Abstract
We have identified a novel mutation in apoA-I (serine 36 to alanine; S36A) in a human subject with severe hypoalphalipoproteinemia. The mutation is located in the N-terminal region of the protein, which has been implicated in several functions, including lipid binding and lecithin:cholesterol acyltransferase (LCAT) activity. In the present study, the S36A protein was produced recombinantly and characterized both structurally and functionally. While the helical content of the mutant protein was lower compared with wild-type (WT) apoA-I, it retained its helical character. The protein stability, measured as the resistance to guanidine-induced denaturation, decreased significantly. Interestingly, native gel electrophoresis, cross-linking, and sedimentation equilibrium analysis showed that the S36A mutant was primarily present as a monomer, notably different from the WT protein, which showed considerable oligomeric forms. Although the ability of S36A apoA-I to solubilize phosphatidylcholine vesicles and bind to lipoprotein surfaces was not altered, a significantly impaired LCAT activation compared with the WT protein was observed. These results implicate a region around S36 in apoA-I self-association, independent of the intact C terminus. Furthermore, the region around S36 in the N-terminus of human apoA-I is necessary for LCAT activation.
Keywords: heart disease, reverse cholesterol transport, structure, lecithin:cholesterol acyltransferase
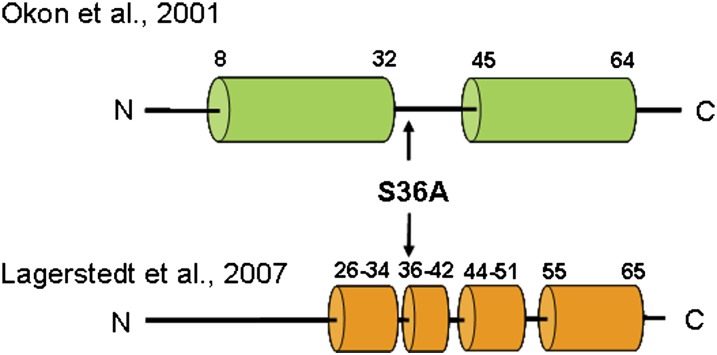
Plasma concentrations of high density lipoprotein (HDL) are a protective lipoprotein inversely correlated with coronary artery disease (CAD) risk. Apolipoprotein A-I (apoA-I), as the main protein component of plasma HDL, represents the major functional domain of HDL, mediating the interaction of HDL with lecithin:cholesterol acyltransferase (LCAT) and ATP binding cassette protein A1 (ABCA1) (reviewed in Refs. 1 and 2). Furthermore, apoA-I must possess structural stability to maintain the structural integrity of HDL, as well as conformational flexibility to accommodate the transformation from lipid-poor to lipid-rich HDL and VLDL-bound forms (3–6). A number of structural studies of lipid-free apoA-I have been published, uniformly demonstrating helix bundle architecture (2, 7, 8). A crystal structure of an N-terminal deletion mutant of apoA-I (Δ1-43) assumed a long, drawn-out horseshoe shape, suggestive of a lipoprotein-bound structure. The limitation of this structure is that the protein was crystallized in the absence of lipid, leading to speculation whether this represents a de facto lipid-bound structure. Furthermore, the protein lacked the N-terminus, highlighting the higher lipid-binding affinity of this region. An NMR solution structure of an N-terminal fragment of apoA-I (9) showed that apoA-I in lipid-mimetic solution contained two helices at the N-terminus (residues 8-32, residues 45-64) (Fig. 1). Based on this information, Scott et al. designed Δ1-43 and Δ1-65 N-terminal deletion mutants of apoA-I and analyzed them in a number of functional studies (10). Both mutants had reduced LCAT activation function (50% and 70% reduction, respectively) but had no impairment of the ability to promote cholesterol efflux. These findings suggest that the first helices (residues 8-64) may be important for LCAT activation. When these proteins were produced in the mouse by adenovirus infection, Δ1-43 behaved at a lower efficiency compared to the wild-type. Therefore, deletion of the first helix does not completely disrupt the structural integrity or the function of the Δ1-43 mutant but makes the protein less efficient. The solution structure determined by Lagerstedt et al. (11) using electron paramagnetic resonance (EPR) spectroscopy showed that the N-terminus (residues 1-65) is not part of the helix bundle but, rather, shorter helices broken by β-structure or no structure (Fig. 1). This is supported in part by the observation of Beckstead et al. (12) who showed that deletion of the N- and C termini (residues 44-186) created a folded, stable, soluble protein.
Fig. 1.
Summary of the structures of apoA-I. The helix boundaries are based on the NMR solution structure (Okon et al. (9)) and EPR analysis (Lagerstedt et al. (11)) of the N-terminal region of apoA-I. apoA-I, apolipoprotein A-I; EPR, electron paramagnetic resonance.
Based on the NMR solution structure, the apoA-I N-terminus includes a proposed N-terminal G* helix (residues 8-32), a “linker region” (residues 33-44), and the class A helix (residues 45-64). The linker region is different from other unstructured regions in that it is longer and therefore not likely to form a turn in the helix bundle. We have shown that the first G* helix is very important in lipid binding (10) and may act in concert with the C terminus to initiate binding to lipid surfaces, as both regions have high lipid-binding affinity (8, 12–16). Others have shown in the so-called “solar flare” and “double super helix” models that the N- and C-terminal helices, including the “linker region,” may interact in homodimers to stabilize the proteins in a lipid-bound state but not necessarily directly interact with lipids (17, 18). Therefore, this region may perform a role in allowing initiation of binding of the N-terminal helix for interaction with lipid or LCAT, in stabilizing a lipid-bound conformation, or in maintaining stability of the helix bundle in the lipid-free state. Interestingly, there have been very few reported mutations in this linker region: 34STOP (functional), A37T (variant-nonfunctional), G39E (variant-nonfunctional) (taken from the Human Gene Mutation Database; www.hgmd.cf.ac.uk) (19). We have recently identified a novel heterozygous mutation (S36A) in a 63-year-old male with HDL deficiency [HDL-C, 0.70 mM (27 mg/dl), less than fifth percentile for age and sex (20)]. We predicted that this mutation was nonfunctional, and the patient exhibited no signs of amyloidosis or CAD. In the current report, we have specifically addressed the effect of the S36A mutation on the structure and function of apoA-I. Our data suggests that S36 is necessary for normal apoA-I quaternary structure in the lipid-free state (self-association) and for function (LCAT activation) in a lipid-bound state. Thus, the S36A mutation does have a significant effect on apoA-I function and may explain the decreased plasma HDL-C in the heterozygous patient.
MATERIALS AND METHODS
Study subject
The subject was a healthy 63-year-old male with a body mass index of 30.1 kg/m2 presenting with a total cholesterol of 3.28 mM (less than fifth percentile), triglycerides, 0.85 mM (less than tenth percentile), LDL-C, 2.19 mM (less than fifth percentile), and HDL-C, 0.70 mM (less than fifth percentile) (20). The study was approved by the University of Ottawa Heart Institute Human Research Ethics Committee and written informed consent was obtained from the subject.
Bacterial expression of human apoA-I
Using a previously described bacterial expression system for apoA-I, S36A mutant and wild-type (WT) apoA-I were cloned and expressed in E. coli (21). For the mutant from the low plasma HDL-C patient, serine 36 was replaced with alanine. All proteins were readily produced by bacteria, and purified proteins were soluble and stable in solution, suggesting an overall folded structure. Mass spectrometry confirmed that the proteins produced were of the expected size (data not shown).
Secondary structure analysis and stability
The ellipticity of apoA-I was measured in a Jasco 810 polarimeter. Far UV scans were performed between 190 and 260 nm using a 0.2 mg/ml protein concentration in a 0.1 cm circular cuvette. Plots are the average of three scans (scan rate 20 nm/min). The ellipticity was converted to molar ellipticity as described in detail previously, and the helical content was estimated by the ellipticity at 222 nm (22). To determine the stability, apoA-I was incubated in guanidine-HCl for 16 h, and the ellipticity was measured at 222 nm.
Phospholipid vesicle solubilization
Dimyristoylphosphatidylcholine (DMPC) was purchased from Avanti Polar Lipids, dissolved in chloroform/methanol (2:1; v:v), and dried. The lipid film was rehydrated in buffer and vortexed for 1 min. Large unilamellar vesicles (LUV) were made by extrusion through a 200 nm filter in the Avanti mini extruder and used the same day to measure the rate of vesicle solubilization. In 1 ml of buffer kept at 24°C, 500 µg of LUVs and 100 µg of protein were mixed, and the change in light scatter was monitored for 30 min at 325 nm in a Shimadzu UV2401 PC spectrophotometer equipped with a TCC-240A Peltier unit.
Fluorescence studies
ApoA-I (25 µg) was incubated with 250 µM 8-anilinonaphthalene-1-sulfonate (ANS, Sigma-Aldrich) in 10 mM sodium phosphate, pH 7.2. ANS was excited at 395 nm, and the fluorescence emission measured between 400 and 550 nm (excitation/emission slit width 5 nm) in a Perkin Elmer LS 55 fluorescence spectrometer. Tryptophan fluorescence was measured by excitation of 25 µg apoA-I at 295 nm, monitoring the emission between 300 and 450 nm.
Lipoprotein binding and lipid efflux
The efflux assay was performed as previously described (20, 21, 23). ApoA-I binding to low density lipoproteins (LDL) was carried out according to Liu et al. (24). LDL (50 µg protein) was incubated with 2 mU of phospholipase C (Sigma-Aldrich), and the absorbance was monitored at 340 nm in a microplate reader (Thermo Labsystems) for 60 min. Incubations were carried out in a 200 µl volume in a 96-well microplate in the absence or presence of various quantities of apoA-I (between 5 and 50 µg).
Electrophoresis analysis
Native polyacrylamide gel electrophoresis (PAGE) was carried out with 4-20% Tris-glycine precast gels (Invitrogen) in the absence of sodium dodecylsulfate (SDS). Gels were loaded with 30 µg apoA-I in sample treatment buffer, run for 4 h, and stained with 0.5% (w/v) naphthol blue black (Sigma-Aldrich). Cross-linking was carried out with dimethyl suberimidate (DMS) in 0.1 M triethanolamine, pH 9.7 (25). ApoA-I (10 µg) was incubated for 2 h with DMS (5, 20, or 50 µg) in 25 µl, after which an equal volume of SDS loading buffer was added, heated for 5 min at 95°C, and analyzed by 4-20% precast SDS-PAGE gels.
Two-dimensional gel electrophoresis
Serum from a pool of normolipidemic males and the S36A apoA-I patient were separated according to charge (agarose, horizontal axis) and size (polyacrylamide gradient, 3% to 20% gel, vertical axis) as previously described (26, 27). ApoA-I was detected with 125I-labeled anti-apoA-I antibody.
Analytical ultracentrifugation
Sedimentation equilibrium experiments were conducted at 22°C in a Beckman XL-I analytical ultracentrifuge using absorbance optics, as described by Laue and Stafford (28). Aliquots (110 µl) of the sample solution were loaded into six sector CFE sample cells, allowing three concentrations to be run simultaneously. Runs were performed at a minimum of three different speeds, and each speed was maintained until there was no significant difference in r2/2 versus absorbance scans taken 2 h apart to ensure that equilibrium was achieved. Sedimentation equilibrium data were evaluated using the NONLIN program, which employs a nonlinear least-squares curve-fitting algorithm described by Johnson et al. (29). The program allows for analysis of both single and multiple data files and can be fit to models containing up to four associating species, depending upon which parameters are permitted to vary during the fitting routine. The protein's partial specific volume and the solvent density were estimated using the Sednterp program (28).
LCAT activity
LCAT activity was assayed in human plasma using the method of Stokke and Norum (30) modified by Lacko et al. (31), as previously described (10). For the in vitro assays, LCAT was purified according to Albers et al. (32), and the cholesterol esterification studies were performed as described previously (10). The reconstituted lipoprotein particles (rHDL) used in these studies, prepared as before (10, 33), were labeled with [1a,2a-3H] cholesterol (PerkinElmer Life Sciences) and with recombinant WT human apoA-I, recombinant S36A human apoA-I, or a 1:1 molar mixture of WT and S36A apoA-I. Two different types of experiments were performed. Different concentrations of particles were incubated with LCAT enzyme for 10 min at 37°C to determine the initial rate constants: apparent Km (appKm) and Vmax, as previously described (10, 33). In the second experiment, the time course of cholesteryl ester (CE) formed was followed over 60 min by incubating rHDL particles at increasing concentrations with LCAT, as previously described (10, 33). For both sets of experiments, the values are the mean (± SD) of quadruplicate measurements. Statistical analysis was performed using Student's t-test.
RESULTS
Secondary structure analysis and stability
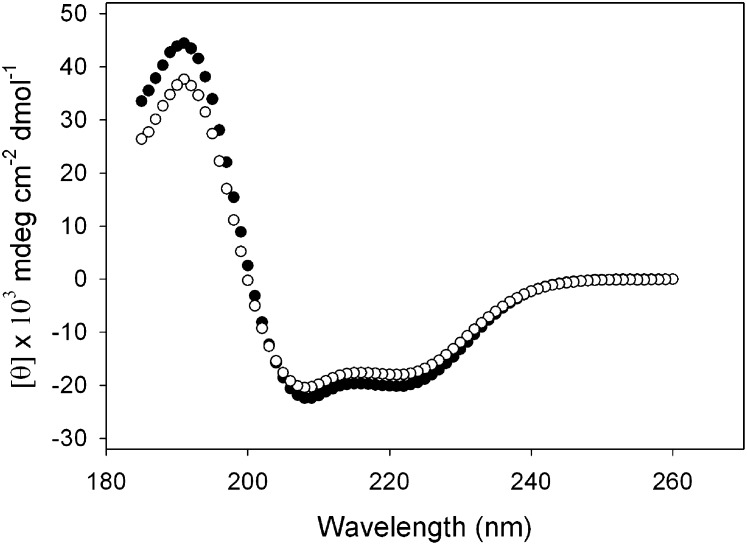
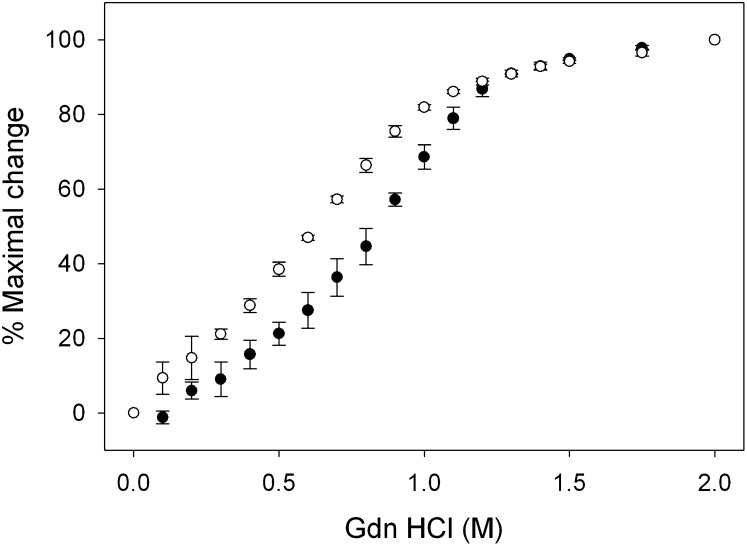
The structural properties of the S36A apoA-I mutant were characterized by circular dichroism (CD). As shown in the far UV scans in Fig. 2, the mutant was largely α-helical, indicated by the distinct troughs at 208 and 222 nm. Calculations based on the ellipticity showed that the helical content of the mutant protein was 54.1 ± 3.0%, which was lower than WT apoA-I (64.1 ± 4.2%), but it retained its overall helical character. The protein was then analyzed for stability by measuring the resistance to guanidine-HCl-induced denaturation to examine if the single amino acid substitution led to perturbations of the protein fold. Increasing concentrations of the denaturant caused the protein to unfold, measured by the change in ellipticity at 222 nm (Fig. 3). WT apoA-I displayed a midpoint of denaturation ([Gdn-HCl]1/2) of 0.82 ± 0.04 M, similar to previously reported values (34). In comparison, the S36A mutant displayed a significantly lower [Gdn-HCl]1/2 value centered on 0.56 ± 0.01 M guanidine-HCl.
Fig. 2.
Far UV CD scan of apoA-I: wild-type apoA-I (closed circles), and S36A apoA-I (open circles). apoA-I, apolipoprotein A-I, CD, circular dichroism; UV, ultraviolet.
Fig. 3.
Resistance of apoA-I to guanidine-HCl induced denaturation. The change in ellipticity was measured at 222 nm, and from the resulting plots the [Gdn-HCl]1/2 was determined for WT apoA-I (closed circles), and S36A apoA-I (open circles). Error bars represent the standard deviation (n = 3). apoA-I, apolipoprotein A-I; WT, wild-type.
Fluorescence studies
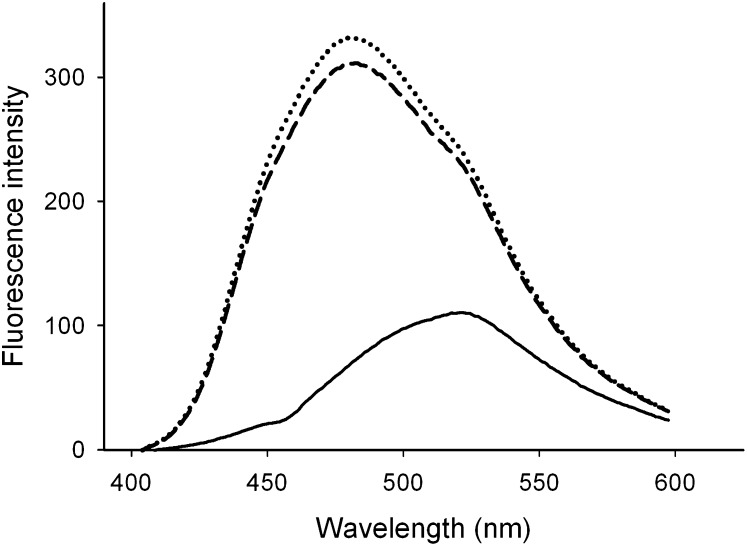
The observed decrease in protein stability provided a possibility that apoA-I helix packing was compromised as a result of the mutation, which could lead to the partial exposure of the hydrophobic interior. Therefore, ANS was used to probe for exposed hydrophobic pockets, which form potential ANS binding sites. The fluorescence intensity of ANS increased 3-fold upon incubation with WT apoA-I, and the emission spectrum showed a strong blue-shift, as shown earlier (7). The ANS emission spectrum in the presence of S36A apoA-I was very similar, and only a marginal increase in the intensity at the wavelength of maximum emission (λmax) was observed (Fig. 4).
Fig. 4.
ANS fluorescence. ANS fluorescence alone (solid line) is significantly enhanced by adding WT (dashed line) or S36A apoA-I (dotted line), accompanied by a blue-shift to 480 nm. ANS, 8-anilinonaphthalene-1-sulfonate; apoA-I, apolipoprotein A-I.
ApoA-I contains four tryptophan residues at positions 8, 50, 72, and 118. To assess for apoA-I structural changes caused by the mutation, the relative exposure to buffer of the tryptophan residues was determined by measuring the maximum wavelength of emission (λmax). However, no differences were observed as both WT and S36A apoA-I displayed a λmax of tryptophan emission ∼341 nm. These results suggest that there was no major perturbation in the environment surrounding these tryptophan residues.
Phospholipid vesicle solubilization
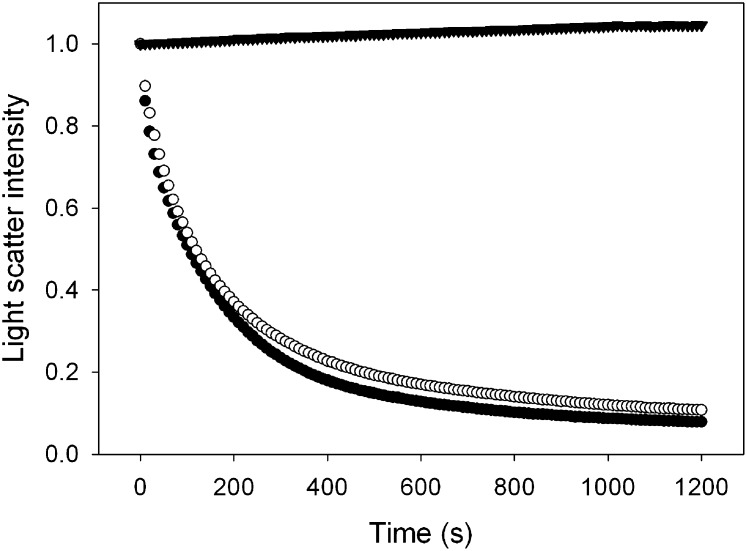
The ability of S36A mutant apoA-I to transform 1, 2-dipalmitoyl-phosphatidylcholine (DPPC) vesicles into discoidal complexes (nanodisks) was not affected. In addition, the size of the newly formed nanodisks was similar to nanodisks prepared with WT protein (based on Native-PAGE, data not shown). The kinetics of the formation of DPPC nanodisks are difficult to analyze; instead, vesicles of DMPC were employed. LUVs of DMPC (200 nm diameter, obtained by extrusion) were incubated in the presence of apoA-I at 24°C, the DMPC gel-liquid crystalline transition temperature. At this temperature, the turbid vesicle suspension was rapidly solubilized, resulting in the appearance of the characteristic small-sized nanodisks. This process was monitored spectrophotometrically at 325 nm, where the decrease in light scatter indicates the disappearance of LUVs. Vesicle solubilization is a rapid process; and a dispersion of 250 μg DMPC vesicles was completely converted by WT apoA-I into nanodisks in about 600 s, with a first-order rate constant k of 9.8 × 10−3 s−1 (Fig. 5). S36A apoA-I was able to induce nanodisk formation at a similar rate (k = 8.3 × 10−3 s−1).
Fig. 5.
DMPC vesicle solubilization. The apoA-I variants were able to transform a turbid dispersion of unilamellar vesicles into nanodisks, as evident by the rapid decrease in light scatter intensity of the sample (measured by the absorbance at 325 nm). Shown are wild-type apoA-I (closed circles) and S36A apoA-I (open circles). The light scatter intensity remained unchanged in the absence of protein (closed triangles). apoA-I, apolipoprotein A-I; DMPC, dimyristoylphosphatidylcholine.
Lipoprotein binding and lipid efflux
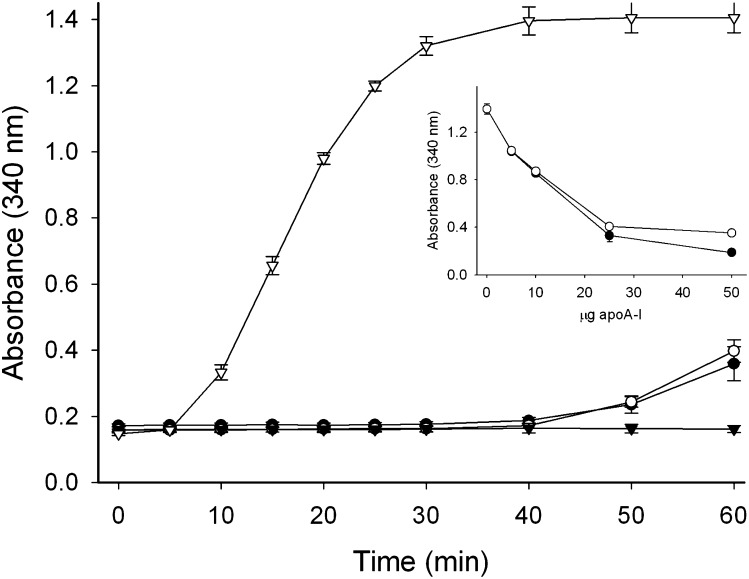
The ability to bind to a lipoprotein surface was tested using a modified LDL surface. Normally, apoA-I does not associate with LDL. However, when the lipoprotein particles are incubated in the presence of phospholipase-C, diacylglycerol appears on the lipoprotein surface resulting from hydrolysis of the phosphocholine headgroup of phosphatidylcholine, which is the main phospholipid on the LDL surface (24). These diacylglycerol-enriched LDL particles are unstable and quickly aggregate, a process that can be monitored by the increase in sample turbidity at 340 nm. When phospholipase-C and LDL were incubated in the presence of WT apoA-I, the solution remained optically clear. This indicated that apoA-I associated with the lipoprotein surface, thereby preventing aggregation. As seen in Fig. 6, 50 µg of apoA-I, either WT or S36A, was sufficient to protect a similar amount of LDL from phospholipase-C-induced aggregation. In an alternative approach, LDL and phospholipase-C were incubated in the presence of limited amounts of apoA-I. In this case, sample turbidity developed because only partial protection was provided and subtle differences between apolipoprotein variants may become evident. Nevertheless, similar absorbance values were obtained for both WT and S36A apoA-I at all protein concentrations tested, indicating a similar capacity to bind to the LDL surface (Fig. 6, inset). This finding showed that there was virtually no difference between mutant and WT apoA-I in the ability to bind to the lipolyzed LDL surface and prevent aggregation.
Fig. 6.
PL-C assay. LDL and phospholipase-C were incubated in the presence of wild-type apoA-I (closed circles) and S36A apoA-I (open circles). Control incubations of LDL (closed triangles) and LDL + phospholipase-C (open triangles) are also shown. Inset: The absorbance of incubations of LDL + phospholipase-C with decreasing amounts of WT apoA-I (closed circles) or S36A apoA-I (open circles). apoA-I, apolipoprotein A-I; PL-C, phospholipase-C; WT, wild-type.
The ability of apoA-I to remove phospholipid and free cholesterol from the surface of cells (termed “lipid efflux”) is dependent on the ATP-binding cassette transporter A1 (ABCA1) and is an essential function of apoA-I. Lipid efflux was measured for WT and S36A mutant apoA-I using standard protocols (20, 21, 23). Lipid efflux was the same for the WT and S36A mutant apoA-I in both a time course assay and with increasing concentrations of apoA-I (data not shown).
ApoA-I oligomerization
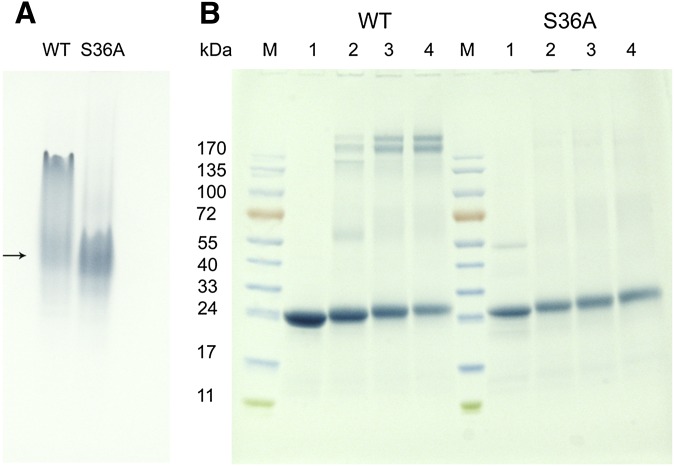
Under lipid-free conditions, human apoA-I is known to self-associate (35), which is dependent on the lipid-binding regions of the N- and C terminus (8, 10, 14, 15, 36). To gain insight into the oligomerization state of S36A apoA-I, the proteins were analyzed by nondenaturing-PAGE. The gel showed that WT apoA-I was present as multiple bands, indicative of protein-protein interactions resulting in the formation of dimers and tetramers (Fig. 7A). The migration pattern of an identical concentration of S36A apoA-I was distinctively different, showing a more homogeneous apoA-I population. This indicates that the mutation led to a decrease in protein-protein interactions resulting in fewer oligomers.
Fig. 7.
PAGE analysis. A: Non denaturing-PAGE of lipid-free apoA-I (in the absence of SDS). Lane 1: 30 μg of WT apoA-I; lane 2: 30 μg S36A apoA-I. The arrow indicates the position of monomeric apoA-I. B: SDS-PAGE of apoA-I cross-linked with DMS. The molecular weight of the marker proteins (M) are shown on the left. Lane 1: 10 μg of apoA-I in the absence of cross-linker. Lane 2: apoA-I (10 μg) and 5 μg DMS, 20 μg DMS (lane 3), and 50 μg DMS (lane 4). WT apoA-I is shown in the left lanes, S36A apoA-I is shown in the right lanes. apoA-I, apolipoprotein A-I; DMS, dimethylsuberimidate; WT, wild-type.
ApoA-I was then crosslinked with DMS and analyzed by SDS-PAGE. This revealed that the majority of the S36A protein was in a monomeric state. However, WT apoA-I showed multiple cross-linked species of dimer, tetramer, and the appearance of a prominent band in the top portion of the gel representing apoA-I octomers (Fig. 7B). These results support the idea that the N-terminus is involved in the propensity to form oligomers and that the single mutation S36A was sufficient to disrupt this.
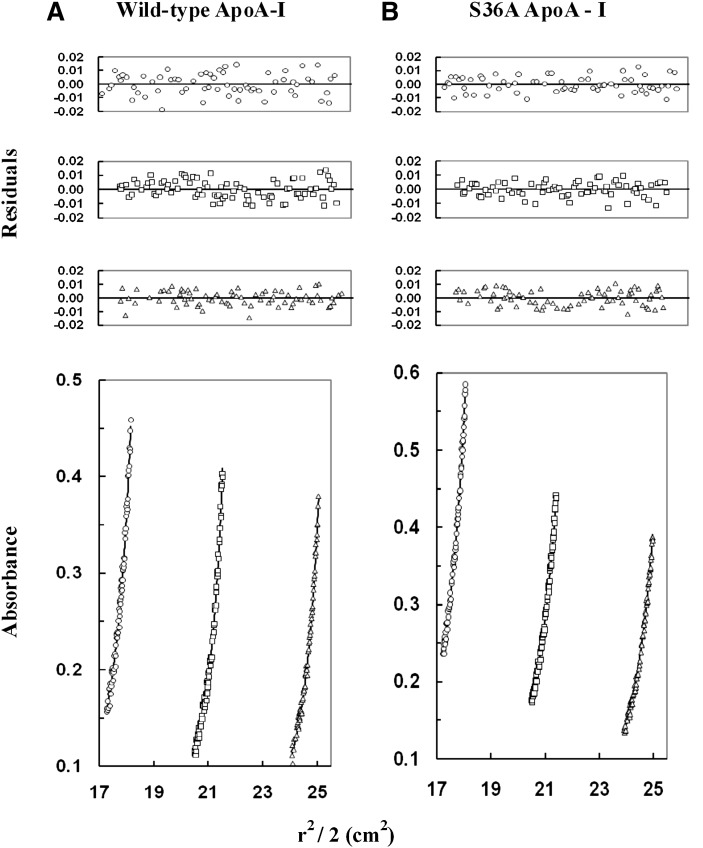
To confirm the oligomeric state of S36A apoA-I, sedimentation equilibrium was performed. Sedimentation equilibrium data were obtained using a Beckman XL-I analytical ultracentrifuge equipped with absorbance optics monitoring the absorbance at 280 nm. The runs were carried out at speeds ranging from 1.6 × 104 rpm to 2.0 × 104 rpm and with loading protein concentrations from 0.6 to 0.3 mg/ml. The combined data from nine data sets were analyzed using the nonlinear least-squares curve-fitting program NONLIN. The data obtained for WT and S36A apoA-I had a global molecular mass average of 36,864 Da and 32,642 Da, respectively (Fig. 8). The modeling of both systems showed evidence of monomer-tetramer with the monomer being predominant, more so in the case of the S36A apoA-I. As well, the calculated association constant (Ka) values were 8.561 × 1014 M−1for WT apoA-I and 6.119 × 1014 M−1for S36A apoA-I (Fig. 8). The global molecular average results suggested that S36A apoA-I clearly had less aggregation than WT apoA-I. The lower level of aggregation in S36A apoA-I was also reflected in the lower calculated Ka value. The molecular mass values obtained are in general agreement with the molecular mass of the S36A protein based on the amino acid sequence (29,316.9 Da; recombinant proteins contain a prosequence) and provide evidence that S36A apoA-I is predominantly monomeric.
Fig. 8.
Sedimentation-equilibrium analysis of wild-type and mutant apoA-I. Wild-type apoA-I and S36A apoA-I were dissolved in 50 mM Tris (pH 7.0), 100 mM sodium chloride, and then centrifuged at 16,000, 18,000, and 20,000 rpm at 20°C (only the data collected at 16,000 rpm are shown). The protein concentration was 0.60 mg/ml (circles), 0.40 mg/ml (squares), or 0.30 mg/ml (triangles) for wild-type apoA-I (A) and for S36A ApoA-I (B). The lower graphs illustrate plots of r2/2 versus absorbance. The symbols represent measured data points, and solid lines represent best-fit curves to a monomer-tetramer model (A, B). The upper graphs illustrate the residuals from the fitting. The random, nonsystematic distribution of the residuals indicates a good fit of the data to the models. apoA-I, apolipoprotein A-I; WT, wild-type.
LCAT activation
To characterize the HDL in the plasma of the patient with the heterozygous mutation for S36A apoA-I, two-dimensional nondenaturing gradient gel electrophoresis (2D-NDGGE) analysis was performed. Plasma lipoproteins were electrophoresed by 2D-NDGGE, transferred to nitrocellulose, and probed with 125I-labeled anti-apoA-I-specific antibody, as previously described (26, 27). Plasma from S36A showed a distinct decrease in preβ- and α3-migrating HDL, but no decrease in other α-migrating HDL, compared with pooled plasma from three healthy male donors (supplementary Fig. I). The LCAT activity of the plasma collected from the patient with the heterozygous mutation for S36A apoA-I was measured. Relative rates of CE generation were significantly different between plasma from the S36A subject and a control subject (supplementary Fig. II).
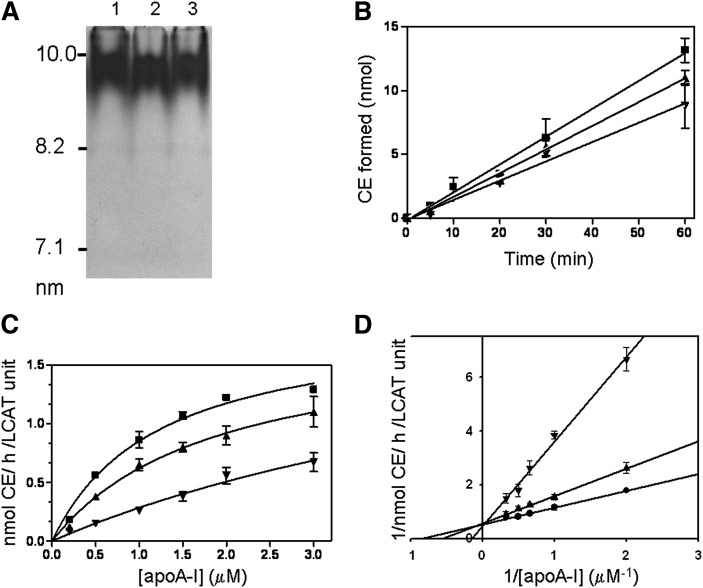
To fully assess the extent of functionality, two separate in vitro LCAT assays were performed. Reconstituted lipoprotein particles (rHDL) containing WT apoA-I (or S36A apoA-I or a 1:1 molar mixture of WT and S36A apoA-I) containing 3H-cholesterol (1-palmitoyl, 2-oleoyl-phosphatidyl choline (POPC):free cholesterol:apoA-I; 160:20:2 molar ratio) were generated and characterized (Fig. 9A).The three rHDL particles were very similar in composition (Table 1). First, rHDL containing WT apoA-I (or S36A apoA-I or a 1:1 molar mixture of WT and S36A apoA-I) was incubated with isolated LCAT for up to 60 min at 37°C. LCAT-generated CE was extracted, isolated by thin layer chromatography, and quantified by scintillation counting. The first-order rates of LCAT-generated CE were determined and found to be significantly decreased for S36A apoA-I compared with WT apoA-I (Fig. 9B). Second, increasing amounts of rHDL containing WT apoA-I, S36A apoA-I, or a 1:1 molar mixture of WT and S36A apoA-I (as above) were incubated with isolated LCAT enzyme for 10 min at 37°C (Fig. 9C). Values obtained were used to generate a Lineweaver-Burk double-reciprocal plot to determine appKm and Vmax (Fig. 9D). Calculated Vmax values were similar for all rHDLs, but appKm values were significantly different (Table 1). The appKm for the S36A rHDL was six times greater than the WT rHDL, demonstrating a lower efficiency of S36A rHDL to activate the LCAT enzyme than WT rHDL. Interestingly, the mixed rHDL had a significant but only moderate increase in appKm, suggesting that the mixed rHDL was less efficient at activating LCAT but that the S36A apoA-I did not act in a dominant fashion inhibiting the WT apoA-I. These results suggest that LCAT activity was significantly impaired in the S36A patient plasma and that the ability of S36A apoA-I to activate LCAT in in vitro assays was significantly impaired.
Fig. 9.
LCAT activation ability is significantly reduced for S36A apoA-I. A: According to McManus et al. (33), we prepared and isolated rHDL lipoprotein particles for the LCAT assay. Native polyacrylamide gradient gel electrophoresis was performed on the rHDL made from wild-type apoA-I (lane 1), S36A apoA-I (lane 2) and a 1:1 molar ratio of wild-type and S36A apoA-I (lane 3). The High Molecular Weight Calibration Kit (GE Healthcare) was used as standards (indicated on the left). B: WT (closed squares) or S36A (inverted triangles) apoA-I or a 1:1 molar mixture of WT and S36A apoA-I (upright triangles) made into reconstituted lipoprotein particles (rHDL) containing 3H-cholesterol was incubated with purified LCAT for up to 60 min at 37°C to ensure first order kinetics. LCAT-generated CE was determined over time and found to be significantly decreased for the S36A rHDL and mixed rHDL compared with WT apoA-I. Values are the mean (± SD) of duplicate experiments with quadruplicate measurements at each data point. C: Increasing concentrations of WT rHDL (closed squares), S36A rHDL (inverted triangles) and mixed rHDL (upright triangles) were incubated with purified LCAT for 10 min to get initial rate constants. D: The double-reciprocal plot of the reciprocal initial velocities versus the reciprocal of the rHDL concentrations was used to calculate the appKm and Vmax for the LCAT activation activity of each of WT rHDL (closed squares), S36A (inverted triangles), and mixed rHDL (upright triangles). apoA-I, apolipoprotein A-I; LCAT, lecithin:cholesterol acyltransferase; WT, wild-type.
TABLE 1.
Physical properties and LCAT kinetic data for reconstituted lipoprotein particles
| Reconstituted Lipoproteina | ApoA-I per Particlea | POPC:FC:ApoA-I Ratio (n = 6)a | appKmb | Vmaxb |
|---|---|---|---|---|
| μM | nmol CE/h/LCAT unit | |||
| WT rHDL | 2 | 87:10:1 | 1.17 ± 0.23 | 1.87 ± 0.08 |
| ±3 ± 0.4 | ||||
| Mixed rHDL | 2 | 87:10:1 | 1.79 ± 0.12c | 1.75 ± 0.09 |
| ±6 ± 1.0 | ||||
| S36A rHDL | 2 | 84:10:1 | 6.88 ± 0.23c | 2.19 ± 0.41 |
| ±4 ± 0.5 |
The reconstituted lipoprotein particles are WT apoA-I, S36A apoA-I, and 1:1 molar mixture of the two proteins. apoA-I, apolipoprotein A-I; LCAT, CE, cholesteryl ester; lecithin:cholesterol acyltransferase; WT, wild-type.
All reconstituted lipoprotein particles were prepared and characterized according to McManus et al. (33).
appKm and Vmax values were determined from Lineweaver-Burk plots.
Differences that are statistically significant (P < 0.05) were calculated by Student's t-test.
DISCUSSION
We have identified a novel mutation in apoA-I (S36A) in a subject with isolated hypoalphalipoproteinemia (20). To complement our previous studies on the role and function of the N-terminus (10), we carried out structure function studies to determine whether this mutation was the cause of the subject's low plasma HDL-C. Mutations in apoA-I have been implicated in amyloidosis disorders and HDL deficiencies (11, 37–41). Systemic amyloidosis results in tissue concentrations of amyloid and many other proteins (including apoA-I), and it was found that certain mutations on apoA-I were the cause. Many mutations in apoA-I have been reported to cause hypoalphalipoproteinemia (Human Mutation Gene Database; www.hgmd.cf.ac.uk). It has been suggested that mutations in apoA-I in the central domains may affect LCAT and that mutations at the N-terminus cause amyloidosis (19, 42). In this exceptional case, the S36A mutation found in the N-terminus affected LCAT activation, but there was no evidence from clinical investigation of amyloidosis in the study subject.
The S36A mutation of apoA-I resulted in a small decrease in the apoA-I helical content (Fig. 2). In contrast, a more pronounced effect was seen when the protein was analyzed for its stability, measured by its resistance to denaturation induced by guanidine-HCl. The [Gdn-HCl]1/2 was decreased from 0.82 M for WT apoA-I to 0.56 M for the S36A mutant (Fig. 3). While the single point mutation at position 36 decreased the protein stability significantly, ANS and Trp fluorescence characteristics were essentially similar for WT and S36A apoA-I (Fig. 4). Thus, the single amino acid change did not have a major effect on the overall helix packing, which should have resulted in a more pronounced change in the fluorescence properties. Instead the mutation may have induced a more localized change in protein structure in the proximity of helix-1. Another group showed that removal of the N-terminal and C-terminal part, creating an apoA-I-(44-186) variant, resulted in a decrease in [Gdn-HCl]1/2 to values observed in our study (12). This report lends support to the idea that the N-terminal helix remains a “loosely attached” part of the helix bundle. It is plausible that the central helices of the apoA-I protein core are responsible for the basic protein stability. The N-terminal helix may be more flexible, providing additional stability to the protein that results in a tighter packing of apoA-I or perhaps in a homodimer formation. We have shown earlier that chicken apoA-I was structurally and functionally analogous to human apoA-I (43) but was monomeric (7, 44). In addition, the [Gdn-HCl]1/2 for chicken apoA-I was substantially lower (0.6 M) (44). It is plausible that the added stability of WT human apoA-I is due to self-association. Therefore, in the experiments described here, the stability of S36A apoA-I may have been decreased by a mutation that limited self-association. Nondenaturing PAGE analysis showed that lipid-free S36A apoA-I displayed a significantly altered electrophoretic mobility compared with WT apoA-I (Fig. 7). While WT apoA-I was present as a smear of higher molecular weight products, S36A apoA-I was visible as a more distinct, low molecular weight band. Cross-linking studies showed that the S36A mutant was predominantly present as a monomer, with some dimers at higher concentrations of crosslinker. On the other hand, WT apoA-I formed dimers, trimers, tetramers, and octamers, similar to what has been previously reported (34, 35). We note that our use of a higher ratio of crosslinker to protein may account for the lack of more dimers and trimers. In addition, sedimentation equilibrium data showed that S36A apoA-I is predominantly monomeric, more so than WT apoA-I (Fig. 8). However, S36A apoA-I is not entirely monomeric, as the C terminus is also responsible for self-association, and this region of the protein remains intact to mediate protein-protein and protein-lipid interactions.
Lipid-free human apoA-I self-associates, and this characteristic has impeded many studies. Proteins that self-associate are poor substrates for crystallization and cannot be used in NMR studies. Much effort has been directed toward generating a mammalian monomeric apoA-I to aid in structural studies. A study using site-directed mutagenesis at multiple sites created a monomeric mouse apoA-I for structural studies (45, 46). We have identified a mutant of apoA-I that is largely monomeric and that may serve as a model protein for further structural studies; we have also described its limitations.
Functional studies of S36A apoA-I have identified an impairment in the LCAT activation activity. This result alone may explain the correlation between the S36A mutation and low HDL-C found in the patient. Indeed, our data show that LCAT activity in the plasma from the S36A heterozygous apoA-I patient is significantly less than in plasma from normolipidemic subjects (supplementary Fig. II). Time course and concentration curve studies of LCAT activity also show that S36A rHDL is significantly impaired compared with WT apoA-I (Fig. 9B, C). The appKm for the S36A rHDL was six times greater than the WT protein, demonstrating a weaker interaction between S36A rHDL with the LCAT enzyme than WT rHDL (Fig. 9D). Interestingly, the mixed rHDL had a significant, but only moderate, increase in appKm, suggesting that the mixed rHDL were less efficient at activating LCAT (as would be the case in plasma) but that the S36A apoA-I did not act in a strongly dominant fashion inhibiting the WT apoA-I. If this is the case, we cannot be certain that mixed populations of HDL in the plasma of the S36A heterozygous patient could lead to hypoalphalipoproteinemia. Therefore, we could not rule out another factor (e.g., S36A apoA-I protein stability) affecting HDL-C levels in plasma. The 2D-gel of the S36A patient plasma may support this contention (supplementary Fig. I). The patient plasma showed a profound decrease in poorly and lightly lipidated apoA-I species (preβ- and α3-migrating HDL) with normal or higher than normal α1- and α2-migrating species. A possible explanation for this unexpected result is that the WT apoA-I present in the plasma of this patient is efficiently transformed into buoyant spherical HDL particles, whereas S36A apoA-I is poorly transformed into a buoyant, spherical HDL particle and removed from plasma. Lack of stability of poorly lipidated S36A apoA-I or lack of maturation of S36A containing-HDL particles to large spherical HDL particles (by impairment of LCAT activity) would lead to enhanced removal from plasma, suggesting that S36A apoA-I stability may indeed be a factor. It is our future plan to introduce S36A apoA-I into an apoA-I-deficient mouse model (by adenovirus) to test the stability of S36A apoA-I directly.
Taking the LCAT data and the fact that a minor mutation in the first N-terminal helix was sufficient to render apoA-I predominantly monomeric, we propose the following model. We can speculate that S36A changes the structure of this region by breaking the amphipathicity of an α-helix or introducing hydrophobicity, even though we have conflicting data on the structure of this region (Fig. 1). Tanaka et al. showed that introduction of the mutation Y18P into apoA-I resulted in an uncovering of an N-terminal lipid-binding region (47). We speculate that this region normally contributes to self-association in the absence of lipid and to lipid binding in the presence of lipid and that the S36A mutation reduces this capacity to self-associate and bind lipid by its preferential intramolecular binding (demonstrated by its predominant monomeric character). We hypothesize that the N-terminal lipid-binding region is involved in binding to and assembly of the lipoprotein substrate and may also act as an anchor along with the C terminus to allow freer movement of the central domain for the purposes of activating LCAT (1). The binding of the N-terminal helix to the lipoprotein substrate is necessary for LCAT activation, as we previously showed (10). However, deletion of the entire helix may have promoted perturbations in the structure of apoA-I that may indirectly impair LCAT activation ability. Our present data confirm these previous observations. However, lipoprotein binding and rHDL particle formation are unimpaired in S36A apoA-I. This could be due to the lack of sensitivity in our lipid-binding assays or to the fact that the C terminus and its lipid-binding properties remain intact. As an alternative, it has been shown that the N-terminus and the C terminus of homodimerized apoA-I on rHDL interact (residues 40 and 239 by intermolecular crosslinking) (1, 17) and that this interaction is constructive to cooperatively anchor apoA-I to the surface of the substrate lipoprotein to allow the central domain of apoA-I to release from the surface of the lipoprotein and interact with LCAT. If the interaction between the termini was impaired, then LCAT activity could also be impaired, especially in the context of a functional homodimer on HDL. Considering the proximity of S36 to the proposed crosslinked residue 40, it is plausible that intermolecular interactions between homodimers may be altered. A S36A/S36A or S36A/WT apoA-I dimer on HDL may simply be not as efficient at activating LCAT, a contention supported by our data (Fig. 9C). Future studies with mouse models may address these issues. These results stress the importance of the N-terminal helix in apoA-I for the structure and function of this critical plasma apolipoprotein. Specifically, these results implicate a region around S36 in apoA-I self-association, independent of the intact C terminus. Furthermore, these results strongly suggest that the N-terminal region of human apoA-I is necessary for LCAT activation and mutation could potentially lead to a plasma HDL deficiency.
Supplementary Material
Acknowledgments
The authors thank Vivian Franklin, Cyril Martin, Jonathan Ma, Janie Desrosiers, and Daniel Tessier for their expert technical assistance.
Footnotes
Abbreviations:
- ABCA1
- ATP binding cassette protein A1
- ANS
- 8-anilinonaphthalene-1-sulfonate
- apoA-I
- apolipoprotein A-I
- CAD
- coronary artery disease
- CE
- cholesteryl ester
- DMPC
- dimyristoylphosphatidylcholine
- DMS
- dimethylsuberimidate
- LCAT
- lecithin:cholesterol acyltransferase
- LUV
- large unilamellar vesicle
- WT
- wild-type
This work was supported by National Institutes of Health, General Medical Sciences Grant SC3GM-089564 (P.M.M.W.); by California State University Long Beach (CSULB) Provost's Undergraduate Student Summer Program and Allergan Undergraduate Summer Research Fellowship (A.B.P); by an Alberta Cancer Care-Alberta Health Services grant (E.G. and C.M.K.); by Canadian Institutes of Health Research Grants MOP-86736 (R.M. and Y.L.M.) and MOP-89972 (R.S.K.); and by Heart and Stroke Foundation of Ontario Grant T5911 (R.M. and Y.L.M.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Thomas M. J., Bhat S., Sorci-Thomas M. G. 2008. Three-dimensional models of HDL apoA-I: implications for its assembly and function. J. Lipid Res. 49: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253. [DOI] [PubMed] [Google Scholar]
- 3.Davidson W. S., Arnvig-McGuire K., Kennedy A., Kosman J., Hazlett T. L., Jonas A. 1999. Structural organization of the N-terminal domain of apolipoprotein A-I: studies of tryptophan mutants. Biochemistry. 38: 14387–14395. [DOI] [PubMed] [Google Scholar]
- 4.Ji Y., Jonas A. 1995. Properties of an N-terminal proteolytic fragment of apolipoprotein AI in solution and in reconstituted high density lipoproteins. J. Biol. Chem. 270: 11290–11297. [DOI] [PubMed] [Google Scholar]
- 5.Marcel Y. L., Kiss R. S. 2003. Structure-function relationships of apolipoprotein A-I: a flexible protein with dynamic lipid associations. Curr. Opin. Lipidol. 14: 151–157. [DOI] [PubMed] [Google Scholar]
- 6.Tricerri M. A., Behling Agree A. K., Sanchez S. A., Bronski J., Jonas A. 2001. Arrangement of apolipoprotein A-I in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry. 40: 5065–5074. [DOI] [PubMed] [Google Scholar]
- 7.Kiss R. S., Kay C. M., Ryan R. O. 1999. Amphipathic alpha-helix bundle organization of lipid-free chicken apolipoprotein A-I. Biochemistry. 38: 4327–4334. [DOI] [PubMed] [Google Scholar]
- 8.Rogers D. P., Roberts L. M., Lebowitz J., Engler J. A., Brouillette C. G. 1998. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 37: 945–955. [DOI] [PubMed] [Google Scholar]
- 9.Okon M., Frank P. G., Marcel Y. L., Cushley R. J. 2001. Secondary structure of human apolipoprotein A-I(1–186) in lipid-mimetic solution. FEBS Lett. 487: 390–396. [DOI] [PubMed] [Google Scholar]
- 10.Scott B. R., McManus D. C., Franklin V., McKenzie A. G., Neville T., Sparks D. L., Marcel Y. L. 2001. The N-terminal globular domain and the first class A amphipathic helix of apolipoprotein A-I are important for lecithin:cholesterol acyltransferase activation and the maturation of high density lipoprotein in vivo. J. Biol. Chem. 276: 48716–48724. [DOI] [PubMed] [Google Scholar]
- 11.Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. 2007. Electron paramagnetic resonance spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apoA-I in solution. J. Biol. Chem. 282: 9143–9149. [DOI] [PubMed] [Google Scholar]
- 12.Beckstead J. A., Block B. L., Bielicki J. K., Kay C. M., Oda M. N., Ryan R. O. 2005. Combined N- and C-terminal truncation of human apolipoprotein A-I yields a folded, functional central domain. Biochemistry. 44: 4591–4599. [DOI] [PubMed] [Google Scholar]
- 13.Koyama M., Tanaka M., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2009. Interaction between the N- and C-terminal domains modulates the stability and lipid binding of apolipoprotein A-I. Biochemistry. 48: 2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers D. P., Brouillette C. G., Engler J. A., Tendian S. W., Roberts L., Mishra V. K., Anantharamaiah G. M., Lund-Katz S., Phillips M. C., Ray M. J. 1997. Truncation of the amino terminus of human apolipoprotein A-I substantially alters only the lipid-free conformation. Biochemistry. 36: 288–300. [DOI] [PubMed] [Google Scholar]
- 15.Rogers D. P., Roberts L. M., Lebowitz J., Datta G., Anantharamaiah G. M., Engler J. A., Brouillette C. G. 1998. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 37: 11714–11725. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H. L., Atkinson D. 2004. Conformation and lipid binding of the N-terminal (1–44) domain of human apolipoprotein A-I. Biochemistry. 43: 13156–13164. [DOI] [PubMed] [Google Scholar]
- 17.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46: 7811–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., Didonato J. A., Hazen S. L. 2009. Double superhelix model of high density lipoprotein. J. Biol. Chem. 284: 36605–36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorci-Thomas M. G., Thomas M. J. 2002. The effects of altered apolipoprotein A-I structure on plasma HDL concentration. Trends Cardiovasc. Med. 12: 121–128. [DOI] [PubMed] [Google Scholar]
- 20.Kiss R. S., Kavaslar N., Okuhira K., Freeman M. W., Walter S., Milne R. W., McPherson R., Marcel Y. L. 2007. Genetic etiology of isolated low HDL syndrome: incidence and heterogeneity of efflux defects. Arterioscler. Thromb. Vasc. Biol. 27: 1139–1145. [DOI] [PubMed] [Google Scholar]
- 21.Bergeron J., Frank P. G., Emmanuel F., Latta M., Zhao Y., Sparks D. L., Rassart E., Denefle P., Marcel Y. L. 1997. Characterization of human apolipoprotein A-I expressed in Escherichia coli. Biochim. Biophys. Acta. 1344: 139–152. [DOI] [PubMed] [Google Scholar]
- 22.Vasquez L. J., Abdullahi G. E., Wan C. P., Weers P. M. 2009. Apolipophorin III lysine modification: Effect on structure and lipid binding. Biochim. Biophys. Acta. 1788: 1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss R. S., Maric J., Marcel Y. L. 2005. Lipid efflux in human and mouse macrophagic cells: evidence for differential regulation of phospholipid and cholesterol efflux. J. Lipid Res. 46: 1877–1887. [DOI] [PubMed] [Google Scholar]
- 24.Liu H., Scraba D. G., Ryan R. O. 1993. Prevention of phospholipase-C induced aggregation of low density lipoprotein by amphipathic apolipoproteins. FEBS Lett. 316: 27–33. [DOI] [PubMed] [Google Scholar]
- 25.Wientzek M., Kay C. M., Oikawa K., Ryan R. O. 1994. Binding of insect apolipophorin III to dimyristoylphosphatidylcholine vesicles. Evidence for a conformational change. J. Biol. Chem. 269: 4605–4612. [PubMed] [Google Scholar]
- 26.Asztalos B. F., Cupples L. A., Demissie S., Horvath K. V., Cox C. E., Batista M. C., Schaefer E. J. 2004. High-density lipoprotein subpopulation profile and coronary heart disease prevalence in male participants of the Framingham Offspring Study. Arterioscler. Thromb. Vasc. Biol. 24: 2181–2187. [DOI] [PubMed] [Google Scholar]
- 27.Krimbou L., Marcil M., Davignon J., Genest J., Jr 2001. Interaction of lecithin:cholesterol acyltransferase (LCAT).alpha 2-macroglobulin complex with low density lipoprotein receptor-related protein (LRP). Evidence for an alpha 2-macroglobulin/LRP receptor-mediated system participating in LCAT clearance. J. Biol. Chem. 276: 33241–33248. [DOI] [PubMed] [Google Scholar]
- 28.Laue T. M., Stafford W. F., III 1999. Modern applications of analytical ultracentrifugation. Annu. Rev. Biophys. Biomol. Struct. 28: 75–100. [DOI] [PubMed] [Google Scholar]
- 29.Johnson M. L., Correia J. J., Yphantis D. A., Halvorson H. R. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36: 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokke K. T., Norum K. R. 1971. Determination of lecithin: cholesterol acyltransfer in human blood plasma. Scand. J. Clin. Lab. Invest. 27: 21–27. [DOI] [PubMed] [Google Scholar]
- 31.Lacko A. G., Reason A. J., Nuckolls C., Kudchodkar B. J., Nair M. P., Sundarrajan G., Pritchard P. H., Morris H. R., Dell A. 1998. Characterization of recombinant human plasma lecithin: cholesterol acyltransferase (LCAT): N-linked carbohydrate structures and catalytic properties. J. Lipid Res. 39: 807–820. [PubMed] [Google Scholar]
- 32.Albers J. J., Cabana V. G., Dee Barden S. Y. 1976. Purification and characterization of human plasma lecithin:cholesterol acyltransferase. Biochemistry. 15: 1084–1087. [DOI] [PubMed] [Google Scholar]
- 33.McManus D. C., Scott B. R., Franklin V., Sparks D. L., Marcel Y. L. 2001. Proteolytic degradation and impaired secretion of an apolipoprotein A-I mutant associated with dominantly inherited hypoalphalipoproteinemia. J. Biol. Chem. 276: 21292–21302. [DOI] [PubMed] [Google Scholar]
- 34.Gwynne J., Brewer B., Jr., Edelhoch H. 1974. The molecular properties of ApoA-I from human high density lipoprotein. J. Biol. Chem. 249: 2411–2416. [PubMed] [Google Scholar]
- 35.Swaney J. B. 1980. Characterization of the high-density lipoprotein and its major apoprotein from human, canine, bovine and chicken plasma. Biochim. Biophys. Acta. 617: 489–502. [DOI] [PubMed] [Google Scholar]
- 36.Burgess J. W., Frank P. G., Franklin V., Liang P., McManus D. C., Desforges M., Rassart E., Marcel Y. L. 1999. Deletion of the C-terminal domain of apolipoprotein A-I impairs cell surface binding and lipid efflux in macrophage. Biochemistry. 38: 14524–14533. [DOI] [PubMed] [Google Scholar]
- 37.Andreola A., Bellotti V., Giorgetti S., Mangione P., Obici L., Stoppini M., Torres J., Monzani E., Merlini G., Sunde M. 2003. Conformational switching and fibrillogenesis in the amyloidogenic fragment of apolipoprotein A-I. J. Biol. Chem. 278: 2444–2451. [DOI] [PubMed] [Google Scholar]
- 38.Genschel J., Haas R., Propsting M. J., Schmidt H. H. 1998. Apolipoprotein A-I induced amyloidosis. FEBS Lett. 430: 145–149. [DOI] [PubMed] [Google Scholar]
- 39.Koldamova R. P., Lefterov I. M., Lefterova M. I., Lazo J. S. 2001. Apolipoprotein A-I directly interacts with amyloid precursor protein and inhibits A beta aggregation and toxicity. Biochemistry. 40: 3553–3560. [DOI] [PubMed] [Google Scholar]
- 40.Mangione P., Sunde M., Giorgetti S., Stoppini M., Esposito G., Gianelli L., Obici L., Asti L., Andreola A., Viglino P., et al. 2001. Amyloid fibrils derived from the apolipoprotein A1 Leu174Ser variant contain elements of ordered helical structure. Protein Sci. 10: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obici L., Franceschini G., Calabresi L., Giorgetti S., Stoppini M., Merlini G., Bellotti V. 2006. Structure, function and amyloidogenic propensity of apolipoprotein A-I. Amyloid. 13: 191–205. [DOI] [PubMed] [Google Scholar]
- 42.Zannis V. I., Chroni A., Krieger M. 2006. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. 84: 276–294. [DOI] [PubMed] [Google Scholar]
- 43.Kiss R. S., Ryan R. O., Francis G. A. 2001. Functional similarities of human and chicken apolipoprotein A-I: dependence on secondary and tertiary rather than primary structure. Biochim. Biophys. Acta. 1531: 251–259. [DOI] [PubMed] [Google Scholar]
- 44.Kiss R. S., Ryan R. O., Hicks L. D., Oikawa K., Kay C. M. 1993. Physical properties of apolipoprotein A-I from the chicken, Gallus domesticus. Biochemistry. 32: 7872–7878. [DOI] [PubMed] [Google Scholar]
- 45.Ren X., Yang Y., Neville T., Hoyt D., Sparks D., Wang J. 2007. A complete backbone spectral assignment of human apolipoprotein AI on a 38 kDa prebetaHDL (Lp1-AI) particle. Biomol. NMR Assign. 1: 69–71. [DOI] [PubMed] [Google Scholar]
- 46.Ren X., Zhao L., Sivashanmugam A., Miao Y., Korando L., Yang Z., Reardon C. A., Getz G. S., Brouillette C. G., Jerome W. G., et al. 2005. Engineering mouse apolipoprotein A-I into a monomeric, active protein useful for structural determination. Biochemistry. 44: 14907–14919. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka M., Dhanasekaran P., Nguyen D., Ohta S., Lund-Katz S., Phillips M. C., Saito H. 2006. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 45: 10351–10358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.