Abstract
Mammalian ceramide synthases 1 to 6 (CerS1–6) generate Cer in an acyl-CoA-dependent manner, and expression of individual CerS has been shown to enhance the synthesis of ceramides with particular acyl chain lengths. However, the contribution of each CerS to steady-state levels of specific Cer species has not been evaluated. We investigated the knockdown of individual CerS in the MCF-7 human breast adenocarcinoma cell line by using small-interfering RNA (siRNA). We found that siRNA-induced downregulation of each CerS resulted in counter-regulation of nontargeted CerS. Additionally, each CerS knockdown produced unique effects on the levels of multiple sphingolipid species. For example, downregulation of CerS2 decreased very long-chain Cer but increased levels of CerS4, CerS5, and CerS6 expression and upregulated long-chain and medium-long-chain sphingolipids. Conversely, CerS6 knockdown decreased C16:0-Cer but increased CerS5 expression and caused non-C16:0 sphingolipids to be upregulated. Knockdown of individual CerS failed to decrease total sphingolipids or upregulate sphingoid bases. Treatment with siRNAs targeting combined CerS, CerS2, CerS5, and CerS6, did not change overall Cer or sphingomyelin mass but caused upregulation of dihydroceramide and hexosylceramide and promoted endoplasmic reticulum stress. These data suggest that sphingolipid metabolism is robustly regulated by both redundancy in CerS-mediated Cer synthesis and counter-regulation of CerS expression.
Keywords: dihydroceramide, hexosylceramide, sphingomyelin, long-chain sphingolipids, very-long-chain sphingolipids, glucosylceramide synthase, short interfering RNA
Sphingolipids are a large and diverse class of lipids whose members serve multiple roles in eukaryotic biology. Sphingolipids are not only components of membrane structure but are essential mediators of cellular functions such as regulation of growth, differentiation, and programmed cell death (1). Ceramide (Cer), or N-acylsphingosine, is a central lipid in sphingolipid metabolism and serves both as a signaling molecule itself and as a precursor for other bioactive sphingolipids, ranging from complex glycosphingolipids to “simpler” lipids such as sphingosine (Sph) and sphingosine-1-phosphate (S1P) (2).
Cer is produced de novo through a series of reactions beginning with the condensation of serine and palmitoyl-CoA by serine palmitoyl-CoA transferase (SPT) (3). The resulting product, 3-ketosdihydrosphingosine, is readily reduced to form dihydrosphingosine (dHSph). dHSph is acylated by the enzyme (dihydro)ceramide synthase (CerS) to form dihydroceramide (dHCer). The long-chain base moiety of this lipid can then be desaturated at the 4–5 position of the sphingoid base backbone to form Cer. Cer is the substrate for numerous enzymes including those that produce complex sphingolipids such as sphingomyelin (SM) and glycosphingolipids. Cer may also be produced via hydrolysis of complex sphingolipids or reacylation of free Sph by CerS through what has been termed the salvage or recycling pathway (4).
The acylation of free sphingoid bases by CerS occurs through an acyl-CoA-dependent process (5). In addition to regulating sphingolipid synthesis, CerS activity has been shown to regulate numerous aspects of cell biology, including cell growth, apoptosis, and autophagy (6–8). The abilities of CerS to control de novo pathways and to salvage pathways of Cer synthesis make the proteins that catalyze these reactions of unique biochemical and biological interest.
Although acyl-CoA-dependent Cer synthesis was first described in 1966, more than 30 years passed before the first genes responsible for dHCer/Cer synthesis were identified in Saccharomyces cerevisiae (9, 10). The first gene was originally termed the longevity assurance gene (LAG1) because of its ability to increase yeast replicative life span (11). Several years later, LAG1 and its homolog, the longevity assurance gene (LAC1) cognate, were shown to be necessary for the synthesis of yeast dHCer (9). A search for genes containing the characteristic Lag1p motif of LAG1 and LAC1 led to the identification of homologs in several organisms, including a human gene product, CerS1 (formerly known as Lag1pHs, Lass1, and UOG1) (12). Overexpression of this gene in mammalian cells led to an increase in CerS activity and Cer (13). Additional mammalian homologs (CerS2–6, formerly known as Lass2–6) were subsequently discovered and shown to regulate the synthesis of Cer as well (14, 15).
Mammalian CerS proteins share many features. Each appears to localize in the endoplasmic reticulum (ER); each has at least five transmembrane domains; and CerS2, CerS5, and CerS6 are N-glycosylated near their N termini (13–15). All CerS, as well as nonmammalian Lag1p homologs, possess a Lag1p homology domain that has been demonstrated to be essential for enzymatic activity in CerS1 and CerS5 (12, 16). All mammalian CerS, except for CerS1, also contain a Hox-like domain the function of which is not known (17). Furthermore, purified CerS5 has been shown in vitro to possess CerS activity, which strongly supports the notion that mammalian CerS proteins possess bona fide acyl-CoA-dependent CerS activity (18).
Each CerS has a preference for a unique range of acyl-CoA groups and therefore controls a particular subset of ceramide species (13–15, 19–21). Long-chain Cer (C14–C18) are formed by CerS1 (C18:0-Cer), CerS5 (C14:0 and C16:0-Cer), and CerS6 (C14:0-Cer and C16:0-Cer). CerS3 has a very broad substrate preference (18–24 carbons) but has also been associated with the synthesis of very-long-chain polyenoic Cer (22). CerS4 also appears to have a broad range of specificity, with a preference for medium-long-chain Cer such as C18:0-Cer and C20:0-Cer (14, 15, 23, 24). CerS2, on the other hand, has a preference for very-long-chain acyl-CoA groups (C22–C26) (15, 25, 26).
Most of the studies investigating the roles of CerS in the regulation of sphingolipids have relied on plasmid-mediated overexpression of CerS proteins to show increased ceramide synthesis as a gain-of-function from these proteins (13–15, 20, 25). How individual CerS contribute to the steady-state sphingolipid pool in mammalian cells and whether specific CerS are necessary for the maintenance of sphingolipids with specific acyl chains are still poorly understood. Furthermore, little is known about the regulation and interregulation of these proteins with regard to maintaining overall sphingolipid homeostasis. Therefore, we hypothesized that knockdown of individual CerS would downregulate specific populations of sphingolipids in a manner consistent with the known acyl-CoA specificities of these enzymes.
In this study, we have used small interfering RNAs (siRNAs) against the six CerS family members in order to specifically reduce the expression of each isoform in MCF-7 breast adenocarcinoma cells. Knockdown of individual CerS resulted in a wide range of effects on nontargeted CerS expression. These effects were associated with both increases and decreases in multiple sphingolipid species, often with little change in total Cer levels. To our surprise, knockdown of some CerS (e.g., CerS5) or knockdown of combined CerS2, CerS5, and CerS6 produced little or no decrease in sphingolipid levels but did cause several lipids, especially dHCer and HexCer, to increase. These data also indicate that changes in CerS activity are met with a shift in sphingolipid metabolism sufficient to allow ceramide and/or sphingoid base levels to be maintained at the expense of accumulating glycosphingolipids.
MATERIALS AND METHODS
Cell culture
MCF-7 human breast adenocarcinoma cells were maintained in RPMI medium (Gibco-Invitrogen, Carlsbad, CA) supplemented with l-glutamine, 10% (v/v) fetal bovine serum. Cells were kept in a humidified incubator at 37°C with 5% Co2.
siRNA transfection
Cells were plated at 4.0 × 105 cells/dish (10 cm dish) and incubated for 24 h. At 24 h, cells were transfected with double-stranded RNA oligomers, using Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Forty-eight h posttransfection, cells were harvested by scraping into ice-cold phosphate-buffered saline (PBS) and pelleted by centrifugation at 250 g. Cell pellets were then frozen at −80°C until used for further analysis. siRNA sequences used in this study are provided in Table 1. We have determined empirically that CerS siRNA oligomers (siCerS) are effective for reducing target expression (>50%) in the 5–20 nM range, and therefore, for most experiments, 5 nM siCerS or negative control siRNA (siControl) was used.
TABLE 1.
siRNA sequences used in this study
| Target | Orientation | Sequence |
|---|---|---|
| Nontargeted control | Sense | AllStars (transfection control)a |
| Antisense | AllStars (transfection control)a | |
| CerS1 | Sense | r(GGU CCU GUA UGC CAC CAG U)dTdT |
| Antisense | r(ACU GGU GGC AUA CAG GAC C)dTdT | |
| CerS2 | Sense | r(GGA ACA GAU CAU CCA CCA U)dTdT |
| Antisense | r(AUG GUG GAU GAU CUG UUC C)dTdT | |
| CerS3 | Sense | r(CCU UCU CAU UUA UAC GUG A)dTdT |
| Antisense | r(UCA CGU AUA AAU GAG AAG G)dTdT | |
| CerS4 | Sense | r(GGA CAU UCG UAG UGA UGU A)dTdT |
| Antisense | r(UAC AUC ACU ACG AAU GUC C)dTdT | |
| CerS5 | Sense | r(ACC CUG UGC ACU CUG UAU U)dTdT |
| Antisense | r(AAU ACA GAG UGC ACA GGG U)dTdT | |
| CerS6 | Sense | r(CGC UGG UCC UUU GUC UUC A)dTdT |
| Antisense | r(UGA AGA CAA AGG ACC AGC G)dTdT |
Qiagen transfection control was used.
Real-time quantitative PCR analysis
RNA was extracted using a Qiagen RNeasy® kit according to the manufacturer's protocol. RNA concentration was determined by using a Quant-iT™ RiboGreen® RNA assay kit (Invitrogen, Valencia, CA). One microgram of RNA was used to produce cDNA, using a SuperScript first-strand synthesis system (Invitrogen). The resultant cDNA was used for real-time quantitative PCR (q-PCR) using a QuantiTect SYBR Green PCR kit (Invitrogen) on an ABI 7300 quantitative (q)-PCR system (Applied Biosystems, Foster City, CA) as described by the manufacturer. Primers used in this study are described in Table 2.
TABLE 2.
Primers used in this study
| Gene | Orientation | Sequence |
|---|---|---|
| β-actin | Forward | 5′- ATT GGC AAT GAG CGG TTC C -3′ |
| Reverse | 5′- GGT AGT TTC GTG GAT GCC ACA -3′ | |
| CerS1 | Forward | 5′- ACG CTA CGC TAT ACA TGG ACA C -3′ |
| Reverse | 5′- AGG AGG AGA CGA TGA GGA TGA G -3′ | |
| CerS2 | Forward | 5′- CCG ATT ACC TGC TGG AGT CAG -3′ |
| Reverse | 5′- GGC GAA GAC GAT GAA GAT GTT G -3′ | |
| CerS3 | Forward | 5′- ACA TTC CAC AAG GCA ACC ATT G -3′ |
| Reverse | 5′- CTC TTG ATT CCG CCG ACT CC -3′ | |
| CerS4 | Forward | 5′- CTT CGT GGC GGT CAT CCT G -3′ |
| Reverse | 5′- TGT AAC AGC AGC ACC AGA GAG -3′ | |
| CerS5 | Forward | 5′- TGT AAC AGC AGC ACC AGA GAG -3′ |
| Reverse | 5′- GCC AGC ACT GTC GGA TGT C -3′ | |
| CerS6 | Forward | 5′- GGG ATC TTA GCC TGG TTC TGG -3′ |
| Reverse | 5′- GCC TCC TCC GTG TTC TTC AG -3′ | |
| CHOP | Forward | 5′- CCT CAC TCT CCA GAT TCC -3′ |
| Reverse | 5′- TGT CAC TTT CCT TTC ATT C -3′ |
Western blot analysis of CerS proteins
Cell pellets were lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100 containing cOmplete Mini protease inhibitor cocktail {Roche Diagnostics, Indianapolis, IN}). Protein concentrations were determined by using the Bradford assay (Bio-Rad, Hercules, CA). Lysates were subjected to Western analysis using antibodies against CerS2 (mouse monoclonal, clone 1A6; Novus Biologicals, Littleton, CO) and CerS6 (mouse monoclonal, clone 5H7; Novus Biologicals).
Ceramide analysis by high-performance liquid chromatography/mass spectrometry
MCF-7 cells were harvested by scraping into 10 ml of ice-cold PBS and pelleted. Lipids were extracted and analyzed by the Lipidomics Core facility at the Medical University of South Carolina, using quantitative high-performance liquid chromatography/mass spectrometry (HPLC/MS), as described previously (27). Our analysis was limited to sphingoid bases, dHCer, Cer, SM, HexCer (consisting of glucosylceramide {GlcCer} and galactosylceramide {GalCer}), and lactosylceramides (LacCer) possessing an 18 carbon sphingoid base backbone and ceramide-containing lipids whose acyl chains were nonhydroxylated and 14 to 26 carbons in length. Sphingolipid levels were normalized to total cellular lipid phosphate, which was determined as described previously (28).
In vitro CerS activity
In vitro CerS activity was performed essentially as described previously (29). Cells were treated as indicated and then harvested by scraping into ice-cold PBS. Cells were pelleted and resuspended in a solution of 20 mM HEPES (pH 7.4), 250 mM sucrose, 2 mM KCl, and 2 mM MgCl2 and then sonicated, and protein content was determined by using the Bradford assay. A reaction mixture (100 μl, final volume) containing 15 μM 17C-dHSph and 50 μM palmitoyl-CoA or lignoceroyl-CoA in 25 mM potassium phosphate buffer (pH 7.4) was prewarmed at 37°C for 5 min, followed by addition of 100 μg of whole-cell lysate to start the reaction. Reaction time was 15 min, after which the reaction mixture was transferred to a glass tube containing 2 ml of extraction solvent (ethyl acetate/2-propanol/water, 60:30:10 {v/v/v}), which stopped the reaction. Lipids were extracted as previously described, and 17C16:0-dHCer or 17C24:0-dHCer content was determined by HPLC/MS (16, 27).
Data presentation and statistical analysis
Heat maps displaying log2-fold change of siCerS over siControl were generated using Matrix2png software (http://chibi.ubc.ca/matrix2png/index.html) (30). Statistical analyses were performed using Microsoft Excel. Statistical significance was determined using a two-tailed, unpaired Student's t-test using a P value of < 0.05 as our criterion for significance.
RESULTS
CerS expression in MCF-7 breast adenocarcinoma cells
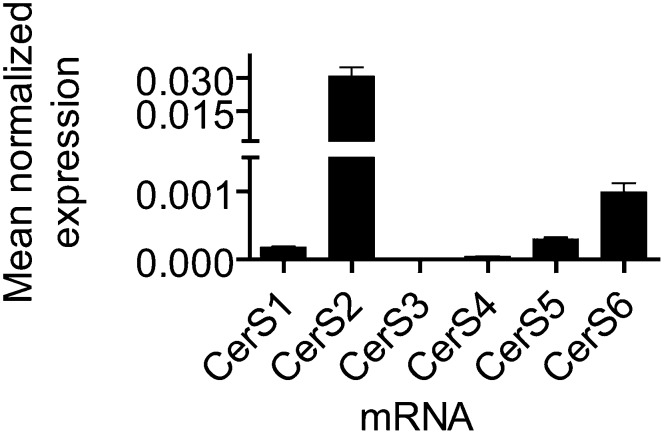
In order to study the roles of CerS in regulating sphingolipid levels, we chose a cell system, MCF-7 breast adenocarcinoma cells, in which multiple CerS transcripts were present and able to be knocked down using siRNA. We were further interested in this cell line because of recent publications demonstrating altered CerS expression in human breast cancer (31–33). We first used real-time q-PCR to determine the expression of CerS1–6 in MCF-7 cells (Fig. 1). The transcript of CerS2 was the most abundant, followed by those of CerS6, CerS5, CerS1, and CerS4. CerS3 mRNA levels were near the limit of detection, which is consistent with reports of CerS3 having a limited tissue distribution and being found mainly in the testis and skin (14, 20). Because CerS2 and CerS6 were the most abundant transcripts, the majority of this report will focus on the effects observed upon siRNA-mediated knockdown of these enzymes. Data concerning the knockdown of the less abundant CerS can be found in the supplementary data section.
Fig. 1.
CerS expression in MCF-7. MCF-7 cells were harvested and RNA was extracted for q-PCR analysis of CerS1-6 expression. q-PCR data are normalized to β-actin mRNA expression and data are mean ± SEM for three independent experiments.
CerS knockdown by targeted siRNA causes multiple changes in nontargeted CerS mRNA levels
We next established the efficacy of siRNA-induced knockdown on the transcript levels of each CerS in MCF-7 cells. As indicated in Fig. 2A, B, knockdown of CerS2 and CerS6, the two most abundant CerS, reduced expression of these targets at the mRNA level. Knockdown of the other CerS (CerS1, CerS3, CerS4, and CerS5) is reported in supplementary Fig. IA–D). Overall, we found that the siRNAs (5 nM) were more effective at reducing the expression of their targets than the siControl.
Fig. 2.
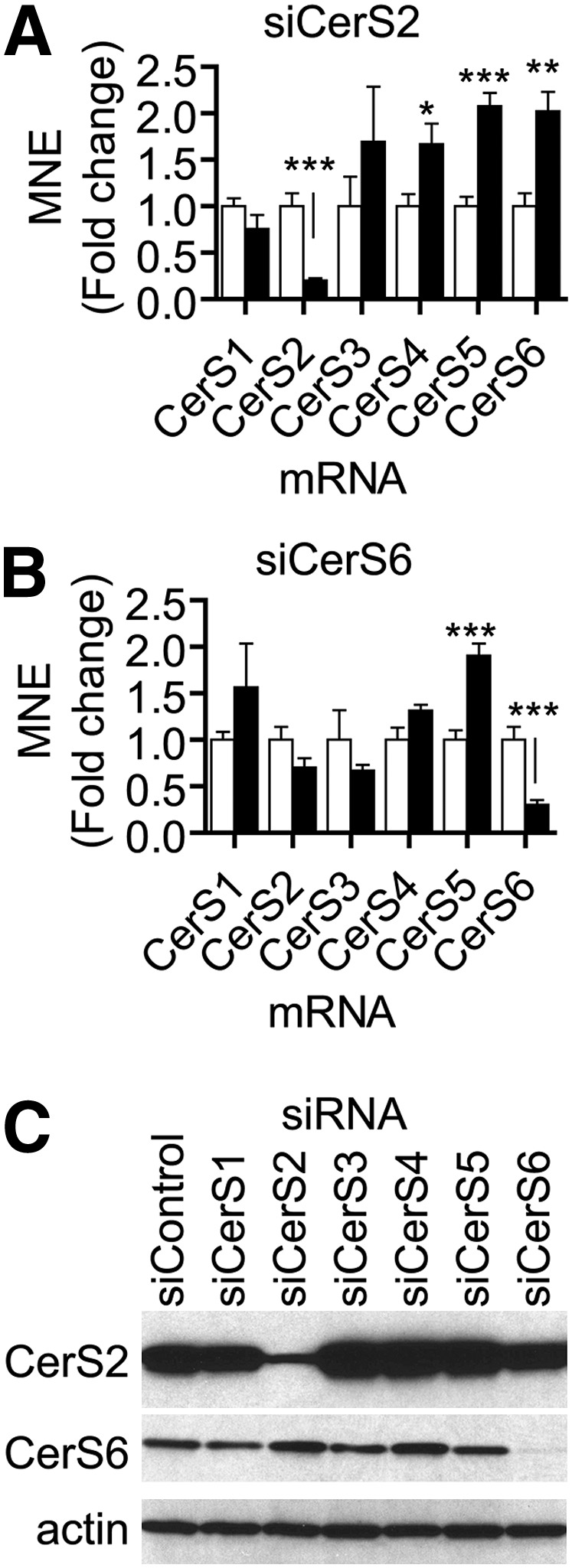
CerS2 and CerS6 downregulation by targeted siRNA causes multiple changes in nontargeted CerS mRNA levels. A and B: MCF-7 cells were transfected with 5 nM siRNA targeted against CerS (black bars) or siControl (white bars) for 48 h. Cells were harvested, and RNA was extracted for q-PCR analysis of expression of CerS1-6. q-PCR data are normalized to β-actin mRNA expression, and data are means ± standard errors of the mean (SEM) for three independent experiments. A: Effects of siCerS2 on CerS1-6 expression. B: Effects of siCerS6 on CerS1-6 expression. C: Western blot analysis of CerS2, CerS6, and β-actin protein expression following transfection with siCerS1-6. CerS2 and CerS6 were detected using monoclonal antibodies specific for these proteins. β-actin protein levels were used as a control for equal gel loading. *, P < 0.05; **, P < 0.01; ***, P < 0.01 versus siControl.
Interestingly, the targeting of certain CerS resulted in the elevation of nontargeted CerS. Knockdown of CerS2 resulted in increased mRNA levels of CerS4, CerS5, and CerS6 (Fig. 2A). siCerS6, on the other hand, upregulated levels of the CerS5 message but did not change levels of the other CerS (Fig. 2B). Both siCerS2 and siCerS4 increased CerS6 mRNA levels (Fig. 2A and supplementary Fig. IC). siCerS5 increased mRNA levels of CerS2 and CerS4 (supplementary Fig. ID). siCerS3, which further decreased the already low levels of its target in MCF-7 cells, also increased CerS4 mRNA (Fig. IB). From these data, we concluded that siRNA-mediated CerS knockdown was effective at the mRNA level and that the targeting of a particular CerS could lead to upregulation of one or more other CerS.
We next tested the ability of siCerS to reduce the protein levels of CerS. Of several commercially available antibodies tested, only those against CerS2 and CerS6 recognized proteins within the range of predicted molecular masses of CerS1–6 (∼40–46 kDa). Fig. 2C shows knockdowns of CerS2 and CerS6 at the protein level. Knockdowns of CerS3, CerS4, and CerS5 showed a slight elevation of CerS2, whereas knockdown of Cer2 or CerS4 showed an elevation in CerS6. These effects were in large part similar to those seen at the mRNA level (Figs. 2A,B and supplementary Fig. I).
Effect of CerS1–6 on acyl chain lengths of sphingolipids
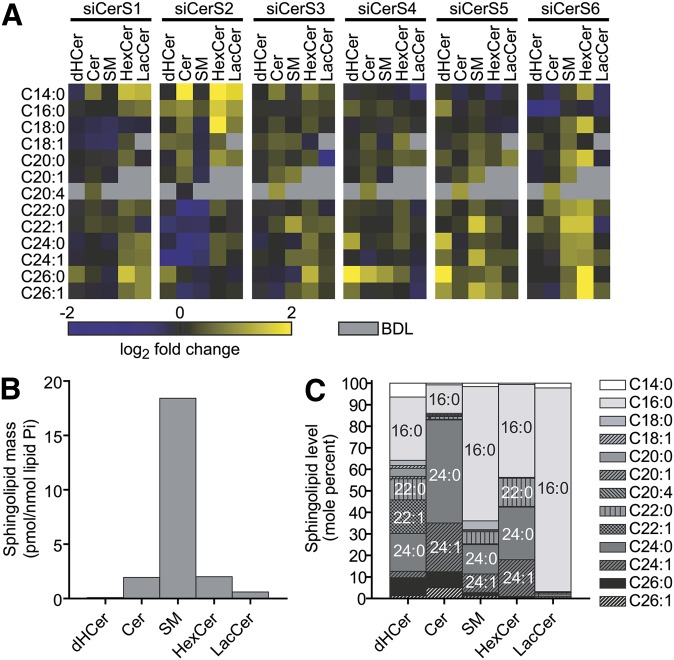
We sought to determine the effects of CerS knockdown on steady-state sphingolipid levels in MCF-7 cells. Following siRNA treatment, we analyzed sphingolipid levels by HPLC/MS and compiled data into a heat map showing log2-fold change of each lipid species in siCerS-treated cells over siControl-treated cells (Fig. 3A). To screen for significant changes in lipids, we used unpaired, two-tailed Student's t-tests to compare each siCerS to siControl (supplementary Fig. I).
Fig. 3.
Knockdown of siCerS1-6 induces changes in multiple sphingolipid species. MCF-7 cells were transfected with 5 nM siRNA targeted against CerS1-6, and sphingolipid masses were determined by HPLC/MS. Sphingolipid levels were normalized to the amount of total lipid phosphate. A: Sphingolipid mass changes are displayed as a heat map of the log2 of the mean fold change versus siControl. Gray boxes indicate that the lipid was below detectable levels (BDL). Data represent three to five independent experiments. B: Mass of each sphingolipid class in siControl-transfected cells. C: Acyl chain distribution of each sphingolipid class displayed as mol%. Data represent the means of three to five independent experiments.
The sphingolipid distribution in control cells was such that the most abundant sphingolipid class analyzed was SM, followed by HexCer, Cer, LacCer, and dHCer (Fig. 3B). The acyl chain distribution of sphingolipids differed across each sphingolipid class. As indicated in Fig. 3C, dHCerS exhibited the most diverse acyl chain profile and contained mostly saturated fatty acids (with the exception of C22:1). Cer was represented mostly by the C16:0, C24:0, and C24:1 species. SM consisted mostly of C16:0-SM, which was the most abundant lipid species observed in our analysis. HexCer was represented predominantly by C16:0, C22:0, C24:0, and C24:1 species, and LacCer contained mostly C16:0 acyl chains. Based on acyl chain distribution alone, one would predict that MCF-7 sphingolipids would be largely dependent on CerS regulating C16:0-Cer (e.g., CerS5 and/or CerS6) and very-long-chain Cer synthesis (e.g., CerS2).
CerS2 downregulation results in shift of sphingolipid distribution to predominantly long-chain species
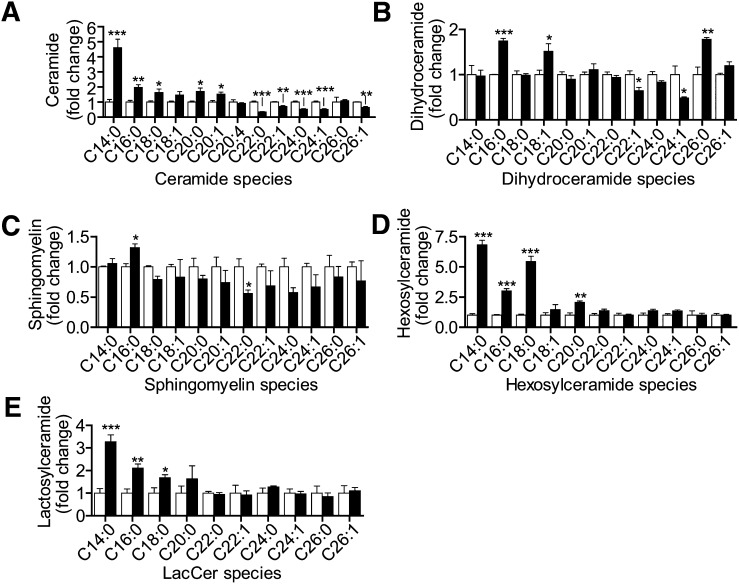
Because CerS2 was the most abundant CerS in MCF-7 cells, we expected that its downregulation would cause the most dramatic changes in sphingolipids. We found that siCerS2 was able to downregulate in vitro C24:0-CerS activity (supplementary Fig. IIIA) and Cer containing C22–C24 saturated and monounsaturated fatty acids (Figs. 3A and 4A). The greatest decrease was in C22:0-Cer (34.1% ± 3.0% of siControl), and the most abundant ceramide, C24:0-Cer, was decreased to 52.2% ± 4.4% of that of siControl. Interestingly, very-long-chain dhCer species were not as affected as very-long-chain Cer (Fig. 4B). There was a trend toward a decrease in several very-long-chain SM species, but only the decrease in C22:0-SM was statistically significant (Fig. 4C). Surprisingly, very-long-chain HexCer and LacCer levels were not significantly 6affected (Fig. 4D, E). Therefore, although very-long-chain Cer were sensitive to CerS2 depletion, very long-chain SM and HexCer were largely unaffected.
Fig. 4.
CerS2 downregulation results in a shift of sphingolipid distribution to predominantly long-chain species, resulting in the accumulation of C16:0-SM and long-chain glycosphingolipids. The effects of siCerS2 (black bars) on Cer (A), dHCer (B), SM (C), HexCer (D), and LacCer (E) compared with siControl (white bars) were determined as described in Fig. 4. Sphingolipid levels are normalized to the amount of total lipid phosphate. Data are means ± SEM for three to five independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.01 versus siControl.
On the other hand, downregulation of CerS2 resulted in large increases in the long-chain Cer species C14:0 and C16:0 (Figs. 3A, 4A), which was similar to the effects seen when this enzyme was downregulated in SMS-KCNR neuroblastoma cells (8). In MCF-7 cells, C14:0-Cer increased by ∼4.5-fold, and C16:0-Cer nearly doubled. The increase in Cer was reflected in a dramatic increase in C14:0- and C16:0-HexCer and LacCer (Fig. 4D, E). C16:0-SM, the most abundant sphingolipid species in our analysis, increased by 35.2% ± 6.4% or ∼4 pmol/nmol lipid phosphate, thus representing the largest sphingolipid change induced by the knockdown of a CerS. However, despite the effects of CerS2 knockdown on C16:0 lipids and CerS5 and CerS6 expression, siCerS2 did not increase the in vitro C16:0-CerS activity (supplementary Fig. IIIB). Therefore, CerS2 is not only a very-long-chain CerS but its absence also causes the accumulation of long-chain sphingolipids.
CerS6 knockdown decreases C16:0-Cer and C16:0-dHCer and increases several SM and HexCer species
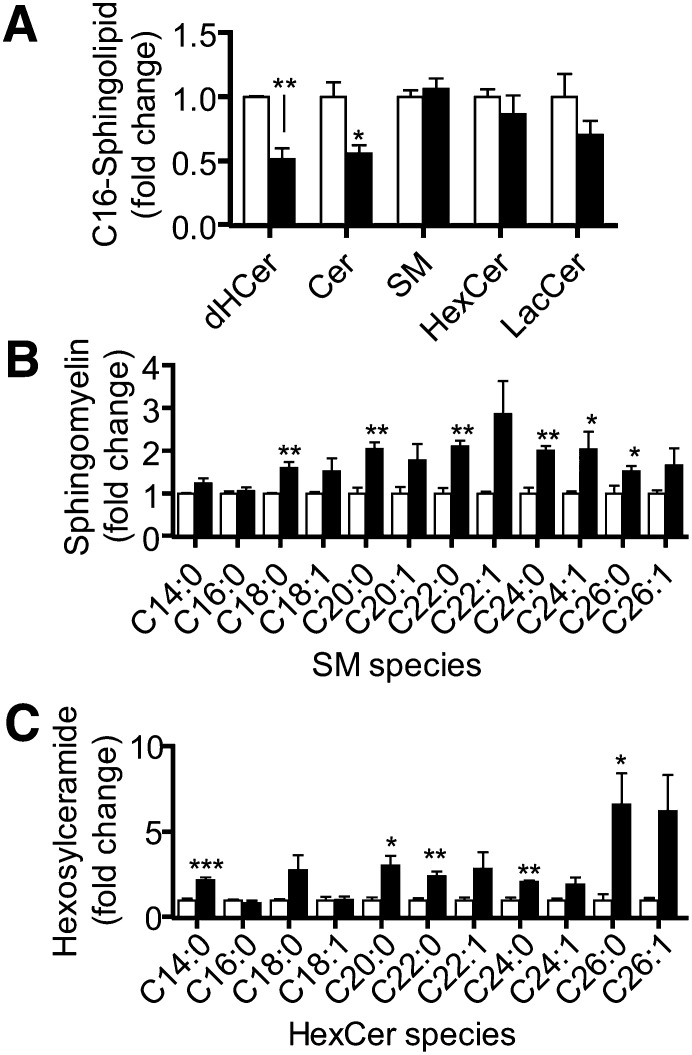
Using siRNA to target CerS6, we were able to significantly downregulate this enzyme at the message and protein levels (Figs. 2F, 3) and decreased the in vitro C16:0-CerS activity (supplementary Fig. IIIC). Consequently, siCerS6 induced a robust decrease in C16:0-containing dHCer and Cer (51.5% ± 8.6% and 56.0% ± 6.7%, respectively) but not SM, HexCer, or LacCer (Fig. 5A). These effects on C16:0-dHCer and Cer are expected based on the previously described substrate specificity for this enzyme (15, 24).
Fig. 5.
siCerS6 decreases C16:0-Cer/dHCer and increases multiple SM and HexCer species. The effects of siCerS6 (black bars) on C16:0-sphingolipids (A), SM species (B), and HexCer species (C) compared to siControl (white bars) were determined as described in Fig. 4. Sphingolipid levels are normalized to the amount of total lipid phosphate. Data are means ± SEM for three to five independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.01 versus siControl.
Like the other siRNAs, siCerS6 downregulation also induced increases in another CerS, as well as in certain sphingolipid species. siCerS6 increased the mRNA levels of CerS5 by approximately 2-fold (Fig. 2F). Regarding sphingolipid changes, significant increases were seen in medium from very-long-chain sphingolipids such as C24:0-SM and C24:0-HexCer, without any detectable increases in C24:0-Cer or C24:0-dHCer (Figs. 3A and 5B, C). C14:0-HexCer was also increased (Fig. 5C). We also examined the in vitro C24:0-CerS activity and found it not to be altered by CerS6 knockdown (supplementary Fig. IIID). Based on the increases in very-long-chain sphingolipids and the decreases in C16:0-dHCer/-Cer, CerS6 knockdown had a clear effect on reducing the amount of C16:0 sphingolipids as a proportion of all acyl chain lengths. Therefore, it is likely that CerS6 is the predominant C16:0-CerS in MCF-7 cells and additionally that it controls the production of very-long-chain sphingolipids.
Downregulation of less abundant CerS
Knockdowns of CerS1, CerS3, CerS4, and CerS5 did not cause robust decreases in any sphingolipid species, but there were several unanticipated increases in several sphingolipids and sphingolipid classes (Fig. 3A). For example, downregulation of CerS1 upregulated several HexCer and LacCer species, especially those possessing a C14:0 acyl chain. A more detailed description of these changes is provided in supplementary Figs. II–VII).
Knockdown of CerS increases total levels of different sphingolipid classes but fails to significantly increase sphingoid base levels
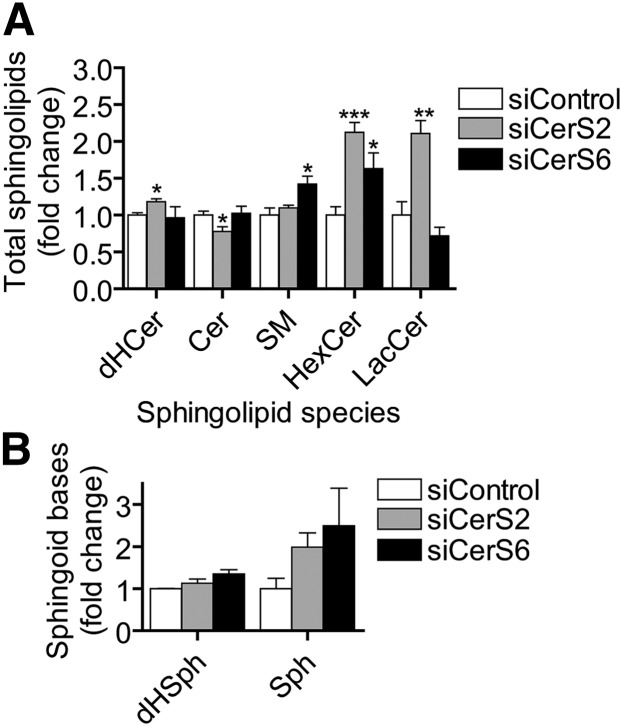
Because many of the decreases in sphingolipids we saw were accompanied by increases in other sphingolipids, we asked whether CerS knockdown affected the total levels of each sphingolipid class (Fig. 6A). The only significant decrease we observed was a slight decrease in ceramide by siCerS2, which can be attributed to the effect of siCerS2 on very-long-chain ceramide (Fig. 6A). CerS6, on the other hand, did not significantly decrease Cer or any other sphingolipid class.
Fig. 6.
Knockdown of CerS2 or CerS6 increases total levels of a particular sphingolipid classes but does not significantly elevate sphingoid bases. MCF-7 cells were transfected with siCerS1-6 or siControl as described in Fig. 3, and total levels of different sphingolipid classes were determined by HPLC/MS. A: Total levels of dHCer, Cer, SM, HexCer, and LacCer were determined following CerS knockdown. B: dHSph and Sph levels were determined following CerS knockdown. Sphingolipid levels are normalized to the amount of total lipid phosphate. Data are means ± SEM for three to five independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.01 versus siControl.
Despite increases in C16:0-SM (Fig. 4C), the most abundant sphingolipid we measured, siCerS2 caused no overall increase in SM (Fig. 6A). This may be explained by the presence of small decreases in multiple very-long-chain SM species being balanced out by the larger change in C16:0-SM. These data also indicate that cells can maintain overall SM levels despite a large shift in acyl chain composition.
Pharmacological inhibition of CerS or genetic deficiency of CerS2 in mice can induce the accumulation of the sphingoid bases dHSph and Sph (34, 35). We therefore asked whether individual knockdowns of CerS would cause an accumulation of these lipids in MCF-7 cells. We analyzed dHSph and Sph levels following CerS1-6 knockdowns and found that none of the knockdowns was capable of significantly increasing sphingoid base levels (Fig. 6B and supplementary Fig. VIII). Of note, several siCerS appeared to have slight stimulatory effects on sphingosine levels; however, none of these values reached our criterion for significance, and further studies using higher doses of siRNA failed to demonstrate Sph increases (data not shown). dHS1P and S1P were near or below detectable levels in the analyses performed and could not be reliably quantified. Based on these data, it is likely that even when CerS are knocked down, there is enough residual activity and redundancy to prevent sphingoid bases from accumulating.
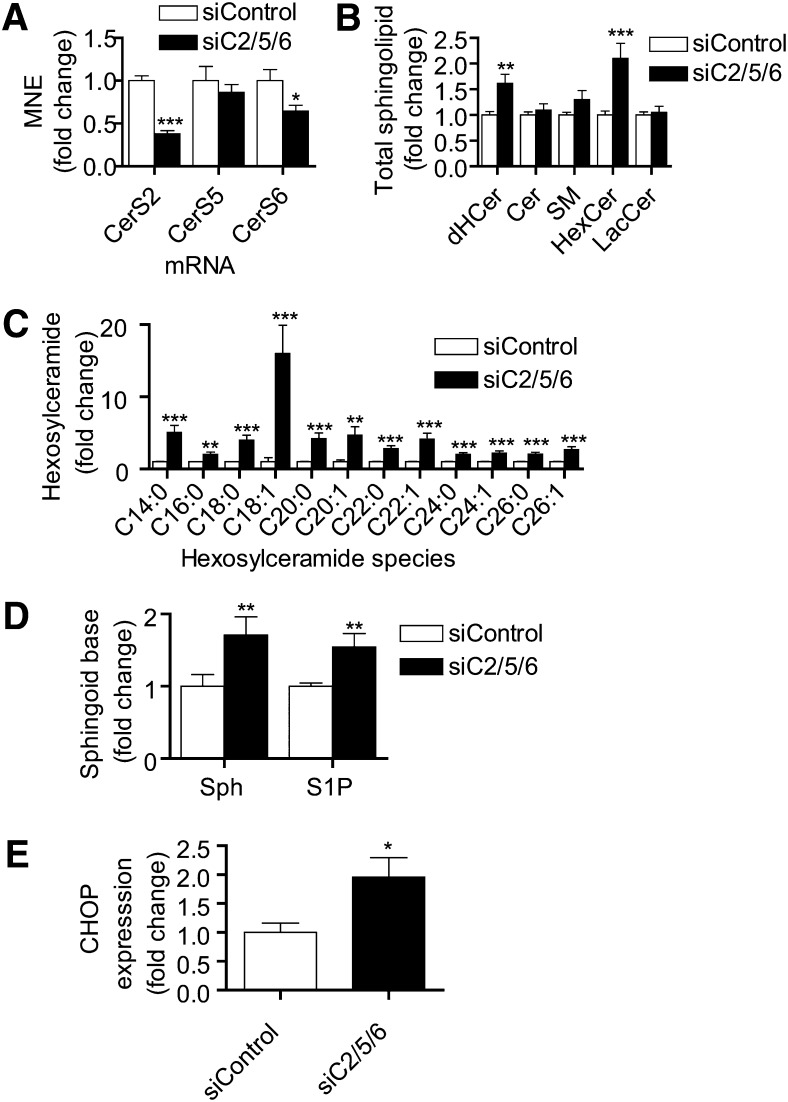
Combined knockdown of CerS2, CerS5, and CerS6 increases HexCer, Sph, and S1P
We observed that knockdown of CerS2 or CerS6 individually caused decreases in particular Cer levels as well as alterations in the expression of other CerS such as CerS5, but only CerS2 knockdown showed an appreciable decrease in total Cer levels. The paucity of effects seen with individual CerS knockdowns on total levels might be explained by compensation from nontargeted CerS, especially when a particular CerS is upregulated (e.g., CerS5 upregulation by siCerS2 and siCerS6). We therefore hypothesized that combined knockdown of the major CerS CerS2 and CerS6, as well as prevention of CerS5 counter-elevation, would cause a detectable decrease in both total and individual ceramide levels due to an overall decrease in CerS activity. We used a treatment that combined siCerS2 (10 nM), siCerS6 (10 nM), and siCerS5 (20 nM) (siCerS2/5/6) and found that siCerS2/5/6 resulted in a decrease in CerS2 and CerS6 expression levels (Fig. 7A). Interestingly, CerS5 mRNA levels, which were increased by either CerS2 or CerS6 alone (Fig. 2A, B), were maintained at control levels by treatment with siCerS2/5/6 (Fig. 7A). Analysis of total lipid changes showed no overall changes in Cer or SM levels (Fig. 7B) but did show a shift of chain length distribution, predominantly among medium-long-chain species (data not shown). Total dHCer, on the other hand, was increased, and this was attributable to increases in long-chain and very-long-chain species. Interestingly, total HexCer was enhanced by more than 2-fold, and every species we analyzed showed a significant increase (Fig. 7C). We also found Sph levels to be elevated, and levels of S1P, although not routinely detectable in MCF-7, were increased following siCerS2/5/6 treatment (Fig. 7D). These data suggest that Cer synthesis was inhibited sufficiently to cause accumulation of sphingoid bases; however, dHSph and dHS1P remained near or below the limit of detection in these samples. Indeed, because multiple CerS are likely metabolizing the same pool of sphingoid base substrates, sufficient knockdown of more than one enzyme is required to see any increase in dHSph or Sph (or their phosphates). These data also suggest that inhibition of multiple CerS is insufficient to drive the total levels of any sphingolipid down. Instead, the increases suggest the presence of a counter-regulatory mechanism that allows the maintenance of Cer and SM levels at the expense of dHCer and HexCer.
Fig. 7.
Treatment of of siCerS2, siCerS5, and siCerS6 combined increases dHCer and HexCer levels and causes elevation of Sph and S1P. The effects of combined treatment of siCerS2 (10 nM), siCerS5 (20 nM), and siCerS6 (10 nM) (siCerS2/5/6) on CerS2, CerS5, and CerS6 expression compared to siControl (40 nM) were analyzed by real-time q-PCR (A). Data are means ± SEM for six independent experiments. B: The effects of siCerS2/5/6 on total levels of dHCer, Cer, SM, HexCer, and LacCer were assessed by HPLC/MS. Data are means ± SEM for six independent experiments. C: Analysis of changes in individual HexCer species in response to siCerS2/5/6. Data are means ± SEM for six independent experiments. D: Changes in Sph and S1P content of cells treated with siCerS2/5/6 or siControl. Data are means ± SEM for six independent experiments. E: Effect of siCerS2/5/6 treatment on CHOP expression. Data are means ± SEM for five independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.01 versus siControl.
Combined knockdown of siCerS2/5/6 upregulates CHOP
Previous work has shown that downregulation of CerS2 or CerS6 can induce ER stress via upregulation of the C/EBP homologous protein (CHOP) (8, 36). We asked whether the siCerS2/5/6 changes in sphingolipids would lead to upregulation of CHOP. We discovered that siCerS2/5/6 indeed caused an upregulation of CHOP expression, indicative of an ER stress response. These data suggest that dysregulation of sphingolipid metabolism, even with preserved total Cer and SM levels, can promote an ER stress response.
DISCUSSION
In this study, we have demonstrated that targeted siRNA knockdown resulted in specific reductions in individual CerS expression. However, in several cases, this was accompanied by increases in mRNA levels of nontargeted CerS, culminating in complex changes in sphingolipid content of cells. Some of our results are in agreement with reported acyl-CoA preferences derived from overexpression and knockdown studies (13–15, 24). For example, we observed an expected decrease in very-long-chain Cer following CerS2 knockdown (Figs. 3A, 4A). With a few exceptions (e.g., C22:0-SM, C24:1-dHCer, and other species), siCerS2 did not significantly reduce SM, HexCer, and LacCer species containing very-long acyl chains (Figs. 3A, 4B–D). Similarly, CerS6 knockdown decreased C16:0-Cer and C16:0-dHCer (Figs. 3A, 5A), and this occurred despite an increase in CerS5 expression (Fig. 2F). Despite reductions in C16:0-dHCer and C16:0-Cer, there were no significant changes in the other C16:0-sphingolipids.
On the other hand, knockdown of less abundant CerS such as CerS1 or CerS5 largely failed to produce the expected decreases in their expected products, C18:0- and C16:0-sphingolipids, respectively (Figs. 4A, 5A). The one exception was a slight reduction in C18:0-SM levels by siCerS1 treatment, and this siRNA decreased C18:1-Cer levels as well (Fig. 3A and supplementary Fig. VIA). CerS4 knockdown was predicted to affect C20:0-Cer, but we failed to observe any changes in this lipid and instead saw a decrease in C24:1-Cer (Fig. 3A and supplementary Fig. VIA). As for CerS3, no major changes were expected because of its low expression in MCF-7 cells (Fig. 1), but we did observe an overall increase in HexCer species (Fig. 3A and supplementary Fig. V, IX).
Knockdown of CerS2 produced several unexpected changes. siCerS2 treatment caused an upregulation of CerS4, CerS5, and CerS6 mRNA levels (Fig. 2B). This was associated with a shift of dHCer and Cer synthesis from very-long-chain species to long-chain species. Interestingly, there was hardly any change in total dHCer or Cer level, suggesting that shifting of synthesis to long-chain species was sufficient to maintain the overall levels of these lipid classes. Of note, CerS2 knockdown did not increase in vitro C16:0-CerS activity (supplementary Fig.IIIB), which supports our previous data (8) indicating that although expression of CerS5 and CerS6 is increased by siCerS2, this does not necessarily translate into increased C16:0-CerS activity. Furthermore, it supports the hypothesis that long-chain Cer/dHCer accumulation is likely due to shunting of sphingoid bases away from CerS2-mediated Cer/dHCer synthesis. Alternatively, the regulation of other enzymes (e.g., forward or reverse Cer activities) may also play a role (8). The mechanism whereby CerS5 and CerS6 mRNA upregulation fails to contribute to increased C16:0-CerS activity is unclear, but there may be a role for ER stress in altering the production or activity of these enzymes (8).
siCerS2 treatment also induced the accumulation of C16:0-SM, long-chain HexCer, and long-chain LacCer, which could be attributed to the accumulation of their Cer precursors (Figs. 3A and 4A–D). Despite an increase in C16:0-SM (∼4 pmol/nmol lipid phosphate), which was the largest mass change we observed in this study, there was no overall increase in SM (Fig. 6A). This may be explained by the presence of small decreases in multiple very-long-chain SM species being balanced out by the larger change in C16:0-SM. These data also indicate that cells can maintain overall SM levels despite a large shift in acyl chain composition. HexCer and LacCer, on the other hand, did exhibit increases in total levels following siCerS2 treatment, suggesting that these lipids are not subject to the same control as Cer and SM. It is surprising that very-long-chain HexCer species do not decrease in response to siCerS2 despite the decrease in Cer levels. This could indicate that steady-state very-long-chain HexCer, consisting of GlcCer and GalCer, are not synthesized and degrade readily enough to show a decrease in response to decreased Cer. Alternatively, the activities of GlcCerS and GalCerS may be altered to enhance or maintain production of HexCer despite decreasing substrate levels.
Other unexpected increases were observed with CerS1, CerS3, CerS4, and CerS5 knockdowns. For CerS1, we found that knockdown caused the increase of nearly all HexCer species, except for C18:0-HexCer (Fig. 3A and supplementary Fig. IVB). One interpretation of these data is that there was an overall increase in HexCer synthesis that allowed cells to maintain a normal level of C18:0-HexCer despite decreased availability of CerS1-derived C18:0-Cer. However, we did not detect a significant decrease in C18:0-Cer, so it is difficult to say whether CerS1 is required for C18:0-Cer synthesis in MCF-7 cells. siCerS3 also caused slight increases in multiple HexCer species that culminated in an increase in total HexCer levels (supplementary Fig. IV). The mechanism of this change is unknown, but it suggests that even though CerS3 is expressed at low levels, it may have a subtle role in regulating HexCer metabolism. siCerS4 caused an increase in C26:0-dHCer/-Cer (supplementary Fig. VIB), and siCerS5 increased C26:1-sphingolipids (supplementary Fig. VIIB). Whether these two latter effects are related is unknown. According to Laviad et al. (25), CerS2 overexpression can increase activity toward C26:0-CoA and C26:1-CoA, so it is possible that CerS2 is somehow contributing to these increases. However, other than the slight increases in CerS2 expression induced by siCerS5, no evidence supports increases in CerS2 activity as a mechanism for elevated C26 sphingolipids. Overall, the changes induced by knockdowns of CerS3–5 were largely unexpected and are not readily explained by the corresponding changes in CerS expression.
Both siCerS6 and siCerS5 caused an unanticipated increase in total SM (Fig. 6A and supplementary Fig. VIIA, respectively), but knockdown of CerS6 had more widespread effects including increased SM and HexCer, as well as elevations in CerS5 message levels (Fig. 2B). In a manner analogous to that of siCerS2, siCerS6 induced a shift in acyl chain compositions of SM (Fig. 5B) and HexCer (Fig. 5C), favoring the increase of nearly all species except C16:0-SM and C16:0-Cer, while total Cer and dHCer remained unchanged (Fig. 6A). Also, the only evidence suggestive of increased CerS5 expression at the sphingolipid level was an increase in C14:0-HexCer (Fig. 5C). The upregulation of CerS5 may indeed be a compensatory mechanism for protecting C16:0-sphingolipid levels from being depleted. Taken together, the effects of CerS6 knockdown likely reflect a shunting of excess sphingoid base substrates into the next most available CerS (e.g., CerS2, CerS5, and others), which leads to the accumulation of non-C16:0-sphingolipids.
The ability of CerS knockdown to cause increases in sphingolipids was exemplified by our attempted simultaneous knockdown with CerS2/5/6 (Fig. 7). The resulting increases in dHCer and HexCer imply that there is a significant response of cells to the reduced CerS expression or activity that involves the accumulation of these lipids (Fig. 7B). Unlike individual knockdowns of CerS2 and CerS6, these data cannot be explained by sphingoid base shunting alone. This notion is supported by the fact that nearly all HexCer species increased (Fig. 7C). It also should be noted that medium-long-chain species (presumably generated collectively by CerS1, CerS4, and CerS5) showed the largest increases and are likely the result of shunting. However, it is also likely that other non-CerS enzymes are being regulated to effect these changes in total dHCer and HexCer while maintaining Cer and SM levels. These results are in contrast to the effects of fumonisin B1 (FB1), an inhibitor of CerS (34), which depletes ceramide levels in MCF-7 cells (our unpublished observations).
According to our data, overall sphingolipid metabolism appears to be regulated such that total Cer levels are maintained at a constant level at the expense of accumulating other lipids, especially HexCer. In order for Cer levels to be maintained, it is possible that substrate supply may be increased by enhanced de novo synthesis of dHSph or augmented Sph production via the salvage pathway. Both of these hypotheses are plausible given the fact that siCerS2/5/6 treatment increased dHCer and Sph levels. However, Sph would have to be salvaged from a sphingolipid that is not included in our analysis in order for there to be a net addition to the sphingolipid pool. Supporting the de novo synthesis hypothesis are data showing that treatment of mice with FB1 upregulates liver expression of the obligatory subunits of SPT, SPTLC1, and SPTLC2 (37). Alternatively, reduced sphingoid base degradation via decreased S1P lyase activity could result in increased S1P and Sph and allow maintenance of Cer/SM levels despite reduced CerS activity (38).
Only a few studies have examined how the loss of these proteins influences sphingolipid metabolism. Laviad et al. (25) showed that loss of CerS2 decreases activity of very-long-chain ceramide synthase activity and levels of very-long-chain ceramides (25). Mizutani et al. (26) knocked down CerS2 in HeLa cells overexpressing fatty acid 2-hydroxylase (FA2H) and found that very-long-chain HexCer were decreased along with a corresponding increase in long-chain HexCer species (26). In that latter study, CerS5 knockdown was shown to downregulate steady-state levels of long-chain HexCer species. More recently, our laboratory demonstrated that CerS2 knockdown in SMS-KCNR neuroblastoma cells increased long-chain Cer and SM (8).
Our understanding of the roles of CerS in sphingolipid metabolism has been advanced very recently by the generation of CerS2 knockout mice (35, 39). These mice exhibit profound defects in very-long-chain sphingolipid synthesis in multiple tissues, and adult mice develop neuronal and hepatic dysfunctions. In the liver, CerS2 deficiency leads to a drop in total Cer, HexCer, and SM levels, and the chain length distribution shifts to one favoring C16:0 and other long-chain fatty acids (39). In the brain and kidney, however, total Cer, HexCer, and SM levels are much less affected by a loss of CerS2. This is likely due to the reduced dependence of these tissues on CerS2 (which is reflected in their sphingolipid acyl chain composition) and the ability of residual CerS (e.g., CerS1 in the brain and CerS5 and CerS6 in the kidney) to compensate for the absence of CerS2 activity. Notably, CerS2 deficiency was also associated with an increase in CerS5 and glucosylceramide synthase GCS expression in the liver (35).
Since their discovery, mammalian CerS proteins have been implicated in many biological processes, most notably in the regulation of cell stress responses (8, 40–43). We have previously shown that downregulation of CerS2 induced ER stress and autophagy in SMS-KCNR neuroblastoma cells and MCF-7 cells (8). Senkal et al. (36) have also shown that CerS6 downregulation results in decreased C16:0-Cer and ER stress via CHOP upregulation in head and neck squamous carcinoma cell lines. In this study, we found that siCerS2/5/6 treatment of MCF-7 cells also produced a characteristic sphingolipid profile (i.e., high HexCer and dHCer levels with an increased proportion of medium-long-chain sphingolipids) that was associated with CHOP upregulation. The evidence that dysregulation of CerS activities elicits changes in both other enzymes of sphingolipid metabolism (including other CerS) as well as stress responses suggests that Cer synthesis is highly regulated and is a determinant of ER homeostasis. The challenge remains to fully triangulate and define the lipids that specifically regulate the changes in CerS expression and the ER stress response.
In this study, we have demonstrated that downregulation of CerS causes multiple changes in sphingolipid metabolism and residual CerS expression. Many changes in sphingolipids reflect the unique acyl-CoA preferences of CerS, but the robust maintenance of Cer and SM levels at the expense of HexCer, as well as the multiple changes seen in nontargeted CerS expression, suggests a high degree of redundancy and interregulation. Taken together, it is clear that CerS are not only important mediators of sphingolipid metabolism, but they are emerging as key regulators of ER homeostasis.
Supplementary Material
Footnotes
Abbreviations:
- Cer
- ceramide
- CerS
- ceramide synthase
- CHOP
- C/EBP homologous protein
- dHCer
- dihydroceramide
- dHSph
- dihydrosphingosine
- ER
- endoplasmic reticulum
- FB1
- fumonisin B1
- GalCer
- galactosylceramide
- GlcCer
- glucosylceramide
- HexCer
- hexosylceramide
- HPLC/MS
- high-performance liquid chromatography/mass spectrometry
- LacCer
- lactosylceramide
- S1P
- sphingosine-1-phosphate
- siRNA
- short interfering RNA
- Sph
- sphingosine
Support for this research was provided by Medical University of South Carolina Department of Pharmaceutical Sciences Training Program in Environmental Stress Signaling (National Institutes of Health/National Institute of Environmental Health Science) Grant 5T32ES012878 and Ruth L. Kirschstein National Research Service Award (NIH/NIEHS) Grant 1 F30 ES016975-01 (T.D.M.); NIH Grant R01 AG016583 (to L.M.O.); National Institutes of Health/National Cancer Institute Grant P01 CA097132 (Y.A.H.); American Heart Association pre-doctoral fellowship Grant 081509E (R.W.J.); and National Institutes of Health Medical Scientist Training Program Grant (GM08716) (R.W.J.). HPLC/MS analysis at the MUSC Lipidomics Core Facility was supported by the Extramural Research Facilities Program of the National Center for Research Resources (National Institutes of Health Grant C06 RR018823). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. This work also received supportin part by a Method to Extend Research in Time (MERIT) Award (L.M.O.) from the Office of Research and Development, Department of Veterans Affairs, Ralph H. Johnson Veterans Affairs Medical Center, Charleston, SC.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight figures.
REFERENCES
- 1.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 2.Taha T. A., Mullen T. D., Obeid L. M. 2006. A house divided: ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim. Biophys. Acta. 1758: 2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884. [DOI] [PubMed] [Google Scholar]
- 4.Kitatani K., Idkowiak-Baldys J., Hannun Y. A. 2008. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell. Signal. 20: 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sribney M. 1966. Enzymatic synthesis of ceramide. Biochim. Biophys. Acta. 125: 542–547. [DOI] [PubMed] [Google Scholar]
- 6.Harel R., Futerman A. H. 1993. Inhibition of sphingolipid synthesis affects axonal outgrowth in cultured hippocampal neurons. J. Biol. Chem. 268: 14476–14481. [PubMed] [Google Scholar]
- 7.Bose R., Verheij M., Haimovitz-Friedman A., Scotto K., Fuks Z., Kolesnick R. 1995. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 82: 405–414. [DOI] [PubMed] [Google Scholar]
- 8.Spassieva S. D., Mullen T. D., Townsend D. M., Obeid L. M. 2009. Disruption of ceramide synthesis by CerS2 downregulation leads to autophagy and the unfolded protein response. Biochem. J. 424: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20: 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. 2001. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell 12: 3417–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Mello N. P., Childress A. M., Franklin D. S., Kale S. P., Pinswasdi C., Jazwinski S. M. 1994. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J. Biol. Chem. 269: 15451–15459. [PubMed] [Google Scholar]
- 12.Jiang J. C., Kirchman P. A., Zagulski M., Hunt J., Jazwinski S. M. 1998. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 8: 1259–1272. [DOI] [PubMed] [Google Scholar]
- 13.Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H., Jr, Futerman A. H. 2002. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-{dihydro}ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 277: 35642–35649. [DOI] [PubMed] [Google Scholar]
- 14.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr, Futerman A. H. 2003. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278: 43452–43459. [DOI] [PubMed] [Google Scholar]
- 15.Mizutani Y., Kihara A., Igarashi Y. 2005. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spassieva S., Seo J. G., Jiang J. C., Bielawski J., Alvarez-Vasquez F., Jazwinski S. M., Hannun Y. A., Obeid L. M. 2006. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J. Biol. Chem. 281: 33931–33938. [DOI] [PubMed] [Google Scholar]
- 17.Mesika A., Ben-Dor S., Laviad E. L., Futerman A. H. 2007. A new functional motif in Hox domain-containing ceramide synthases: identification of a novel region flanking the Hox and TLC domains essential for activity. J. Biol. Chem. 282: 27366–27373. [DOI] [PubMed] [Google Scholar]
- 18.Lahiri S., Futerman A. H. 2005. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl CoA as acyl donor. J. Biol. Chem. 280: 33735–33738. [DOI] [PubMed] [Google Scholar]
- 19.Pewzner-Jung Y., Ben-Dor S., Futerman A. H. 2006. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281: 25001–25005. [DOI] [PubMed] [Google Scholar]
- 20.Mizutani Y., Kihara A., Igarashi Y. 2006. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 398: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillas I., Jiang J. C., Vionnet C., Roubaty C., Uldry D., Chuard R., Wang J., Jazwinski S. M., Conzelmann A. 2003. Human homologues of LAG1 reconstitute acyl-CoA-dependent ceramide synthesis in yeast. J. Biol. Chem. 278: 37083–37091. [DOI] [PubMed] [Google Scholar]
- 22.Rabionet M., van der Spoel A. C., Chuang C. C., von Tumpling-Radosta B., Litjens M., Bouwmeester D., Hellbusch C. C., Korner C., Wiegandt H., Gorgas K., et al. 2008. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J. Biol. Chem. 283: 13357–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min J., Mesika A., Sivaguru M., Van Veldhoven P. P., Alexander H., Futerman A. H., Alexander S. 2007. (Dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Mol. Cancer Res. 5: 801–812. [DOI] [PubMed] [Google Scholar]
- 24.Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., Futerman A. H. 2007. Kinetic characterization of mammalian ceramide synthases: determination of K(m) values towards sphinganine. FEBS Lett. 581: 5289–5294. [DOI] [PubMed] [Google Scholar]
- 25.Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H., Jr, Futerman A. H. 2008. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283: 5677–5684. [DOI] [PubMed] [Google Scholar]
- 26.Mizutani Y., Kihara A., Chiba H., Tojo H., Igarashi Y. 2008. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J. Lipid Res. 49: 2356–2364. [DOI] [PubMed] [Google Scholar]
- 27.Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. 2006. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods 39: 82–91. [DOI] [PubMed] [Google Scholar]
- 28.Van Veldhoven P. P., Bell R. M. 1988. Effect of harvesting methods, growth conditions and growth phase on diacylglycerol levels in cultured human adherent cells. Biochim. Biophys. Acta. 959: 185–196. [DOI] [PubMed] [Google Scholar]
- 29.Spassieva S., Bielawski J., Anelli V., Obeid L. M. 2007. Combination of c(17) sphingoid base homologues and mass spectrometry analysis as a new approach to study sphingolipid metabolism. Methods Enzymol. 434: 233–241. [DOI] [PubMed] [Google Scholar]
- 30.Pavlidis P., Noble W. S. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics 19: 295–296. [DOI] [PubMed] [Google Scholar]
- 31.Ruckhaberle E., Rody A., Engels K., Gaetje R., von Minckwitz G., Schiffmann S., Grosch S., Geisslinger G., Holtrich U., Karn T., et al. 2008. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res. Treat. 112: 41–52. [DOI] [PubMed] [Google Scholar]
- 32.Schiffmann S., Sandner J., Birod K., Wobst I., Angioni C., Ruckhaberle E., Kaufmann M., Ackermann H., Lotsch J., Schmidt H., et al. 2009. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis 30: 745–752. [DOI] [PubMed] [Google Scholar]
- 33.Erez-Roman R., Pienik R., Futerman A. H. 2010. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem. Biophys. Res. Commun. 391: 219–223. [DOI] [PubMed] [Google Scholar]
- 34.Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H., Jr 1991. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266: 14486–14490. [PubMed] [Google Scholar]
- 35.Pewzner-Jung Y., Park H., Laviad E. L., Silva L. C., Lahiri S., Stiban J., Erez-Roman R., Brugger B., Sachsenheimer T., Wieland F., et al. 2010. A critical role for ceramide synthase 2 in liver homeostasis: I. Alterations in lipid metabolic pathways. J. Biol. Chem. 285: 10902–10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senkal C. E., Ponnusamy S., Bielawski J., Hannun Y. A., Ogretmen B. 2010. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 24: 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Q., Suzuki H., Sharma N., Sharma R. P. 2006. Ceramide synthase inhibition by fumonisin B1 treatment activates sphingolipid-metabolizing systems in mouse liver. Toxicol. Sci. 94: 388–397. [DOI] [PubMed] [Google Scholar]
- 38.Bektas M., Allende M. L., Lee B. G., Chen W., Amar M. J., Remaley A. T., Saba J. D., Proia R. L. 2010. S1P lyase deficiency disrupts lipid homeostasis in liver. J. Biol. Chem. 285: 10880–10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imgrund S., Hartmann D., Farwanah H., Eckhardt M., Sandhoff R., Degen J., Gieselmann V., Sandhoff K., Willecke K. 2009. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 284: 33549–33560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koybasi S., Senkal C. E., Sundararaj K., Spassieva S., Bielawski J., Osta W., Day T. A., Jiang J. C., Jazwinski S. M., Hannun Y. A., et al. 2004. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 279: 44311–44319. [DOI] [PubMed] [Google Scholar]
- 41.Senkal C. E., Ponnusamy S., Rossi M. J., Bialewski J., Sinha D., Jiang J. C., Jazwinski S. M., Hannun Y. A., Ogretmen B. 2007. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther. 6: 712–722. [DOI] [PubMed] [Google Scholar]
- 42.Jin J., Hou Q., Mullen T. D., Zeidan Y. H., Bielawski J., Kraveka J. M., Bielawska A., Obeid L. M., Hannun Y. A., Hsu Y. T. 2008. Ceramide generated by sphingomyelin hydrolysis and the salvage pathway is involved in hypoxia/reoxygenation-induced Bax redistribution to mitochondria in NT-2 cells. J. Biol. Chem. 283: 26509–26517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White-Gilbertson S., Mullen T., Senkal C., Lu P., Ogretmen B., Obeid L., Voelkel-Johnson C. 2009. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene. 28: 1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.