Abstract
To establish successful infection, a retrovirus must insert a DNA replica of its genome into host cell chromosomal DNA1,2. This process is carried out by the intasome, a nucleoprotein complex comprised of a tetramer of integrase (IN) assembled on the viral DNA ends3,4. The intasome engages chromosomal DNA within a target capture complex to carry out strand transfer, irreversibly joining the viral and cellular DNA molecules. Although several intasome/transpososome structures from the DDE(D) recombinase superfamily were reported4-6, the mechanics of target DNA capture and strand transfer by these enzymes have not been established. Herein, we report crystal structures of the intasome from prototype foamy virus in complex with target DNA, elucidating the pre-integration target DNA capture and post-catalytic strand transfer intermediates of the retroviral integration process. The cleft between IN dimers within the intasome accommodates chromosomal DNA in a severely bent conformation, allowing widely spaced IN active sites to access the scissile phosphodiester bonds. Our results elucidate the structural basis for retroviral DNA integration and moreover provide a framework for the design of INs with altered target sequences.
To elucidate how the retroviral integration machinery engages chromosomal DNA, we co-crystallized the prototype foamy virus (PFV) intasome with a model target DNA (tDNA) construct (Fig. 1a), which was designed on the basis of the PFV integration site consensus7,8. Inclusion of Mg2+ allowed strand transfer to occur during crystallization experiments (Supplementary Fig. S1), resulting in crystals of the post-catalytic strand transfer complex (STC), while pre-catalytic target capture complex (TCC) crystals were obtained in the absence of the essential catalytic metal TCCApo). Furthermore, using a viral DNA mimic lacking the reactive 3′-hydroxyl group enabled us to grow crystals of the catalytically trapped complex (TCCddA) in the presence of Mg2+, which considerably extended their diffraction limit. The STC, TCCApo, and TCCddA structures were refined to 2.81, 3.32, and 2.97 Å resolution, respectively (Supplementary Table 1, Supplementary Fig. 2).
Figure 1. Crystal structure of the PFV STC.
a, Sequence of the model tDNA. The arrows point to the sites of strand transfer with a 4-bp stagger (boxed). Nucleotide numbers are indicated for both strands. The green diamond indicates the 2-fold symmetry axis. b-c, Surface (b) and cartoon (c) representations of the STC crystal structure, viewed along or perpendicular to the crystallographic two-fold axis, respectively. IN active site carboxylates are shown in red. Protein domains (NTD, N-terminal domain; CTD, C-terminal domain; CCD, catalytic core domain; NED, NTD extension domain) as well as viral (v) and target (t) DNAs are indicated.
As predicted earlier4, the tDNA is accommodated within the cleft between the halves of the symmetric intasome (Fig. 1b, c and Supplementary Movie 1). The intasome does not undergo dramatic rearrangements (Supplementary Fig. 3) but induces severe bending of the tDNA duplex (Fig. 2a and Supplementary Movie 2). Crucially, this binding mode provides the active sites of the inner IN subunits, separated as far as 26.5 Å in the TCC structures, with direct access to the scissile phosphodiester bonds within tDNA (Supplementary Figs 2a-c, 4 and 5). Superposition of the TCCddA structure and the Mn2+-bound form of the intasome4 positions the 3′-hydroxyl group of viral DNA, coordinated to metal B of the active site, for in-line Sn2 nucleophilic substitution at the phosphorus atom of the scissile phosphodiester in tDNA (Fig. 2b). Notably, the sugar moiety of the tDNA nucleotide at the site of strand transfer (cytosine 0) adopts different conformations in the TCC and STC structures. Primarily due to an ~110° rotation around the deoxyribose C4-C5 bond, the viral DNA-tDNA phosphodiester shifts from its pre-strand transfer position by 2.3 Å and thus is ejected from the active site in the STC structure (Fig. 2b). Crucially, such a conformational change would prevent a reversal of the strand transfer reaction.
Figure 2. Details of DNA conformations, recognition, and active site mechanics during strand transfer.
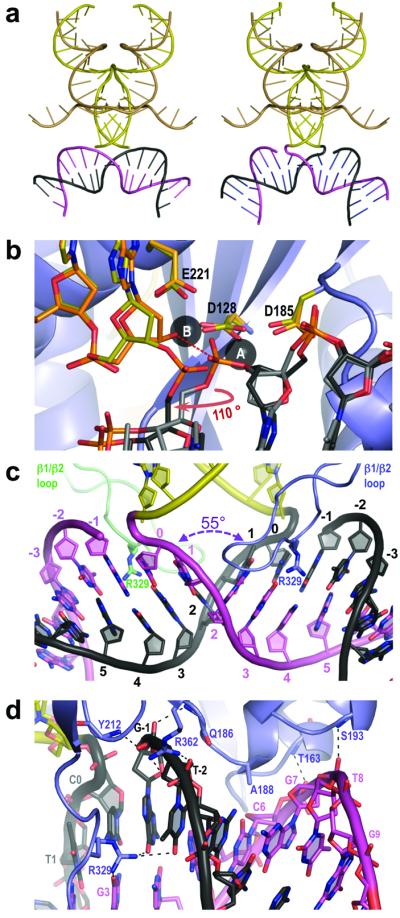
a, Cartoon representation of the viral and target DNAs in the TCCApo (left panel) and STC (right panel) structures. b, Mechanics of DNA strand transfer. Superposition of common elements within TCCddA, STC, and Mn2+-loaded intasome4 (PDB ID 3L2S). Carbon atoms of viral DNA and IN active site residues are painted orange (3L2S) and yellow (STC), respectively. c, Close in view of the integration site in the STC structure. d, IN interactions with the tDNA backbone. Dashed lines indicate hydrogen bonds.
Overall, the conformations of the synapsed DNA molecules within the TCC and STC structures are very similar (Fig. 2a and Supplementary Movie 2), and will be discussed in the context of the STC. At the centre of the integration site, the major groove of the target is widened to 26.3 Å, and the minor groove is compressed to 9.6 Å due to a −55° roll between base pairs involving thymine 1 and adenine 2, resulting in the complete unstacking of the two consecutive base pairs (Fig. 2c). Remarkably, this severe DNA kinking does not involve direct interactions between unstacked base pairs and protein. The TCC and STC are stabilized by 8 rigid hydrogen bonds between amide groups of the protein main chain (residues Thr163, Gln186, Ser193, and Tyr212 from each inner IN monomer) and the tDNA phosphodiester backbone in addition to a pair of salt bridges involving inner chain CTD Arg362 residues (Fig. 2d). As can be expected from the relatively low degree of integration site sequence selectivity of retroviruses for chromosomal DNA9-11, interactions between IN and tDNA bases are sparse. Nonetheless, two close interactions could be identified within the TC/STC structures. Firstly, the side chain of Arg329, based on the loop connecting IN CTD β1 and β2 strands (β1/β2 loop), is hydrogen bonded to guanine 3, guanine −1, and thymine −2 bases within the expanded major groove of the tDNA (Fig. 2c, d and Supplementary Fig. 2d). Secondly, the methyl group of Ala188, at the beginning of α2 helix of the inner IN chain CCD, is involved in a Van der Waals interaction with the O2 atom of cytosine 6 within the minor groove (Fig. 2d).
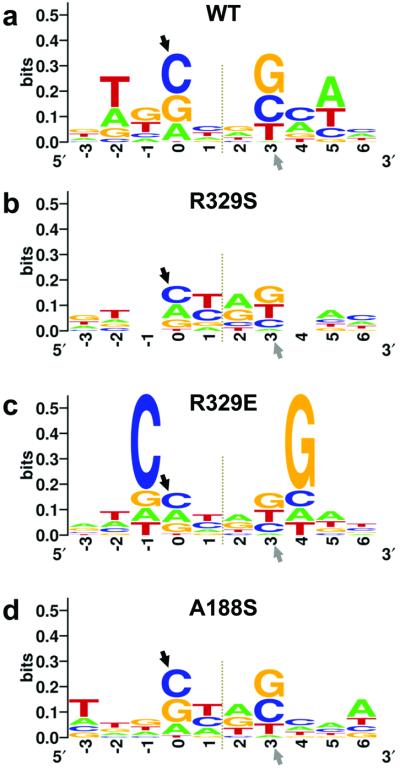
Due to their naturally low extent of base stacking, pyrimidine (Y) – purine (R) dinucleotide steps are known to be the most flexible, followed by YY (or RR) and, the least deformable, RY steps12. Concordantly, PFV integration sites are significantly enriched in YR dinucleotides at positions 1 and 2 (Supplementary Fig. 6). The length of the β1/β2 loop and the presence of Arg at equivalent 329 positions are invariant among Spumavirus INs, underscoring the importance of the interactions observed in the crystals. Substitution of PFV IN Arg329 for Ser, a residue with smaller side chain, is expected to abolish hydrogen bonding with the tDNA bases as well as reduce the geometric fit between the intasome and the functional tDNA conformation. While not affecting intasome assembly and strand transfer activity (Supplementary Fig. 7), the R329S mutation significantly increased the bias of PFV IN against the rigid RY dinucleotides at positions 1 and 2 of the resulting integration sites (Fig. 3a, b and Supplementary Fig. 6), confirming the role of Arg329 in tDNA bending. A more drastic mutation, R329E, greatly reduced strand transfer activity of the intasome (Supplementary Fig. 7). Sequencing of the residual R329E strand transfer products revealed a striking preference for guanosine at position 4 (and a symmetric preference for cytosine at position −1) of these integration sites (p <10−32) (Fig. 3c). Interaction of the mutant Glu residue with a cytidine base13 at position −1 of the integration site likely explains the marked tDNA sequence preferences of the R329E intasome. Strong selectivity towards chromosomal DNA sequence would be expected to limit the available pool of integration sites and therefore reduce viral fitness. Arg, a residue with a flexible, protonated side chain, is able to form a plethora of hydrogen bond interactions within the major groove of DNA13, helping to offset the energetic penalty associated with tDNA bending, while introducing only minor sequence preferences for positions 4 and 5 (Fig. 3a).
Figure 3. Sequence analysis of strand transfer reaction products.
a-d, Sequence logos representing nucleotide frequencies at PFV integration sites in vitro using wild type (a), R329S (b), R329E (c), or A188S (d) INs. Target DNA sequences as joined to the viral DNA ends were used in the alignments. Arrowheads indicate sites of strand transfer, and dotted lines symmetry axes of the integration site consensi; the strict two-fold symmetry is imposed by the use of intasomes containing pairs of U5 viral DNA ends.
In retroviral INs, the positions equivalent to PFV 188 are invariably occupied by small amino acids, typically Ala, Pro, or Ser, suggesting that a close contact between α2 and the minor groove of tDNA is a common feature of retroviral TC/STCs. In line with prior observations14,15, substitution of PFV IN Ala188 (a residue structurally equivalent to HIV-1 Ser119) for Asp ablated strand transfer activity (Supplementary Fig. 7). A188S, a less drastic mutation, did not affect the level of strand transfer activity, but yielded a significant bias for adenosine at tDNA position 6 (p =10−4) (Fig. 3d), likely due to hydrogen bonding of the mutant side chain to N3 of the adenine base.
One puzzling feature of the PFV integration site consensus is a pronounced bias against thymidine at position 0 and the symmetrical avoidance of adenosine at position 3 (Fig. 3a). Furthermore, the bias against integration immediately upstream of a thymidine appears to be a common feature of retroviruses9-11. Modelling a T:A base pair at the site of integration reveals that the C5 methyl group of thymine would be too close to the phosphate group of the target phosphodiester bond, approaching it at 3.4 Å in the STC (Supplementary Fig. 8), and thus likely interfering with the mechanics of transesterification.
The conformation of tDNA within the STCs is fully consistent with earlier observations that pre-bending of tDNA promotes retroviral integration16,17. Of note, in vitro PFV IN appears more selective for flexible target sequences compared to what is observed during viral infections (Supplementary Fig. 6). We speculate that chromosomal DNA packaging into nucleosomes and the presence of other DNA bending factors in cells contribute to this subtle difference. In the context of integration into chromatin, the NTDs and CTDs of the outer IN subunits, disordered in our structures, might participate in interactions with nucleosomal DNA and/or the histone octamer. The TCC and STC structures presented herein elucidate the structural basis for retroviral DNA integration and indirect recognition of the optimal tDNA sequence. As such, they provide a framework for the design of INs with altered target sequences and will boost ongoing efforts to create site-specific retroviral vector systems for future applications in gene therapy18,19. Furthermore, the PFV intasome4 as well as TCC and STC structures reported herein will afford building of reliable models for the respective intermediates of the HIV integration process20 to aid improvement of existing, as well as the discovery of novel approaches to block viral replication.
METHODS SUMMARY
PFV intasomes, assembled with wild type full-length IN and a mimic of the pre-processed U5 viral DNA containing or lacking the reactive 3′-hydroxyl group were co-crystallized with a self-complementary tDNA oligonucleotide 5′-CCCGAGGCA CGTGCTAGCACGTGCCTCGGG. Inclusion of the natural 3′-hydroxyl in the viral DNA construct allowed strand transfer to occur during crystallization in the presence of MgCl2 (Supplementary Fig. 1). Although the intasome can engage the tDNA construct at multiple sites, the constraints imposed by crystal symmetry and lattice contacts likely accounted for the selective crystallization of the symmetric complex (compare lanes 3 and 4 in Supplementary Fig. 1c). In both TCC and STC crystals, the asymmetric units contained half of the intasome structure (IN chains A and B, viral DNA strands C and D) and one strand of tDNA (chain T); the complete biological assemblies were generated by crystallographic two-fold symmetry operations. Sixteen and eighteen base pairs of tDNA could be built into the TCC and STC electron density maps, respectively. Similar to the original PFV intasome crystals4, the N- and C-terminal domains of the outer IN subunits (chains B and B′) were disordered and are hence absent in the final models. Crystallographic and refinement statistics for the seven resulting models are summarized in Supplementary Table 1. The models had good geometry with 96.7 and 0% (STC), 92.9 and 0.4% (TCCddA), and 91.8 and 1.2% (TCCApo) of amino acid residues in most preferred and disallowed regions of the Ramachandran plot, respectively.
METHODS
Intasome preparation, crystallization, and structure determination
Full-length wild type PFV IN and its mutant forms were produced according to established procedures4. Synthetic DNA was purchased from Eurogentec (Seraing, Belgium) and Midland Certified (Midland, TX). Donor DNA was obtained by annealing HPLC-purified synthetic oligonucleotides 5′-TGCGAAATTCCATGACA (reactive strand) or its 3′-deoxy derivative 5′-TGCGAAATTCCATGAC[2′,3′-ddA] and 5′-ATTGTCATGGAATTTCGCA (non-transferred strand). The intasomes, assembled by dialysis of IN/donor DNA mixtures as previously described4, were purified by chromatography through HiLoad 16/60 Superdex-200 column (GE Healthcare) in 320 mM NaCl, 20 mM Bis-Tris Propane (BTP)-HCl, pH 7.45. Intasome preparations, mixed with 1.5-molar excess of self-annealed tDNA oligonucleotide 5′-CCCGAGGCACGTGCTAGCACGTG CCTCGGG, were dialyzed overnight against excess of 225 mM NaCl, 2 mM dithiotreitol (DTT), 25 μM ZnCl2, 20 mM BTP-HCl, pH 7.45 and concentrated to 6 mg/ml using Vivaspin-4 devices with a 10-kDa cut-off.
Hanging drop vapour diffusion crystallization experiments were setup by mixing 1 μl of protein-DNA complex solution with 1 μl reservoir solution. The reservoir contained 20% (+/−)-2-methyl-2,4-pentanediol (MPD), 40 mM MgCl2 and 50 mM sodium cacodylate, pH 5.4 (STC); 38% PEG400, 180 mM Li2SO4, 20 mM MgCl2, 100 mM Tris-HCl, pH 7.5 (TCCddA); or 34% PEG400, 180 mM Li2SO4 and 100 mM Tris-HCl, pH 7.5 (TCCApo). Crystals, grown at 18°C, appeared within 2-3 days and grew to a size of 100-400 μm within 2-4 weeks. Crystals, cryoprotected in 30% MPD, 3 mM MgCl2, 150 mM NaCl and 30 mM sodium cacodylate-NaOH, pH 5.9 (STC); 40% PEG400, 150 mM NaCl, 150 mM Li2SO4, 15 mM MgCl2 and 100 mM Tris-HCl, pH 7.4 (TCCddA); or 35% PEG400, 150 mM NaCl, 150 mM Li2SO4 and 0.1 M Tris-HCl, pH 7.5 (TCCApo), were frozen by quick submersion in liquid nitrogen.
Diffraction data, collected at beam line I02 of the Diamond Light Source (Oxfordshire, UK) at 100 K, were integrated with XDS21, merged and scaled using Scala of the CCP4 suite22. Crystals belonged to space group P41212 and as they were nearly isomorphous to intasome crystals obtained in the absence of tDNA4, the structures were solved by isomorphous replacement using the intasome coordinates from PDB entry 3L2R (ref. 4). The tDNA strands were built into the resulting Fo-Fc difference maps. The final structures, refined using Refmac23, were validated with MolProbity24. The models had good geometry with 96.7 and 0% (STC), 92.9 and 0.4% (TCCddA), and 91.8 and 1.2% (TCCApo) of amino acid residues in most preferred and disallowed regions of the Ramachandran plot, respectively. X-Ray data collection and refinement statistics are given in Supplementary Table 1; examples of final weighted 2Fo-Fc electron density maps are given in Supplementary Fig. 2. Structure figures were generated using PyMOL (http://www.pymol.org).
Strand transfer assays and integration product sequencing
Strand transfer assays used WT and mutant intasomes purified by size exclusion chromatography through a Superdex 200HR 10/30 column (GE Healthcare) in 320 mM NaCl, 20 mM BTP-HCl, pH 7.45 (Supplementary Fig. 7) and quantified by spectrophotometry at 260 nm. Strand transfer assays contained 300 ng supercoiled pGEM9-Zf(−) (Promega) tDNA and 1.6 or 0.4 pmol intasome in 40 μl of 115 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 4 μM ZnCl2, 25 mM BTP-HCl, pH 7.45. Reactions were incubated at 37°C for 30 min and stopped by addition of 0.5% SDS and 25 mM EDTA. DNA products deproteinized by digestion with 20 μg proteinase K for 30 min at 37°C and precipitated with ethanol were separated in 1.5% agarose gels and visualized by staining with ethidium bromide. For sequence analyses8, concerted strand transfer products isolated from agarose gels were treated with phi29 DNA polymerase (New England Biolabs) in the presence of 450 μM each dNTP, 5′-phosphorylated using T4 polynucleotide kinase (New England Biolabs) and ligated with a blunt-ended DNA fragment spanning the Tn5 kanamycin resistance gene flanked by KpnI restriction sites. The kanamycin resistance cassette was generated by PCR using primers 5′-GGCGGGTACCAGAAAGCAGGTAGCTTGCAGTGG and 5′-GGCGGGTACCCGAAGAACTCCAGCATGAGATCC (KpnI sites underlined) and pCP15 (ref. 25) as a template. Escherichia coli TOP10 cells (Invitrogen), transformed with the ligation products, were selected with 35 μg/ml kanamycin. Plasmids isolated from individual clones were analysed by digestion with KpnI (New England Biolabs), and those releasing DNA fragments of expected sizes (~2,900 and 1,200 bp) were sequenced using primers annealing close to the ends of the kanamycin resistance cassette (5′-TACTTTGCAGGGCTTCCCAACC and 5′-CGAAATGA CCGACCAAGCGACG). Because strand transfer products were converted to fully double-stranded form prior to blunt-end ligation, the remainder of clones (21%) could be discarded as cloning artefacts. Of all sequenced clones 99.3% contained pairs of donor DNA fragments joined to tDNA. Only unique clones from each E. coli transformation were used in further analyses. Deletions of various sizes (8-920 bp) accounting for 9.3% of all clones are explained by multiple integration events into a single target plasmid. The expected tDNA sequence duplication size of 4-bp was observed in the majority of remaining clones (98.5%), and only these were used in final sequence alignments (65 clones for WT, 49 for R329S, 45 for R329E, and 58 for A188S). Sequence logos were generated using WebLogo26.
Supplementary Material
Acknowledgements
We thank Dr. Alan Engelman and Dr. Fred Dyda for critical reading of the manuscript, Dr. Thomas Sorensen and Juan Sanchez-Weatherby of the Diamond Light Source beamline I02 for assistance with X-ray data collection, and Dr. Jeremy Moore for expert help with crystallization screening and the in-house X-ray generator. This work was funded by the UK Medical Research Council.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Atomic coordinates and structure factors for STC, TCCddA and TCCApo have been deposited with the Protein Data Bank under accession codes 3OS0, 3OS1 and 3OS2, respectively. Raw diffraction images are available upon request.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Craigie R. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. ASM Press; 2002. pp. 613–630. [Google Scholar]
- 2.Lewinski MK, Bushman FD. Retroviral DNA integration--mechanism and consequences. Adv. Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Mizuuchi M, Burke TR, Jr., Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 6.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trobridge GD, et al. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valkov E, et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RC, Stella S, Heiss JK. In: Protein-nucleic acid interactions. Rice PA, Correll CC, editors. Ch. 8. RSC Publishing; 2008. pp. 176–220. [Google Scholar]
- 13.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konsavage WM, Jr., Burkholder S, Sudol M, Harper AL, Katzman M. A substitution in rous sarcoma virus integrase that separates its two biologically relevant enzymatic activities. J. Virol. 2005;79:4691–4699. doi: 10.1128/JVI.79.8.4691-4699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J. Virol. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim KI, Klimczak R, Yu JH, Schaffer DV. Specific insertions of zinc finger domains into Gag-Pol yield engineered retroviral vectors with selective integration properties. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12475–12480. doi: 10.1073/pnas.1001402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan L, et al. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W., Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.