volatile thiols from cysteinylated precursors during fermentation. The researchers used targeted gene deletion to remove these candidate carbon–sulphur lyases from the wine and laboratory yeast strains, and they identified four genes that potentially contribute to the release of these important aroma molecules.
Swiegers et al (2007) then engineered a wine yeast, VIN13, to constitutively express a carbon–sulphur lyase gene, tnaA, from E. coli. Sensory analysis revealed that, compared with its non-engineered relative, this transgenic yeast, VIN13 (CSL1), had a positive impact on the release of volatile thiols from a Sauvignon Blanc grape juice. The authors commented that wine assessors preferred the VIN13 (CSL1)-derived experimental wines to the relatively neutral VIN13-derived wines.
A similar approach has been used to engineer yeasts for the enhanced production of fruity esters (Lilly et al, 2006a) and to increase the production of higher, fusel alcohols (Lilly et al, 2006b)—all of which contribute to the flavour profiles of wines. Although this work is in the early stages of development, it shows the value of yeast molecular biology, and the amazing resources that come with it.
Wine alcohol content is of growing importance to the wine industry. In some wine regions, it has been increasing during recent decades (Godden & Muhlack, 2010). The main reason for this increase is that grapegrowers tend to leave fruit on the vine as long as possible to increase fruity characters—which develop as berries mature—and reduce undesirable ‘green' characters. This practice, however, produces fruit with a higher sugar content, which translates to higher ethanol concentrations in the wine.
A recent review by Kutyna et al (2010) discusses several metabolic engineering strategies that have been explored to generate wine yeasts that can divert some carbon metabolism away from ethanol production, with the aim of decreasing ethanol yields during vinification. Understanding the central metabolism of yeast and the genes that drive it has been crucial to this work. Candidate genes that are likely to influence ethanol yields can be identified from a range of sources, including the SGD, and then manipulated and cloned as required. Several laboratories have targeted the glycerol-3-phosphate dehydrogenase isozymes GPD1 and GPD2, which divert carbon from glycolysis to glycerol production (Michnick et al, 1997; Remize et al, 1999; de Barros Lopes et al, 2000).
Increased expression of either of the GPD paralogues increased glycerol and decreased ethanol yields. However, increased Gpd activity also led to increased amounts of acetic acid in the fermentation product. This was probably owing to rectification—by one or more of the five aldehyde dehydrogenase isozymes—of a redox imbalance that resulted from excessive Gpd-driven oxidation of NADH. Aldehyde dehydrogenase isozymes drive the oxidation of acetaldehyde to acetic acid with concomitant reduction of coenzymes NAD+ or NADP, depending on which isozyme is involved (Navarro-Aviño et al, 1999). This might be good for a yeast cell struggling with an imposed redox imbalance, but an increase in acetic acid production is not good news for winemakers; excessive vinegar is not desirable in wine. This problem was alleviated by knocking out one of the five aldehyde dehydrogenase isozymes, ALD6 (Eglinton et al, 2002; Cambon et al, 2006).
Similar approaches have targeted S. cerevisiae pyruvate decarboxylase isozymes, alcohol dehydrogenase isozymes and glycerol transporters, leading to increased glycerol yields and reduced ethanol production (Kutyna et al, 2010). However, while there are probably several good candidate ‘low-ethanol' wine yeast strains sitting in various labs around the world, none have been tested in commercial-scale, industrial fermentations. This is largely because consumers are generally unaccepting of genetically modified organisms (GMOs) in foods and beverages.
Another area of ongoing research in wine yeast molecular biology is the development of strains that flocculate—that is, form clumps—at the end of fermentation. This facilitates the process of settling them out of suspension and separating them from the wine, thereby reducing the need for clarification. The timing of flocculation is crucial; it must not happen too early, as yeast in large flocs are inefficient at sugar utilization and can generate suboptimal—stuck or sluggish—fermentations (Pretorius, 2000).
Generally, wine yeasts are not good at flocculation; they do not form large clumps that settle out of suspension. Many years of research using laboratory strains of S. cerevisiae led to the identification and characterization of genes that encode cell-surface glycoproteins—including lectin-like flocculins—that cause, among other things, flocculation and subsequent settling to the bottom of the fermentation vessel (Pretorius, 2000).
Recent findings have identified a problem with extrapolating basic research on laboratory strains to those used in industry; yeasts domesticated for different purposes have different phenotypes. Work by Govender et al (2008) on the flocculation genes FLO1, FLO5 and FLO11, for example, demonstrated the potential ability of engineered ADH2- or HSP30-promoter/FLO gene combinations to switch on flocculation at the end of fermentation; ADH2 and HSP30 are both upregulated in stationary-phase cells, so their promoters are suitable candidates to drive the expression of genes in later stages of wine fermentation.
The results of this work were promising, but, when they were carried over to wine yeast, the findings were rather different. There were even substantial differences between wine yeast strains, leading the authors to caution that “optimisation of the flocculation pattern of individual commercial strains will have to be based on a strain-by-strain approach” (Govender et al, 2010). Nonetheless, controlled expression of FLO genes at the end of fermentation remains a plausible technique for improving the performance of wine yeast, but the strategies required to achieve a desirable outcome might be more complex than was originally thought.
While the complexity of biological systems is a cause for excitement and wonder to most biologists, it can make engineering novel strains for industrial applications trickier than molecular biology and biotechnology textbooks might suggest. For those of us working on industrial yeast strains, it might be pertinent to directly tackle the issue of complexity and use systems biology approaches to better understand the workings of yeast metabolism. This should lead to more accurate modelling of metabolic processes for better-informed manipulations, to achieve targeted, predictable outcomes.
However, molecular biologists face one important obstacle to this progress: near worldwide refusal to permit the use of GMOs in the production of foods and beverages…
S. cerevisiae has been at the forefront of ‘-omics' research. This provides us with enormous opportunities to improve understanding of wine yeast complexity, which, in turn, will inform the design of new strains for industrial applications. Increased and improved knowledge from a huge number of studies investigating strains of S. cerevisiae at the various -omic levels gives wine yeast scientists a head start in this field (Borneman et al, 2007; Petranovic & Vemuri, 2009).
One of the most interesting developments has come from the sequencing of a wine yeast genome, and its comparison with the genomes of a laboratory strain and an opportunistic pathogenic S. cerevisiae (Borneman et al, 2008). The authors found a difference of about 0.6% in sequence information between the wine yeast and the other strains. They also found, perhaps more importantly, 100 kb of additional genome sequence in the former; enough to carry at least 27 genes. Open reading frames (ORFs) in the additional sequences do not resemble anything found in other strains of S. cerevisiae, but seem to be similar to genes found in distant fungal relatives. BLAST searches have indicated that some of the genes that are specific to wine yeast are similar to those encoding cell-wall proteins. This might contribute to the greater robustness of wine yeast, compared with laboratory strains. Other genes might encode proteins associated with amino acid uptake, which is significant in the context of wine sensory attributes; amino acid metabolism is central to the production of many sensorially important volatile aroma compounds.
Novo et al (2009) published similar findings from a different wine yeast strain (EC1118) and suggested that the extra sequence was probably the result of horiziontal gene transfer. Further work using functional genetics—to determine the effects of knocking out and overexpressing the ORFs—should enable characterization of the phenotypes of these ORFs, determine their relevance in the context of winemaking and might also reveal their origins.
There have also been numerous studies describing transcriptomic, proteomic and metabolomic analyses of wine-yeast fermentations. This work is beginning to provide insights into wine-yeast fermentations, but it is still early days. It should also be noted that much of the -omics work on wine yeast has used resources and databases that are based on laboratory strains. It is now clear that there are genomic differences between wine and lab strains of S. cerevisiae, and these might affect -omics data acquisition and analysis. For example, gene-array chips based on the reference laboratory strain S288c will not include the additional ORFs found in wine strains. This does not suggest that earlier work is invalid, but that there are likely to be gaps in it.
As the various -omics fields progress, it should be possible to build systems-based mathematical models of metabolism that will facilitate the in silico design of new wine yeast strains (Borneman et al, 2007). In parallel, we see the emergence of synthetic biology where, yet again, S. cerevisiae is a key player. It should not be too long before we have customised S. cerevisiae genomic components—regulatory elements to control the expression of targeted genes, or cassettes carrying genes encoding metabolic pathways to shape wine-relevant traits, for example—available ‘off the shelf' for designing, building and refining metabolic processes in our wine yeast. But are consumers ready for this brave and exciting new world?
The engineered wine yeast strains described in this paper show the potential of novel yeast strain development to improve wine quality. But molecular biologists face a major obstacle to this progress: near world-wide refusal to permit the use of GMOs in the production of foods and beverages, at least in ‘developed' countries (Gross, 2009; Pretorius & Høj, 2005). Wine industries in most parts of the world have eschewed the use of GMOs in commercial winemaking, leaving most new-generation wine yeasts on the laboratory shelf, where they await more enlightened times.
Two genetically modified wine yeast strains have been released to market in a limited number of countries including the USA, Canada and Moldova: ML01 and 522EC−. ML01, a transgenic wine yeast, has genes that enable it to perform malolactic fermentation (MLF), a deacidifying secondary fermentation in which malic acid—present in grape juice—is decarboxylated to lactic acid. MLF is usually performed by the lactic acid bacterium Oenococcus oeni after alcoholic fermentation. However, this bacterium is rather fastidious, being inhibited by a range of conditions that are typical of fermented grape juice—low pH, high alcohol content, poor nutrient availability and the presence of sulphur dioxide—and can become ‘stuck' or take considerable time to complete fermentation (Davis et al, 1985). In addition, lacitic acid bacteria can produce a range of biogenic amines, which are associated with health risks (Lonvaud-Funel, 2001).
A wine yeast that completes both primary and secondary fermentations should therefore have great potential in the wine industry. The genetically modified wine yeast ML01 carries two foreign genes—the Schizosaccharomyces pombe malate transporter gene (mae1) and the O. oeni malolactic enzyme gene (mleA)—which are both chromosomally integrated and regulated by the S. cerevisiae PGK1 promoter and terminator (Husnik et al, 2006). This enables the host wine yeast to perform MLF, in parallel with alcoholic fermentation.
The researchers went to great lengths to ensure the safety of ML01. The transgenes came from microorganisms found in wine, there were no antibiotic resistance genes or vector sequences carried by the yeast and transcriptome and proteome analysis showed no important differences in gene expression profiles between the genetically modified strain and its parent. The FDA granted ‘Generally Regarded As Safe' status to ML01, but it has not been widely adopted, even in countries where it is approved for use. This is largely owing to concerns about export markets that do not tolerate GMOs. In fact, wine industries in many countries have banned the use of GMOs in wine production, in order to avoid jeopardizing their exports.
© AWRI, illustration by Geoffrey Reed Communications, Bridgehead, Australia
The genetically modified wine yeast 522EC− was engineered to reduce the risk of ethyl carbamate production during fermentation. Ethyl carbamate, a potential carcinogen, is the product of yeast-derived urea reacting with ethanol. It is usually produced at such low levels—if at all—that it is not a cause for concern, but it sometimes can make an appearance in some wine-producing regions.
S. cerevisiae is able to degrade urea before it is secreted and release ammonia instead, thereby reducing the risk of generating ethyl carbamate. This is achieved by the action of an enzyme encoded by DUR1,2, but this gene is repressed by nitrogen and therefore downregulated throughout much of wine fermentation. Coulon et al (2006) placed a copy of DUR1,2 behind a constitutive (PGK1) S. cerevisiae promoter, which led to a reduction in ethyl carbamate yields. Interestingly, this genetically modified yeast is self or cis cloned; it carries no foreign DNA and therefore is not transgenic. Nonetheless, because it was generated by using techniques that involved the manipulation of DNA in vitro, the regulations of many countries classify it as a GMO. Again, to the best of our knowledge, this yeast is not being used in the industry. This might be because ethyl carbamate production is not a widespread problem, but it probably also reflects the influence of GMO bans and the reluctance of winemakers to risk losing market share in countries that harbour strong anti-GMO sentiment.
Who knows what bottled masterpieces await us as we sculpt novel yeast strains in the laboratory using molecular, systems and synthetic biology
Winemaking, science and technology have interwoven histories and have grown together over the millennia, benefiting from each other. Although science is an important part of an oenologist's training and scientific methods and equipment are routinely employed in the winery, winemakers are not scientists per se. They are, perhaps more appropriately regarded as artisans, with the emphasis on the ‘art'. As for many human endeavours, the Arts progress with developments in technology; think of the use of acrylic paint in the fine arts since its introduction in the 1950s, or David Hockney's use of a Polaroid camera to create photocollages. In the way that acrylic paint and photography have provided more options to artists, enabling them to broaden their horizons, yeast science and technology is adding to the winemaker's palette. Who knows what bottled masterpieces await us as we sculpt novel yeast strains in the laboratory using molecular, systems and synthetic biology. The only real obstacle that we face is consumer acceptance of GMOs; we can only hope that rationality will eventually prevail.
Science & Society Series on Food and Science.
This article is part of the EMBO reports Science & Society series on ‘food and science' to highlight the role of natural and social sciences in understanding our relationship with food. We hope that the series serves a delightful menu of interesting articles for our readers.
Paul J. Chambers
Isak S. Pretorius
Acknowledgments
Research at the Australian Wine Research Institute (AWRI) is financially supported by Australia's grapegrowers and winemakers through their investment body, the Grape and Wine Research Corporation, with matching funding from the Australian Government. Systems biology research at the AWRI uses resources provided as part of the National Collaborative Research Infrastructure Strategy (NCRIS), an initiative of the Australian Government, in addition to funds from the South Australian State Government. AWRI's collaborating partners within this NCRIS-funded initiative—which is overseen by Bioplatforms Australia—are Genomics Australia, Proteomics Australia, Metabolomics Australia (of which the Microbial Metabolomics unit is housed at the AWRI) and Bioinformatics Australia.The AWRI is part of the Wine Innovation Cluster in Adelaide.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barnett J (1998) A history of research on yeasts: Work by chemists and biologists, 1789–1850. Yeast 14: 1439–1451 [DOI] [PubMed] [Google Scholar]
- Barnett J (2000) A history of research on yeasts: Louis Pasteur and his contemporaries, 1850–1880. Yeast 16: 755–771 [DOI] [PubMed] [Google Scholar]
- Barnett J (2007) A history of research on yeasts: Foundations of yeast genetics. Yeast 24: 799–845 [DOI] [PubMed] [Google Scholar]
- Barnett J, Lichtenthaler F (2001) A history of research on yeasts: Emil Fischer, Eduard Buchner and their contemporaries, 1880–1900. Yeast 18: 363–388 [DOI] [PubMed] [Google Scholar]
- Beggs J (1978) Transformation of yeast by a replicating hybrid plasmid. Nature 275: 104–109 [DOI] [PubMed] [Google Scholar]
- Borneman AR, Chambers PJ, Pretorius IS (2007) Yeast systems biology: modelling the winemaker's art. Trends Biotechnol 25: 349–355 [DOI] [PubMed] [Google Scholar]
- Borneman AR, Forgan A, Pretorius IS, Chambers PJ (2008) Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res 8: 1185–1195 [DOI] [PubMed] [Google Scholar]
- Cambon B, Monteil V, Remize F, Camarasa C, Dequin S (2006) Effects of GPD1 overexpression in Saccharomyces cerevisiae commercial wine yeast strains lacking ALD6 genes. Appl Environ Microbiol 72: 4688–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon J, Husnik JI, Inglis DL, van der Merwe GK, Lonvaud A, Erasmus DJ, van Vuuren HJJ (2006) Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine. Am J Enol Vitic 57: 113–124 [Google Scholar]
- Davis C, Wibowo D, Eschenbruch R, Lee T, Fleet G (1985) Practical implications of malolactic fermentation—A review. Am J Enol Vitic 36: 290–301 [Google Scholar]
- de Barros Lopes M, ur-Rehman A, Gockowiak H, Heinrich A, Langridge P, Henschke P (2000) Fermentation properties of a wine yeast over-expressing the Saccharomyces cerevisiae glycerol 3-phosphate dehydrogenase gene (GPD2). Aust J Grape Wine Res 6: 208–215 [Google Scholar]
- Dubourdieu D, Tominaga T, Masneuf I, Gachons C, Murat M (2006) The role of yeasts in grape flavor development during fermentation: The example of Sauvignon Blanc. Am J Enol Vitic 57: 81–88 [Google Scholar]
- Eglinton J, Heinrich A, Pollnitz A, Langridge P, Henschke P, De Barros Lopes M (2002) Decreasing acetic acid accumulation by a glycerol overproducing strain of Saccharomyces cerevisiae by deleting the ALD6 aldehyde dehydrogenase gene. Yeast 19: 295–301 [DOI] [PubMed] [Google Scholar]
- Giaever G et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Godden P, Muhlack R (2010) Trends in the composition of Australian wine 1984–2008. Aust NZ Grapegrower Winemaker 558: 47–61 [Google Scholar]
- Goffeau A et al. (1996) Life with 6000 genes. Science 274: 563–567 [DOI] [PubMed] [Google Scholar]
- Govender P, Bester M, Bauer FF (2010) FLO gene-dependent phenotypes in industrial wine yeast strains. Appl Microbiol Biotechnol 86: 931–945 [DOI] [PubMed] [Google Scholar]
- Govender P, Domingo J, Bester M, Pretorius IS, Bauer FF (2008) Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl Environ Microbiol 74: 6041–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M (2009) European dissent over GM crops. Curr Biol 19: 267–268 [DOI] [PubMed] [Google Scholar]
- Hinnen A, Hicks J, Fink G (1978) Transformation of yeast. Proc Natl Acad Sci USA 75: 1929–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K, Klein M, Swiegers J, Hayasaka Y, Elsey G, Fleet G et al. (2005) Genetic determinants of volatile-thiol release by Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol 71: 5420–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik J, Volschenk H, Bauer J, Colavizza D, Luo Z, Van Vuuren HJ (2006) Metabolic engineering of malolactic wine yeast. Metab Eng 8: 315–323 [DOI] [PubMed] [Google Scholar]
- Kutyna D, Varela C, Henschke P, Chambers P, Stanley G (2010) Microbiological approaches to lowering ethanol concentration in wine. Trends Food Sci Technol 21: 293–302 [Google Scholar]
- Lambrechts MG, Pretorius IS (2000) Yeast and its importance to wine aroma—A review. S Afr J Enol Vitic 21: 97–129 [Google Scholar]
- Lilly M, Bauer FF, Lambrechts MG, Swiegers JH, Cozzolino D, Pretorius IS (2006a) The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 23: 641–659 [DOI] [PubMed] [Google Scholar]
- Lilly M, Bauer FF, Styger G, Lambrechts MG, Pretorius IS (2006b) The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res 6: 726–743 [DOI] [PubMed] [Google Scholar]
- Lonvaud-Funel A (2001) Biogenic amines in wines: role of lactic acid bacteria. FEMS Microbiol Lett 199: 9–13 [DOI] [PubMed] [Google Scholar]
- Michnick S, Roustan JL, Remize F, Barre P, Dequin S (1997) Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol 3-phosphate dehydrogenase. Yeast 13: 783–793 [DOI] [PubMed] [Google Scholar]
- Navarro-Aviño J, Prasad R, Miralles V, Benito R, Serrano R (1999) A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast 15: 829–842 [DOI] [PubMed] [Google Scholar]
- Novo M et al. (2009) Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. PNAS 106: 16333–16338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic D, Vemuri G (2009) Impact of yeast systems biology on industrial biotechnology. J Biotechnol 144: 204–211 [DOI] [PubMed] [Google Scholar]
- Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16: 675–729 [DOI] [PubMed] [Google Scholar]
- Pretorius IS, du Toit M, van Rensburg P (2003) Designer yeasts for the fermentation industry of the 21st Century. Food Technol Biotechnol 41: 3–10 [Google Scholar]
- Pretorius IS, Høj PB (2005) Grape and wine biotechnology: Challenges, opportunities and potential benefits. Aust J Grape Wine Res 11: 83–108 [Google Scholar]
- Remize F, Roustan JL, Sablayrolles JM, Barre P, Dequin S (1999) Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in By-product formation and to a stimulation of fermentation rate in stationary phase. Appl Environ Microbiol 65: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis R, Brown P (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Swiegers JH et al. (2007) Engineering volatile thiol release in Saccharomyces cerevisiae for improved wine aroma. Yeast 24: 561–574 [DOI] [PubMed] [Google Scholar]
- Swiegers JH, Bartowsky EJ, Henschke PA, Pretorius IS (2005) Yeast and bacterial modulation of wine aroma and flavour. Aust J Grape Wine Res 11: 139–173 [Google Scholar]
- Swiegers J et al. (2009) The influence of yeast on the aroma of Sauvignon Blanc wine. Food Microbiol 26: 204–211 [DOI] [PubMed] [Google Scholar]
- Swiegers J, Pretorius IS (2007) Modulation of volatile sulfur compounds by wine yeast. Appl Microbiol Biotechnol 74: 954–960 [DOI] [PubMed] [Google Scholar]
- Tominaga T, Gachons C, Dubourdieu D (1998) A new type of flavor precursors in Vitis vinifera L. cv. Sauvignon Blanc: S-Cysteine conjugates. J Agric Food Chem 46: 5215–5219 [Google Scholar]
- Winzeler E, Shoemaker D, Astromoff A, Liang H, Anderson K, Andre B et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]