Summary
We consider joint spatial modelling of areal multivariate categorical data assuming a multiway contingency table for the variables, modelled by using a log-linear model, and connected across units by using spatial random effects. With no distinction regarding whether variables are response or explanatory, we do not limit inference to conditional probabilities, as in customary spatial logistic regression. With joint probabilities we can calculate arbitrary marginal and conditional probabilities without having to refit models to investigate different hypotheses. Flexible aggregation allows us to investigate subgroups of interest; flexible conditioning enables not only the study of outcomes given risk factors but also retrospective study of risk factors given outcomes. A benefit of joint spatial modelling is the opportunity to reveal disparities in health in a richer fashion, e.g. across space for any particular group of cells, across groups of cells at a particular location, and, hence, potential space–group interaction. We illustrate with an analysis of birth records for the state of North Carolina and compare with spatial logistic regression.
Keywords: Conditionally auto-regressive models, Disease mapping, Health disparities, Hierarchical models, Log-linear models, Multilevel models, Spatial random effects
1. Introduction
1.1. Overview
Our concern here is with spatial categorical data that are available at areal unit scale. A common setting where counts that are associated with such units arise is disease mapping, and we have witnessed the development of an extensive literature in this area in recent years. In these applications, patients’ data are assigned to areal units because of limitations of collection or to protect subjects’ confidentiality sufficiently. However, disease maps of raw, local mortality estimates in areal units have well-known deficiencies: geographically large but sparsely populated areas often visually dominate the map but provide the least reliable estimates, especially for rare diseases (Gelman and Price, 1999). Researchers have addressed these deficiencies by using methods that smooth, spatially or otherwise, raw estimates by ‘borrowing strength’ across areas. Bayesian methods for spatial disease mapping formally combine information on observed events in an area with information on nearby rates as well as overall rates of disease to estimate area-specific rates of disease. Such models were introduced by Clayton and Kaldor (1987) and placed in a fully Bayesian setting by Besag et al. (1991). They have been reviewed by Mollié (1996) and Wakefield et al. (2001).
Spatial smoothing in such models is typically achieved by using a conditionally auto-regressive prior on areal unit level random effects (Besag et al., 1991), where a unit’s rate estimate is smoothed on the basis of its neighbours’ rates, and with ‘neighbours’ typically defined as the adjacent units. Such models are often used to analyse aggregated data (i.e. data that are summed over individuals and groups to achieve areal unit totals). If interest lies in subgroups, covariates can be introduced to yield area-specific rates by group. However, since this approach constrains all groups to share the same spatial random effects, it can be unsatisfying for subgroup analysis. An alternative is to fit separate models for each subgroup, but this may lead to an excessive number of parameters, sparse areal counts for some models and awkwardness if we seek to recombine groups. It would also fail to provide the type of integrated model that seems necessary to a comprehensive investigation of the aetiology of health disparities.
Instead, we begin with a joint association model. Some variables may be viewed as potential responses and others as explanatory (as, typically, in public health data), but the joint specification makes no such distinction (as is common in economics and public opinion data). In particular, we work with joint distributions of the cells in a multiway contingency table and do not specify a response variable. Hence, we do not confine ourselves to explaining a particular outcome given a particular set of risk factors (i.e. to conditional probabilities). Rather, once we have joint probabilities, we can calculate arbitrary marginal and conditional probabilities, i.e. we can aggregate or condition in any fashion, enabling a rich range of inference without having to reconsider and refit models to investigate other potential hypotheses. Flexible aggregation allows investigation of groups of interest, whereas flexible conditioning enables study of single as well as multiple outcomes given risk factors along with consideration of incidence of risk factors given outcomes, perhaps in the presence of other risk factors or outcomes. Conditioning enables sensitivity, specificity and related inference that may be appropriate for retrospective data examination. Additionally, our approach avoids potential Simpson’s paradox issues that can arise under collapsing of categories (for example, see Simpson (1951) and Agresti (2002)).
Our approach assumes that observations within areal units retain individual level characteristics but without precise locations. We further assume that the characteristics are categorical (or have been made categorical) so that, for the areal unit, we can envision a contingency table and associated model that describes the cell probabilities (or, equivalently, the joint distribution). These probabilities might be driven by features of the areal unit, and, additionally, we expect spatial dependence in the sense that these joint distributions will be similar for areal units that are close to each other.
In implementing this approach, we specify spatially varying log-linear models: for each areal unit, we imagine Poisson-distributed cell counts driven by a local log-linear model. Some of the parameters in the log-linear model will be allowed to vary spatially, and they may also be explained by using areal unit level covariate information, i.e. contextual variables associated with these units, such as household median family income, percentage home ownership or crime rate. These contextual variables may be useful in explaining differences in cell probabilities across units. An attractive advantage of the log-linear model approach is that, with a multiway table, the number of cells in the table will typically be far greater than the number of parameters in the log-linear model. With ordinal data classifications, we can draw on suitable classes of ordinal categorical data models (Agresti, 2002) which, again, achieve dimension reduction. Hence, through log-linear modelling, we can introduce cell level spatial dependence across areal units without requiring a distinct spatial random effect for every cell at every location.
A further benefit of such modelling is its utility in addressing the complex issue of how to assess disparities in health at a meaningful geographic scale while protecting patients’ confidentiality and accommodating both sparsely and densely populated areas. We can study disparities across space for any grouping of cells, in addition to being able to study disparities across groups of cells. In fact, our approach can tease out space by risk factor interactions. In the context of disparities in health, such general analysis has not been previously considered but is consistent with the US’s ‘Healthy people’ 2010 initiative, whose two goals include the elimination of disparities in health, including those that occur by race, gender or geographic location (US Department of Health and Human Services, 2000).
1.2. Motivating data set
We developed our disaggregation model for application to North Carolina birth record data. These data include information on every documented live birth in the state since 1990, including maternal age, birth weight, clinical estimate of gestation, plurality, maternal complications, congenital anomalies, maternal tobacco and alcohol use, number of living children, number of children born alive now dead and a variety of maternal and paternal demographic variables. We restricted our analysis to singleton (i.e. one fetus) live births with no congenital anomalies born to mothers aged 15–44 years who self-identified their race as either African American or white, for the period 1999–2003. These restrictions eliminate from the analysis adverse birth outcomes that are driven by the physiology of pregnancy (more than one fetus, very young mothers or advanced maternal age) or the pathophysiology of fetal conditions (congenital anomalies).
A total of 462 362 births were included in our analysis, including 52 697 (11.4%) adverse birth outcomes (i.e. infants that were low birth weight (LBW) (7.0%) or preterm (9.3%) or both (4.9%)). More precisely, we take as areal units the 100 counties in North Carolina with the following M = 5 binary classification variables: X for maternal race, African American (1) or white (0); Y for birth weight outcome, LBW (1, less than 2500 g) or not LBW (0, 2500 g or heavier); Z for infant sex male (0) or female (1); W for maternal use of tobacco during pregnancy, yes (1) or no (0); V for preterm birth (1, less than 37 weeks of gestation) or not (0, 37 weeks of gestation or longer). So we have L = 25 = 32 cells in the contingency table. Many hypotheses can be entertained involving these five variables. Any particular one would involve computing functions of cell probabilities and comparing these functions within a county or across counties. In presenting results, we focus on the putative outcome ‘term LBW’, which refers to women who carry their pregnancies to term (i.e. gestation at least 37 weeks) and, nevertheless, have LBW infants (i.e. under 2500 g). Most of these infants would be classified as ‘small for gestational age’, which is a category of particular concern to both obstetricians and paediatricians. Lastly, we note that the methodology can be extended to a richer set of variables and that we could work at other areal scales, e.g. zip codes or census units.
1.3. Format
The format of the paper is as follows. In Section 2, we provide formal details for our disaggregation modelling approach and consider marginal and conditional probabilities from the model. Assuming interest in a regression to explain outcomes, in Section 3 we compare our approach with customary logistic spatial regression models. We then illustrate our approach with an analysis of data drawn from the North Carolina birth record data in Section 4. Finally, we provide a summary and discuss possible extensions in Section 5.
2. Modelling details and properties
In this section, we formalize the disaggregation model that we employ for the balance of the paper. We also discuss marginalization and conditioning under this model.
2.1. The disaggregation model
Consider the subjects in areal unit s as cross-classified by M categorical variables with a total of L classification cells. To connect with our motivating data set let denote the population count of births in cell l in county s. The L × 1 data vector for county s, n(s), consists of entries { }. Let denote the total count for areal unit s. Also, define as the vector of expected cell counts for county s.
We assume that, at the first stage, the data { } are conditionally independent such that . We model the expected cell counts as linear on the log-scale, i.e.
| (1) |
with Xl denoting the design vector for the lth cell, βs denoting the coefficients in the log-linear model for areal unit s and r ≤L. In practice, we do not use saturated models but rather models with fewer parameters than the number of cells, so r < L. Customarily, second-order interactions and perhaps a few three-way interactions are introduced, but rarely higher order interactions. With regard to these βs, we shall sometimes use notation such that μ denotes the overall (or ‘grand’) mean term and γs denote the main effects and interaction terms. We also assume the usual constraints on the γs, i.e. that they sum to 0 over any index. For instance, in analysing the illustrative data set, if we used a model with all two-way interactions, we would have a 1 + 5 + 10 = 16-dimensional β-vector consisting of μ and γs. With the assumed constraints on the γs, each Xl-vector would have ±1 as entries.
Collecting to a vector, we may write the first level of our model as
| (2) |
noting that the XT-matrix is L × r and does not depend on areal unit s, i.e. the design matrix is the same for each areal unit. We make this assumption to ensure that the βs have the same interpretation across s, so that it is sensible to introduce spatial random effects into some components of βs.
We intentionally model the rather than the cell probabilities , to work with a product Poisson likelihood rather than a multinomial. The cell probabilities are induced by the γs; to extract them for each county we need to normalize by . Hence, when fitting the hierarchical model by using Markov chain Monte Carlo techniques, at each iteration, we calculate the and normalize to obtain posterior samples of the pl(s).
We next add a second stage to our model, in the spirit of the multilevel models of Raudenbush and Bryk (2002). We set
| (3) |
where ηtu is the parameter that is associated with the areal unit covariate wsu. In our illustrative setting, ws1 might be median family income, ws2 might be percentage home ownership, ws3 might be crime rate, etc. The random effects , t = 1, 2, …, r, are assumed to be spatially structured. To do so, we introduce an overall intercept for each t, , and write so that the are centred at 0. (Implicitly, this means that ws does not include an intercept.) We address below the question of whether or not to include a φ for each t as well as the modelling for the φs that we do include. However, for now, employing notation that includes all φs, we can write βs as
| (4) |
where φ0 is the r × 1 vector of intercepts, η is an r × q matrix of second-level model parameters, ws is a q × 1 column vector of second-level covariates and φ(s) is an r × 1 vector of spatial random effects.
Inserting equation (4) into equation (2) and expanding provides
| (5) |
The interpretation of equation (5) is attractive. We explain the λs through the individual level characteristics, through the local contextual variables (recall that the first entry in every X-vector is a 1), through interactions between individual characteristics and local covariates, with spatial adjustment to the intercept (again, since the first entry in every X-vector is a 1), and also spatial adjustment to the coefficients that are associated with individual characteristics. Thus, we achieve an extremely flexible explanatory model.
We note that we could develop an analogue of equation (5) without introducing a log-linear model. An advantage of the log-linear approach is that we must explain only r βs rather than L λs where r will usually be much smaller than L. Hence, we must introduce only at most r sets of random effects.
The usual modelling intent using equation (5) is to incorporate spatial dependence between cell probabilities and to effect spatial smoothing of these probabilities, so we model the as purely spatial effects. (Heterogeneous random effects can be envisioned also, as in many disease mapping applications (for example, see Banerjee et al. (2004) and references therein). However, they are difficult to separate from the spatial random effects; the prior specifications essentially drive the inference.) Hence, as priors for the we use customary independent intrinsic conditionally auto-regressive models (Besag, 1974; Besag et al., 1991, Banerjee et al., 2004), though dependence in these random effects could be achieved with multivariate conditionally auto-regressive priors, such as in Gelfand and Vounatsou (2003) or Jin et al. (2005). We do not use multivariate conditionally auto-regressive priors here, expecting that introducing such dependence will not gain much with regard to inference about desired probabilities since these probabilities arise as complicated non-linear functions of these effects. Details of the model implementation are provided in Appendix A.
We also remark that a slightly more general form is possible: we need not require the same set of covariates to explain each βst. This generalized form is given in Raudenbush and Bryk (2002). Thus, in equation (3), q would change to qt and we could change the above notation accordingly. Of course, we can always work with the full set of covariates of interest and let the data tell us which ηtus are significant.
Finally, we comment that ordinal classifications can be accommodated. Using typical models for such classifications (see Agresti(2002)), the associated Xlt s would introduce appropriate scores.
2.2. Marginal and conditional probabilities
It is straightforward to write down general forms for any marginal and conditional probabilities under the above modelling. In general, the marginal probabilities that are associated with the collection of cells L′ ⊂ L are
Conditional probabilities arise as
Relative risks and odds ratios can be computed similarly.
For instance, in the illustrative example that was introduced in Section 1.2 and is described in detail below in Section 4, the probability of an LBW infant born to an African American mother in areal unit s is . The conditional probability in areal unit s of LBW given an African American mother is
Aggregation and conditioning could occur over s as well, in addition to that over i, j, k, m and n.
In considering marginal and conditional probabilities, we might ask under what spatial random-effects specifications will they be spatially varying? As in the examples above, marginalizations of interest will typically be over a set of the M categorical variables in the M-way table. Conditional events of interest will also typically involve probabilities in both numerator and denominator that marginalize over categories.
In this regard, we turn to the question of which should be chosen to be spatially varying. Suppose, for the moment, that we have no contextual variables so . It is clear that, if βs1 = μ(s) is the grand mean term, for any probability, will cancel out in the ratio. If we introduce spatial effects only on this overall ‘intercept’, we shall not achieve spatially varying probabilities. In other words, an intercept random effect for each areal unit will not work and, in fact, there is no reason to introduce them. However, if we introduce for every other t, we ensure that any conditional probability will be spatially varying. However, r-dimensional spatial random-effects vectors might be more than the data can explain and might introduce problems of identifiability and unstable computation. Below, we illustrate, by specifying spatial effects for the intercept, all main effects and all two-way interactions. (The reader can verify that this guarantees spatially varying relative risks and odds ratios.)
3. Connection with customary logistic spatial regression
In many categorical settings interest focuses on modelling an outcome variable given a set of explanatory variables. In this regard, if we have a log-linear model with, say, m = 3 and we identify the binary outcome variable X as the response with Y and Z as covariates, then log (p1jk/p0jk) is the logistic regression that is associated with the conditional event that X = 1 given Y = j and Z = k. In fitting this model, we look at the counts n1jk as the number of successes in n:jk Bernoulli trials at level Y = j, Z = k, each with conditional probability p1|jk = p1jk/(p0jk + p1jk). In situations where Z = 1 denotes occurrence of disease, the p1|jk are usually small and the n:jk large so a Poisson approximation for the distribution of n1jk is adopted with parameter λ̃jk = n:jkp1|jk, i.e. λ̃jk is the expected number of cases of disease given Y = j and Z = k. With the definition, λijk = n:jkpijk, we have λ̃jk = λ1jk/(λ0jk + λ1jk). The familiar disease mapping setting indexes the λ̃ by location and produces spatial modelling for the resulting λ̃(s).
3.1. The usual disease mapping model
Arguably the most widely used model to explain areal unit counts arises in the setting of disease mapping using the log-relative-risk in a Poisson regression with random effects, i.e.
| (6) |
| (7) |
(See, for example, Besag et al. (1991), Banerjee et al. (2004) or Waller and Gotway (2004).) In the context of the example in Section 1.2, we would identify an outcome, such as LBW among term births (again, ‘term LBW’ birth), leading to O(s) as the observed number of term LBW births in county s. Then, E(s) is the expected number of term LBW births in county s, θ(s) is the relative risk of a term LBW in county s and ws are county level covariates (analogous to our earlier ws, and with corresponding coefficients ξ).
To model this conditional outcome with the usual disease mapping model, the analyst must restrict the data to only term births, i.e. the binary birth weight outcome would be conditioned on the set of term births; all preterm births would be discarded. By contrast, the joint approach uses all of the data to fit the model; hence all of the data bear on the conditional inference and, in fact, on any inference of interest. Continuing, the Us are county-specific random effects that are spatially structured via a conditionally auto-regressive prior and result in spatial smoothing towards a local mean rate. The O(s) are regarded as random and conditionally independent given the θ(s) and E(s). The E(s) are regarded as fixed and known. When possible the E(s) are obtained externally to the data set but in many applications in the literature are calculated via internal standardization (Fleiss et al., 2003). When this is so, the data are used on both the ‘left-hand side’ and the ‘right-hand side’ of the model. By contrast, in our modelling, we work with λs rather than λ̃s and the full vector of counts n(s), eliminating the need for standardization.
3.2. Modelling multiple groups
In practice, overall rates as well as rates for specific subgroups are of interest. How would the model in equations (6) and (7) be used to infer about rates for subgroups within areal units? There appear to be two approaches:
introduce an analysis-of-variance style of model incorporating suitable main and interaction effects in the model for the log(θ)s in equation (7) to capture the subgroups of interest; or
fit a separate model for each subgroup.
The former would introduce one set of spatial effects as in equation (7), arguably too restrictive though frequently used (e.g. in Waller et al. (1997)). The latter would introduce a set of spatial random effects for each subgroup.
Option (b) raises numerous questions and concerns, primarily due to modelling λ̃ rather than λ. For example, should the be independent across l? In this regard, our modelling, by introducing multiple spatial random effects for each cell count, as discussed in Section 2.2, automatically introduces dependence to the λ̃. Should we adopt a distinct ξ for each subgroup or a common ξ (to connect the groups)? How do we make comparisons across groups, e.g. rates of risk or odds ratios? Further, to investigate aggregated rates across groups we would have to fit a new model for every aggregation. With a large number of cells, we would fit models to data sets having many cells with sparse counts, leading to potentially unstable fitting.
Whether we create the specification under (a) or (b), the models would provide conditional probabilities of outcome given subgroup. These are the only probabilities that can be studied. Retrospective analyses for risk factors given presence (or absence) of outcome are precluded. In addition, by converting log-linear models to (essentially) logistic regression models, some effects will disappear; we cannot assess their explanatory contribution. Finally, one must specify, a priori, which subgroups to study and which variable(s) or functions of variables to regard as an ‘outcome’ (as is the case in Section 3.1 with the outcome term LBW birth). These questions and concerns seem to us either not to arise or to be more easily accommodated under the disaggregated model proposed. Finally, in using all of the available categorical information, the disaggregated model produces improved inference for the λ̃s relative to a model of the form given in equations (6) and (7), as we demonstrate in Section 4.
An additional benefit deserves comment. A primary reason for investigating many public health databases is to illuminate disparities. Within the disaggregation framework, we can straightforwardly study disparities. Let L′ denote a particular subgroup of the L cells with probabilities . Then we can directly study spatial disparities by examining this set across the areal units. Next, suppose that we seek to compare subgroups. Let L″ be a second sub-group with probabilities . We can compare these with the locally (at each unit) and make comparison across space (to detect group–space interaction). Such interactions would suggest that the spatial random effects are serving as surrogates for an environmental variable with spatial structure not included in our model (i.e. not in the ws) but interacting with some aspect that defines the subgroup. By averaging, in a suitable fashion, we can make partial spatial or non-spatial comparisons. Again, such detailed disparity analysis is precluded by customary modelling approaches.
4. Analysis of the motivating data set
4.1. Applying the disaggregation approach
As a point of departure, we began our analysis by ignoring the role of county and fitting a non-spatial model for the entire state of North Carolina. For our resulting (M =) 5-way table, we first fit a saturated log-linear model with 32 parameters. With L = 32 cells, many cell probabilities will be small (i.e. non-zero) and there is little reason for concern with more than 460 000 births. With smaller data sets, we might have to confine ourselves to fewer variables. In any event, no cell will have 0 counts across all counties. If a particular outcome were that rare, we would collapse over it. Moreover, in this regard, the benefit of Bayesian hierarchical modelling emerges: our model will borrow strength across counties to infer about cell probabilities for any particular county. In any case, we could reduce our model by eliminating non-significant parameters to (XYWV, XZV, YZV), in which all 22 terms were either significant or compelled by the hierarchy principle. With such a large data set, not surprisingly, this baseline model emerged from both classical and Bayesian fitting. The number of parameters was reduced by a third; more pronounced reduction would be expected with a larger M.
We used this baseline log-linear model (XYWV, XZV, YZV) as the starting point for our spatial analysis. In addition, we considered county level household median income data from the census for inclusion (as ws with an associated η). We constructed four variants from this model:
a baseline model with neither spatial random effects nor county level covariates (model 0);
a model without spatial random effects but with county level covariates (model A);
a model with spatial random effects but without county level covariates (model B);
a model with both spatial random effects and county level covariates (model AB).
For spatial random effects, we placed spatial structure on terms up to two-way interaction terms (i.e. the three-way and four-way interaction terms did not receive spatial structure). In total, 15 of the 22 terms ( and ) received spatial random effects in the form of independent conditionally auto-regressive prior distributions, whereas the grand mean terms (μ(s)) received independent vague, normal priors. We completed the model specification in accord with the details that are provided in Appendix A, and we fitted all four models in WinBUGS (http://www.mrc-bsu.cam.ac.uk/bugs/welcome.shtml). (Run times are slow, between 2 and 7 days for the spatial models, owing to the size of the data set and the very large number of spatial random effects. Much faster run times could be achieved by using C code.)
To inform model selection, we used four simple measures, with the only stipulation being that they operate in the space of the ps (since these are the objects of inferential concern). The measures that are presented are as follows: MSE is the mean over all LS = 3200 cells of the squared errors, based on the observed probability and the modelled value ; log(p) is the sum over all cells of ; χ2 is the sum over all cells of , where O denotes observed and E expected counts; and KL is the sum over all cells of , the Kullback–Leibler distance (Kullback and Leibler, 1951) of fitted against observed, all with standard adjustment for observed 0 cell counts. Table 1 also provides median values and corresponding 95% credible intervals (CIs) based on samples from the posterior. We found that models B and AB, the two with spatial structure, performed substantially better than the models without spatial structure (models 0 and A) by using these measures and we would expect this to be so by using any other appropriate criterion. Even with penalization for model complexity, models 0 and A would not be competitive.
Table 1.
Model fit metrics for four models
| Metric | Results for the following models: |
||||
|---|---|---|---|---|---|
| Model 0 | Model A | Model B | Model AB | ||
| MSE | Median × 10−4 | 261.3 | 243.6 | 5.10 | 5.14 |
| 95% CI × 10−4 | (260.9, 261.8) | (243.0, 244.1) | (4.49, 5.89) | (4.52, 5.97) | |
| log(p) | Median | 8376 | 8665 | 2956 | 2964 |
| 95% CI | (8326, 8429) | (8593, 8741) | (2867, 3053) | (2877, 3062) | |
| χ2 | Median | 71650 | 64310 | 3205 | 3226 |
| 95% CI | (71300, 72010) | (63960, 64670) | (3092, 3331) | (3113, 3351) | |
| KL | Median | 14.92 | 14.38 | 1.098 | 1.108 |
| 95% CI | (14.89, 14.95) | (14.36, 14.41) | (1.062, 1.139) | (1.072, 1.150) | |
Table 2 provides main effect parameter estimates from the two spatial models (B and AB) and shows the substantial similarity of their results. We note that all main effect parameters obtain statistical significance, on the basis of 95% CIs, in the models. For illustration of our methodology, we employ the more parsimonious model B in what follows.
Table 2.
First-level effect estimates from two models
| Effect† | Results for the following models: |
||
|---|---|---|---|
| Model B | Model AB | ||
| Grand mean | Median | −3.463 | −3.812 |
| 95% CI | (−4.015, −3.255) | (−3.931, −3.521) | |
| MR | Median | −0.7312 | −0.7377 |
| 95% CI | (−0.7541, −0.7100) | (−0.7993, −0.6712) | |
| LBW | Median | −0.7652 | −0.7686 |
| 95% CI | (−0.7789, −0.7526) | (−0.7831, −0.7548) | |
| IS | Median | −0.01868 | −0.01873 |
| 95% CI | (−0.02726, −0.01031) | (−0.02791, −0.00969) | |
| MTU | Median | −0.6741 | −0.7329 |
| 95% CI | (−0.6900, −0.6591) | (−0.7649, −0.6982) | |
| PT | Median | −0.5493 | −0.5512 |
| 95% CI | (−0.5633, −0.5351) | (−0.5702, −0.533) | |
| MR × LBW | Median | 0.1819 | 0.1837 |
| 95% CI | (0.1700, 0.1935) | (0.1716, 0.1961) | |
| MR × IS | Median | −0.01263 | −0.01264 |
| 95% CI | (−0.01875, −0.00658) | (−0.01884, −0.00623) | |
| MR × MTU | Median | −0.1924 | −0.1579 |
| 95% CI | (−0.2076, −0.1785) | (−0.1788, −0.137) | |
| MR × PT | Median | 0.008638 | 0.01439 |
| 95% CI | (−0.00458, 0.02165) | (0.001109, 0.02778) | |
| LBW × IS | Median | −0.08882 | −0.08856 |
| 95% CI | (−0.09646, −0.08107) | (−0.09666, −0.08044) | |
| LBW × MTU | Median | 0.1844 | 0.1886 |
| 95% CI | (0.1729, 0.1956) | (0.1777, 0.2000) | |
| LBW × PT | Median | 0.9077 | 0.9161 |
| 95% CI | (0.8952, 0.9208) | (0.9025, 0.9298) | |
| IS × MTU | Median | 0.004782 | 0.00427 |
| 95% CI | (5.49 × 10−6, 0.009607) | (−6.73 × 10−4, 0.009278) | |
| IS × PT | Median | 0.07691 | 0.07704 |
| 95% CI | (0.06922, 0.08475) | (0.06903, 0.08492) | |
| MTU × PT | Median | −0.04761 | −0.0391 |
| 95% CI | (−0.05984, −0.03526) | (−0.05055, −0.02775) | |
| MR × LBW × MTU | Median | −0.05651 | −0.05609 |
| 95% CI | (−0.06663, −0.04676) | (−0.06584, −0.04614) | |
| MR × LBW × PT | Median | −0.02172 | −0.0208 |
| 95% CI | (−0.03464, −0.00935) | (−0.03100, −0.01065) | |
| MR × IS × PT | Median | −0.01193 | −0.01199 |
| 95% CI | (−0.01747, −0.00628) | (−0.01749, −0.00645) | |
| MR × MTU × PT | Median | 0.02444 | 0.0245 |
| 95% CI | (0.013, 0.03611) | (0.0143, 0.03482) | |
| LBW × IS × PT | Median | 0.03118 | 0.03121 |
| 95% CI | (0.02373, 0.03869) | (0.02393, 0.03868) | |
| LBW × MTU × PT | Median | −0.06719 | −0.06723 |
| 95% CI | (−0.0799, −0.05373) | (−0.07728, −0.05704) | |
| MR × LBW × MTU × PT | Median | 0.01125 | 0.01104 |
| 95% CI | (8.48 × 10−4, 0.02155) | (0.001786, 0.02126) | |
MR, maternal race; IS, infant’s sex; MTU, maternal tobacco use; PT, preterm.
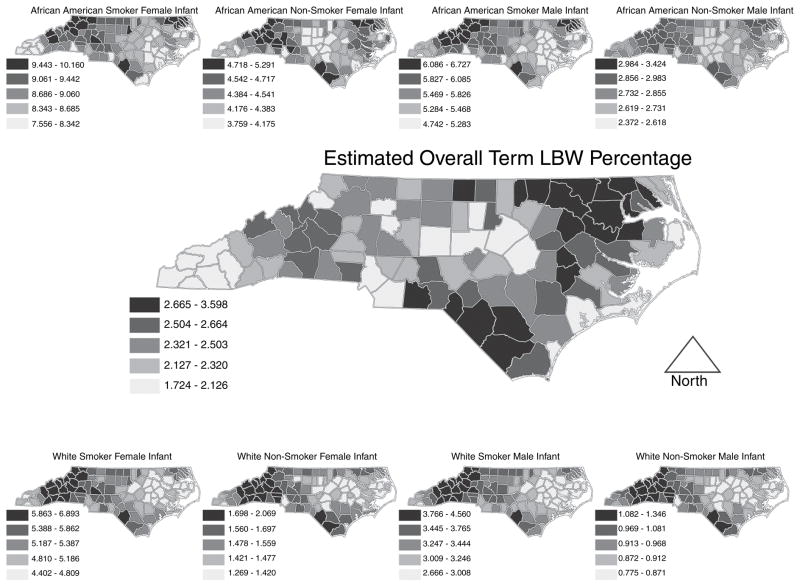
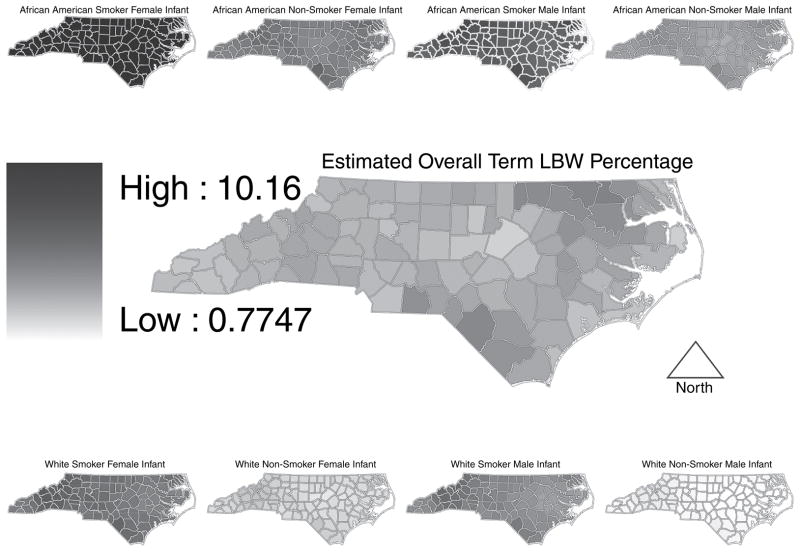
We next turn to other quantities of interest that can be built from the disaggregated probabilities. Fig. 1 shows estimated incidence of term LBW in its centre map, flanked by analogous maps for the eight race × infant sex × tobacco use subgroups. In Fig. 1 internally derived quintiles determine the cut points of the categories for each map, so, despite being on very different scales, the darkest shading denotes, in each map, the 20 counties with highest incidence, permitting us to make a particular type of relative comparison. Fig. 2 displays the same estimates of term LBW incidence, but with ‘global’ quintiles based on the rates over all eight of the race × infant sex × tobacco use subgroups, permitting absolute comparison across maps. Figs 1 and 2 together help to illuminate the story. Fig. 1 is most effective for seeing within-map variation whereas Fig. 2 reveals across-map variation. Though the flanking maps in Fig. 2 do not individually reveal much variation, a comparison between them is striking.
Fig. 1.
Overall term LBW incidence (centre) flanked by the eight race × infant sex × tobacco use combinations: in these maps, internally derived quintiles determine the cut points of the categories for each map
Fig. 2.
Overall term LBW incidence (centre) flanked by the eight race × infant sex× tobacco use combinations: in these maps, a continuous colour scale corresponds to the overall range of values
There are several striking features in Figs 1 and 2 that demonstrate the utility of our disaggregation model. In Fig. 1, the large central map of overall term LBW incidence shows that counties in eastern North Carolina generally suffer higher overall incidence. However, the eight flanking maps of the race × infant sex × tobacco use subgroups tell a different story. Counties in north-western North Carolina tend to have higher incidence rates for each subgroup. The overall incidence map is subject to confounding with respect to county level composition of the eight subgroups, i.e., in the overall map, a county’s rate is a weighted average of the eight subgroup rates, but the precise weighting differs from county to county in North Carolina, leading to the Simpson’s paradox situation (Agresti, 2002) that emerges in the ensemble of maps.
The notable visual impression from Fig. 2 is the lack of overlap among the eight flanking subgroup maps. For example, the counties in the upper leftmost map (for female infants born to African American mothers who used tobacco during pregnancy) are entirely in the upper quintile, whereas nearly every county in the lower rightmost map (for male infants born to white mothers who did not use tobacco during pregnancy) belongs to the lowest quintile. In this vein, the centre map in Fig. 2 shows that the variability in overall county term LBW birth incidence is rather small compared with the variability across the eight subgroup maps, and that within-map variability is small compared with across-map variability.
In addition, Fig. 2 recapitulates the parameter estimates from Table 2, particularly for the interaction of maternal race and LBW (MR × LBW). Table 2 shows that the magnitude of this effect (0.1819 in model B) is similar to the magnitude of the effect for the interaction of LBW and maternal tobacco use (LBW × MTU; 0.1844 in model B). Comparing the range of values in
the leftmost map in the lower row with the map second from the left in the upper row and
the map second from the right in the lower row with the rightmost map in the upper row
demonstrates what these parameter estimates also show: the effect of maternal race on LBW incidence is about the same as the effect of maternal tobacco use. Such conclusions might be difficult to obtain with other modelling approaches.
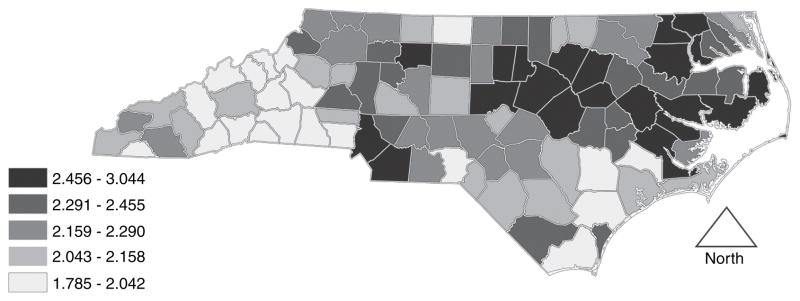
Although Figs 1 and 2 suggest differences across racial groups for adverse birth outcome incidence, our modelling framework is sufficiently flexible to support direct measures of racial disparity easily. For this, Fig. 3 depicts county level odds ratios OR of a term LBW birth for African Americans versus whites, . For these ORs a value of 1.0 would reflect no disparity, and Fig. 3 therefore documents the large (estimated county ORs ranged from 1.8 to 3.0, with median 2.2) and geographically pervasive burden in term LBW incidence borne by African Americans versus whites. In particular, areas in central and eastern North Carolina (mostly) suffer from greater-than-average levels of disparity. Other measures of disparity, such as those advocated by the Centers for Disease Control and Prevention (Keppel et al., 2005), may also be built from the model output.
Fig. 3.
Odds ratio OR of LBW for African Americans versus whites: this OR may be regarded as a measure of racial disparity in term LBW incidence, with the null value of 1.0 reflecting no disparity and values greater than 1.0 demonstrating the excess burden borne by African Americans
By viewing Fig. 3 in conjunction with Fig. 1 and/or Fig. 2, we see that
disparity can be high even where incidence of term LBW is relatively low for both African American and white mothers,
disparity can be low even where term LBW incidence is relatively high for both African American and white mothers and
disparity can (of course) be high where African American mothers have relatively high term LBW incidence while white mothers have relatively low incidence.
For (a) and (b), policy makers might have competing interests and trade-offs to consider in crafting intervention strategies, whereas for (c) intervention strategies might be focused on a subset of the county population. Such intervention strategies based on Figs 1–3 could consider whether to target the geographic areas with the worst overall outcomes, the subpopulations with the worst outcomes, areas with the worst racial disparity or some combination of these.
4.2. Comparing the conditional and disaggregation approaches
Finally, we compare our approach with the conditional model given in Section 3.1. We consider two different outcomes for this comparison:
term LBW, as in Section 4.1, and
LBW.
For fits to the North Carolina birth record data, the county level 95% CIs from the conditional model are wider than the corresponding 95% CIs from the disaggregation model for both outcomes. For term LBW, the county level CIs are on average about 30% wider whereas for LBW the CIs are on average about 25% wider, and in many cases more than 40% wider (for both outcomes). Further, for the four largest counties (each with over 20 000 births), where we would expect both models to perform well, the CIs are still about 15% wider for the usual model for term LBW and over 25% wider for LBW. Finally, we recall that, for term LBW, fitting the conditional model excluded preterm births from the analysis, whereas the disaggregation approach allows us to use all the data. This may explain the even poorer performance of this model for this outcome.
To support further the advantages of the disaggregation model, we investigated a simulated data set. To place the simulation on more familiar terrain for the usual model, we did not include preterm birth as an outcome, focusing only on the other four dichotomous variables (maternal race, birth weight outcome, infant sex and maternal tobacco use). Simulated data were generated from a known model similar to the 5 years of North Carolina data and with spatially varying cell probabilities, i.e. cell counts for the 100 × 24 = 1600 categories were generated. The disaggregation model was fitted to these cell counts whereas the usual model was fitted to totals marginalized over the demographic variables. Model performance was based on estimation of the 100 known county level probabilities of LBW. As expected, both models achieved roughly 95% empirical coverage: 93/100 for the disaggregation model and 97/100 for the conditional model. However, the CIs were roughly 50% wider for the usual model compared with the disaggregation model. Also, the disaggregation model had an MSE that was less than half of the MSE from the usual model. In other words, the interval estimates for the usual model were too long and not well centred. Lastly, turning to the 100 × 16 = 1600 underlying cell probabilities which the disaggregation model estimates, satisfyingly, we found 1528 of 1600 CIs (95.5%) contained the true cell probability.
5. Summary and extensions
We have argued that, in the case where we have observations assigned to areal units but which retain individual characteristics, there are numerous advantages to modelling at the individual level through the use of log-linear models incorporating spatial random effects. In particular, we can spatially investigate arbitrary marginal and conditional probabilities. In the context of public health databases, this enables us to illuminate disparities in a richer fashion than previously available.
In this analysis, we studied term LBW in North Carolina with such a model. We observed patterns that might have been difficult or impossible to study with other techniques, such as the higher overall rates in eastern North Carolina yet higher rates for each subgroup in western North Carolina. This same model was used to provide a picture of racial disparity in term LBW, and to classify areas of high disparity into subcategories that might call for different intervention strategies.
In future work, we shall apply this modelling approach to contingency tables of much higher dimension and to tables involving ordinal classifications. In addition, there will be interest in how selected probabilities and disparities change not only across space but also evolve in time, for instance, under an annual model for the birth record data from Section 1.2. This will lead us to dynamic versions of our approach, perhaps through space–time auto-regressive random-effects specifications.
Finally, there are data sets where we do have the geocoded locations that are associated with the individuals. This would encourage us to work at the point level rather than the areal unit level. Conceptually, we would envision a spatial process of log-linear models. Practically, we would have one multinomial trial at each of locations rather than n multinomial trials ignoring location or using only areal unit information. Structured spatial dependence enables us to handle the former case, and investigation of this approach is currently work in progress.
Acknowledgments
We thank Sharon Edwards for preparing the North Carolina birth record database for analysis and Joshua Tootoo for preparing the maps displaying output from the analysis. We also thank Dr Geeta Swamy and Simone Gray for their contributions to the analysis. This publication was made possible by grant 5P20-RR-020782-03 from the National Center for Research Resources, a component of the National Institutes of Health, and by grant 5P30-ES-011961-04 from the National Institute of Environmental Health Sciences, National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Appendix A: Model fitting
We build our fully Bayesian model as follows. For prior distributions, we use for t = 1, …, r; ηtu ∼ N(0, 1002) for t = 1, …, r and u = 1, …, q, and for certain t, as given in Section 1.2. For the t without spatial structure, , with the exception of the grand mean term. For the grand mean term, each parameter received an N(0, 1002) prior. The hyperpriors for the τt s are τt ∼ gamma(0.5, 0.005), which is discussed in Wakefield et al. (2001). The joint posterior is
writing (φ0, φ(s), η) for λ. After simplification, the likelihood term is
The resulting full conditional distributions are as follows:
;
, where δs is the set of neighbours of s, defined by adjacency so that ωsv = 1 if v ∈ δs (and otherwise ωsv = 0) with the convention that ωss = 0, where , and where ;
;
, based on Mollié (1996).
We implemented our fully Bayesian model by using Markov chain Monte Carlo methods (Gilks et al., 1996). We ran three overdispersed, random, parallel chains of 15 000 Markov chain Monte Carlo simulations, discarding the first 5 000 iterations from each chain as burn-in and retaining every 10th iteration after burn-in (to reduce potential problems with auto-correlation), for a total of 3 × 10 000 = 30 000 samples for posterior inference. There were no problems with convergence on the basis of Gelman–Rubin statistics and visual inspection of the mixing of the chains (Gelman and Rubin, 1992).
References
- Agresti A. Categorical Data Analysis. New York: Wiley; 2002. [Google Scholar]
- Banerjee S, Carlin BP, Gelfand AE. Hierarchical Modeling and Analysis for Spatial Data. Boca Raton: Chapman and Hall–CRC; 2004. [Google Scholar]
- Besag J. Spatial interaction and the statistical analysis of lattice systems (with discussion) J R Statist Soc B. 1974;36:192–236. [Google Scholar]
- Besag J, York J, Mollié A. Bayesian image restoration, with two applications in spatial statistics. Ann Inst Statist Math. 1991;43:1–20. [Google Scholar]
- Clayton D, Kaldor J. Empirical Bayes estimates of age-standardized relative risks for use in disease mapping. Biometrics. 1987;43:671–681. [PubMed] [Google Scholar]
- Fleiss JL, Levin B, Paik M. Statistical Methods for Rates and Proportions. 3. New York: Wiley; 2003. [Google Scholar]
- Gelfand AE, Vounatsou P. Proper multivariate conditional autoregressive models for spatial data analysis. Biostatistics. 2003;4:11–15. doi: 10.1093/biostatistics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Gelman A, Price PN. All maps of parameter estimates are misleading. Statist Med. 1999;18:3221–3234. doi: 10.1002/(sici)1097-0258(19991215)18:23<3221::aid-sim312>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rubin D. Inference from iterative simulation using multiple sequences. Statist Sci. 1992;7:457–472. [Google Scholar]
- Gilks WR, Richardson S, Spiegelhalter DJ, editors. Markov Chain Monte Carlo in Practice. London: Chapman and Hall; 1996. [Google Scholar]
- Jin X, Carlin BP, Banerjee S. Generalized hierarchical multivariate CAR models for areal data. Biometrics. 2005;61:950–961. doi: 10.1111/j.1541-0420.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- Keppel K, Pamuk E, Lynch J, Carter-Pokras O, Kim I, Mays V, Pearcy J, Schoenbach V, Weissman J. Methodological issues in measuring health disparities. Vitl Hlth Statist, ser 2. 2005;(141) [PMC free article] [PubMed] [Google Scholar]
- Kullback S, Leibler RA. On information and sufficiency. Ann Math Statist. 1951;22:79–86. [Google Scholar]
- Mollié A. Bayesian mapping of disease. In: Gilks WR, Richardson S, Spiegelhalter DJ, editors. Markov Chain Monte Carlo in Practice. London: Chapman and Hall; 1996. pp. 359–379. [Google Scholar]
- Raudenbush SW, Bryk AS. Hierarchical Linear Models: Applications and Data Analysis Methods. Thousand Oaks: Sage; 2002. [Google Scholar]
- Simpson EH. The interpretation of interaction in contingency tables. J R Statist Soc B. 1951;13:238–241. [Google Scholar]
- US Department of Health and Human Services. Healthy People 2010: Understanding and Improving Health. 2. Washington DC: US Government Printing Office; 2000. [Google Scholar]
- Wakefield J, Best N, Waller L. Bayesian approaches to disease mapping. In: Elliott P, Wakefield J, Best N, Briggs D, editors. Spatial Epidemiology: Methods and Applications. Oxford: Oxford University Press; 2001. pp. 104–127. [Google Scholar]
- Waller LA, Carlin BP, Xia H, Gelfand AE. Hierarchical spatio-temporal mapping of disease rates. J Am Statist Ass. 1997;92:607–617. [Google Scholar]
- Waller L, Gotway C. Applied Spatial Statistics for Public Health Data. New York: Wiley; 2004. [Google Scholar]