Abstract
Proprotein convertase subtilisin-like/kexin type 9 (PCSK9) regulates LDL cholesterol levels by inhibiting LDL receptor (LDLr)-mediated cellular LDL uptake. We have identified a fragment antigen-binding (Fab) 1D05 which binds PCSK9 with nanomolar affinity. The fully human antibody 1D05-IgG2 completely blocks the inhibitory effects of wild-type PCSK9 and two gain-of-function human PCSK9 mutants, S127R and D374Y. The crystal structure of 1D05-Fab bound to PCSK9 reveals that 1D05-Fab binds to an epitope on the PCSK9 catalytic domain which includes the entire LDLr EGF(A) binding site. Notably, the 1D05-Fab CDR-H3 and CDR-H2 loops structurally mimic the EGF(A) domain of LDLr. In a transgenic mouse model (CETP/LDLr-hemi), in which plasma lipid and PCSK9 profiles are comparable to those of humans, 1D05-IgG2 reduces plasma LDL cholesterol to 40% and raises hepatic LDLr protein levels approximately fivefold. Similarly, in healthy rhesus monkeys, 1D05-IgG2 effectively reduced LDL cholesterol 20%–50% for over 2 weeks, despite its relatively short terminal half-life (t1/2 = 3.2 days). Importantly, the decrease in circulating LDL cholesterol corresponds closely to the reduction in free PCSK9 levels. Together these results clearly demonstrate that the LDL-lowering effect of the neutralizing anti-PCSK9 1D05-IgG2 antibody is mediated by reducing the amount of PCSK9 that can bind to the LDLr.
Keywords: antibodies, cholesterol, metabolism, drug therapy, low density lipoprotein metabolism, lipoprotein receptors, proprotein convertase subtilisin-like/kexin type 9
Proprotein convertase subtilisin-like/kexin type 9 (PCSK9) has recently emerged as a major regulator of plasma low-density lipoprotein cholesterol (LDLc) and is a promising therapeutic target for treating coronary heart disease (CHD). The link between PCSK9 and CHD stems from genetic studies which indicate that individuals carrying rare dominant missense mutations (putatively, gain-of-function mutants) in the PCSK9 gene exhibit severe hypercholesterolemia and elevated risk of CHD (1, 2). Conversely, approximately 2%–3% of the human population is heterozygous for specific truncation or missense mutations in the PCSK9 gene (putatively, loss-of-function mutants) and have 15%–40% reduction in plasma LDLc and 50%–90% reduction in the risk of CHD over 15 years (3–5). A recent genome-wide association study further established a link between a single nucleotide polymorphism at a locus near PCSK9 with early onset myocardial infarction (6).
There is extensive evidence that plasma PCSK9 raises LDLc levels by binding to cell surface LDL receptor (LDLr) protein and directing LDLr to lysosomes for degradation (7–11). Consistent with this mechanism, inhibition of PCSK9 by recombinant LDLr fragments (12–14) or by mono- or polyclonal antibodies (15, 16) restored LDLc uptake in cells. Furthermore, intravenous (iv) injection of a monoclonal antibody (MAb) that disrupted the PCSK9/LDLr interaction (15) or small interfering RNAs targeting liver PCSK9 (17) was found to reduce plasma LDLc in mice and nonhuman primates. Collectively, these results support PCSK9 as an attractive and viable target for therapeutic intervention against hypercholesterolemia.
We previously characterized a fragment antigen-binding (Fab) from a human MAb, 1G08, which binds to the C-terminal domain of PCSK9 and partially inhibits its effect on LDLc uptake in vitro. Interestingly, binding of the 1G08 Fab to PCSK9 does not affect the PCSK9/LDLr interaction but inhibits PCSK9 internalization (18).
Using a human combinatorial antibody phage display library, we have now identified a Fab, 1D05, which binds to the catalytic domain of PCSK9 with nanomolar affinity. The 1D05-Fab antibody and corresponding human antibody 1D05-IgG2 completely block the interaction between PCSK9 and LDLr and the inhibitory effect of PCSK9 on cellular LDLc uptake. Moreover, administration of 1D05-IgG2 leads to sustained reduction of plasma LDLc in a mouse model with human-like lipid and PCSK9 profiles, and in rhesus monkeys. The crystal structure of the PCSK9/1D05-Fab complex reveals that 1D05 acts as a structural mimic of the EGF(A) domain of LDLr and sterically prevents PCSK9 from binding to the receptor. Finally, by using highly sensitive dissociation-enhanced lanthanide fluorescence immunoassays (DELFIAs), which selectively detect total or antibody-free PCSK9, we demonstrate that the LDL-lowering effect of the 1D05-IgG2 antibody closely follows the reduction in antibody-free-PCSK9 levels and the increase in percentage of PCSK9 bound to 1D05-IgG2.
EXPERIMENTAL PROCEDURES
PCSK9 and PCSK9ΔC protein expression and purification
Full-length human and mouse PCSK9-V5-His proteins, as well as the PCSK9 types carrying mutations S127R and D374Y, respectively, were expressed and purified in stably transfected HEK293 cells as described previously (8). The human LDLr ectodomain was purchased from R&D Systems (Minneapolis, MN). Expression and purification of human PCSK9ΔC (residues 53–451) was performed as previously described (12).
Isolation of anti-PCSK9 antibody 1D05
Human combinatorial antibody phage display libraries (19) were panned against recombinant murine PCSK9-V5-His proteins immobilized on Nunc Maxisorp plates, and PCSK9 binding clones were identified by ELISA. Initial expression and purification of 1D05-Fab procedures were performed as previously described (18).
1D05-Fab clone heavy chain variable (VH) regions were fused in frame with the IgG2m4 constant region (20, 21), whereas the light chain variable (VL) regions fused with corresponding constant regions (e.g., a κ variable region matched with a κ constant region) using In-Fusion technology (Clontech, Mountain View, CA). The 1D05-IgG2m4 antibody was expressed in HEK293 cells, and secreted antibodies were purified from culture medium by using standard protein A/G affinity chromatography (Pierce, Rockford, IL).
Surface plasmon resonance
All surface plasmon resonance experiments were performed using a Biacore 2000 instrument as described previously (18). Briefly, polyclonal anti-human IgG2 antibody was covalently coupled to the surface of a CM5 sensor chip, and 1D05-IgG2 was captured. Association and dissociation binding rates of soluble human, rhesus, and mouse PCSK9-V5-His proteins were then determined and used to calculate the affinity constant KD. Specifically, kinetic constants for the on-rate (ka) and for the off-rate (kd) were determined and used to calculate the apparent equilibrium affinity constant, KD, using the equation KD = kd/ka.
PCSK9ΔC/1D05-Fab complex formation and crystallization
Purified PCSK9ΔC and 1D05-Fab were mixed at a molar ratio of 1:1.5 and incubated for 2 h at 4°C. The mixture was then loaded onto a Superdex G-200 size-exclusion chromatography column equilibrated in a solution containing 40 mM Tris, pH 8.0, 100 mM NaCl, 5% glycerol, 20 µM CaCl2; and fractions containing the complex, as assayed by SDS-PAGE, were collected, concentrated to 10 mg/ml by ultrafiltration, using Vivaspin 15R HY concentrators (Vivascience), and used immediately for crystallization.
Crystallization experiments were performed at room temperature by the hanging-drop and sitting-drop vapor diffusion methods. Thin plate-like crystals were obtained in 100 mM sodium citrate, pH 6.5, 13% polyethylene glycol 6000 (PEG 6000). The presence of both components of the complex in the crystals was confirmed by silver staining of SDS gels of washed crystals. For data collection, crystals were transferred to a stabilizing solution [20 mM Tris, pH 8.0, 50 mM sodium citrate, pH 6.5, 100 mM NaCl, 5% glycerol, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 1 µM CaCl2, 25% PEG 6000], then transferred into a cryoprotectant solution (20 mM Tris, pH 8.0, 50 mM sodium citrate, pH 6.5, 100 mM NaCl, 20% glycerol, 1 mM TCEP, 1 µM CaCl2, 35% PEG 6000) for 2 min, and finally placed directly into liquid nitrogen. Diffraction data were collected at a temperature of 100 K at the ID14-EH2 beam-line by the European Synchrotron Radiation Facility (ESRF) (Grenoble, France). Data were processed with MOSFLM software and scaled with SCALA software (22). The structure of the PCSK9ΔC/1D05-Fab complex was determined to a 2.7-Å resolution by the molecular replacement method with PHASER software (22) in MR_AUTO mode and with five separate search models. Specifically, Protein Data Bank code 2w2m (12) was used as the search model for PCSK9ΔC; homology models of the variable and constant domains of the antibody VL and VH chains, generated by the SWISS-MODEL server (23), were used for searching the 1D05-Fab antibody. The structure was improved by multiple rounds of automated model building with ARP/wARP (22) and manual model building with COOT (24), and refinement with REFMAC (22) software, using a maximum likelihood target function and individual B-factor refinements. The final model (Table 1) contains residues 61–152 from the PCSK9 prodomain and residues 155–157 and 178–422 from the PCSK9 catalytic domain, as well as residues 1–212 from the Fab light chain and residues 1–236 from the Fab heavy chain. In addition, one Ca2+ ion bound to PCSK9 and 209 water molecules were identified. There was no electron density present for PCSK9 residues 53–60, 153–154, 158–177, and 423–451, nor for 1D05 Fab residues 237–255 of the heavy chain and residue 213 of the light chain. These residues were therefore excluded from the refinement. The structure satisfies all the criteria set by PROCHECK software (25) for a structure refined at this resolution. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 2xtj.
TABLE 1.
Binding rates of 1D05-IgG2 to PCSK9 proteins from different species
| Antigen | On-rate | Off-rate | KD (nM) | N |
|---|---|---|---|---|
| Human PCSK9 | 7.21 ± 1.60 | 3.16 ± 0.16 | 4.4 ± 1.4 | 3 |
| Rhesus PCSK9 | 11.3 | 4.34 | 3.89 | 1 |
| Mouse PCSK9 | 6.72 ± 0.61 | 2.15 ± 0.16 | 3.2 ± 0.06 | 2 |
Kinetic constants for on-rate [ka (M−1•s−1 × E + 04)] and off-rate [kd (s−1 × E − 04)] were determined and used to calculate the apparent equilibrium affinity constant, KD, using the equation KD = kd/ka. Data are shown as mean ± SEM.
Isolation of LDLc and labeling of LDLc, LDLr, and PCSK9
LDLc was isolated from healthy human volunteers as previously described (8). Labeling of PCSK9 with Alexa Fluor 647 (AF 647-PCSK9), LDL with AF 546 (AF 546-LDL), and LDLr labeled with Eu3+ (Eu3+-LDLr) (8044)-DTA (Perkin-Elmer, Waltham, MA) was performed as previously reported (26).
LDLr/PCSK9 interaction studies by time-resolved fluorescence resonance energy transfer assay
Time-resolved fluorescence resonance energy transfer (TR-FRET) experiments were performed as previously described, using 4 nM Eu3+-LDLr ectodomain and 10 nM AF 647-PCSK9 (18).
LDLc uptake
Cellular uptake of AF 546-LDL was studied in HEK293 cells and HepG2 cells as described previously (8).
In vivo studies in mice
Cholesteryl ester transfer protein (CETP)/LDLr-hemi mice, which are hemizygous for overexpression of the human CETP and heterozygous for LDLr, were generated by crossing mice overexpressing ApoAI promoter-driven human CETP (B6;SJL-Tg[CETP]; Taconic, Hudson, NY) with LDLr null mice (B6.129S7-LDLrtm1Her/J; Jackson Laboratory, Bar Harbor, ME). Following brother-sister mating, mice homozygous for both the human CETP and the LDLrTm1 alleles were selected and then crossed onto a C57BL6/J background to generate mice hemizygous for both alleles. The background of the homozygous mice was B6, 129S7 N = 3. The background of the hemizygous mice was B6, 129 N = 4.
Each animal received a single iv injection of purified 1D05-IgG2 at the indicated dose through the tail vein. Blood samples were collected at designated time points after dosing and stored at −70°C until analysis.
In vivo studies in rhesus monkeys
Male rhesus monkeys (Macaca mulatta), naïve to biologics, were used in the study. Each animal (n = 3) received a single iv injection of 1D05-IgG2, 3 mg/kg of body weight via the cephalic vein. Blood samples were collected from the saphenous/femoral vessels at designated time points after dosing and stored at −70°C until analysis.
All animal studies were conducted in conformity with the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC) at Merck Research Laboratories.
Western blot analysis of mouse liver LDLr
Crude protein extracts were prepared from mouse liver using radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology Inc., Santa Cruz, CA) with PMSF, sodium orthovanadate, and proteinase inhibitors. A total of 50 µg of proteins per lane were loaded in NuPAGE 4%–12% bis-Tris gels (Life Technologies, Carlsbad, CA). Mouse LDLr protein was detected using FITC-labeled anti-LDLr goat IgG (R&D Systems, Minneapolis, MI). Tubulin protein was detected with anti-tubulin antibody (Sigma, St Louis, MO) to normalize protein loading. Proteins of interest were quantified by densitometric scanning of the autoradiogram.
Lipid analysis
To generate lipoprotein profiles, plasma was fractionated by chromatography using a Superose-6 size-exclusion column (GE LifeSciences, Inc.) with an Ultimate 3000 series HPLC system (Dionex Corporation, Sunnyvale, CA). Total cholesterol levels in the column effluent were continuously measured using an in-line mixture with an enzymatic and colorimetric cholesterol detection reagent (Total Cholesterol E, Waco TX), followed by spectrophotometric detection of the reaction products at an absorbance of 600 nm. LDLc and HDLc were eluted from the column. The concentration for each lipoprotein fraction was calculated by extrapolating the ratio of corresponding peak areas to total peak areas and multiplying by the total cholesterol concentration measured in each sample.
Total cholesterol was measured with a 1:1 mixture of diluted plasma and cholesterol E reagent, using a plate reader. Cholesterol standards were provided in the kit at 200 mg/dl and were serially diluted to generate a standard curve.
Bioanalyses of total and free PCSK9 concentrations in plasma
To assess mouse plasma PCSK9 levels, we established a PCSK9 dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA) value. High-binding 96-well plates (catalog no. 4HBX; ThermoLabsystems, Helsinki, Finland) were coated with anti-mouse PCSK9 (anti-mPCSK9) antibody A at 4°C overnight. Plates were blocked with a blocking solution (TBS, 1% BSA, 0.05% Tween-20) and washed before purified mPCSK9 protein (as standard) or diluted mouse plasma samples were added. PCSK9 was detected in the samples by using biotinylated anti-mPCSK9 Fab B. Europium-labeled streptavidin (Perkin Elmer, Waltham, MA) and a DELFIA enhancer (Perkin Elmer, Waltham, MA) were then added to quantify PCSK9 with a europium plate reader. The PCSK9 detection sensitivity of this assay is ∼100 pM, with an average technical variance of 10%. Serial dilutions of mouse plasma samples showed that the assay can tolerate ∼50% mouse serum or plasma without affecting the assay signal-to-noise ratio.
A DELFIA was also used to detect mouse plasma PCSK9 which was not bound to antibody (free PCSK9) following 1D05-IgG2 treatment. In this assay, instead of anti-PCSK9 Fab B, biotinylated 1D05-IgG2 was used as the detection antibody to selectively measure 1D05-free PCSK9.
Similarly, using two different antibody pairs, we also established two DELFIAs for measuring total and free PCSK9 in rhesus plasma samples. Importantly, antibodies used in these assays recognize both rhesus and human PCSK9. Therefore, these assays can also be used in clinical studies.
Determination of the percentage of PCSK9 bound to 1D05-IgG2
The following formulae were used to determine plasma PCSK9 bound to 1D05-IgG2 and percentage of antibody-captured PCSK9: 1D05-bound PCSK9 = total PCSK9 – free PCSK9; and percentage of antibody-captured PCSK9 = 100 × (1D05-bound PCSK9/total PCSK9).
Bioanalysis of 1D05-IgG2 concentrations in serum
The serum levels of 1D05-IgG2 were determined with an anti-human IgG immunoassay using Gyrolab microfluidic CD technology (Gyros, Sweden). Briefly, anti-human κ-chain-specific antibody (BD Biosciences, San Jose, CA) was used to capture 1D05-IgG2 from rhesus serum. After a wash step, an AF 647-labeled anti-human Fc-specific antibody (Southern Biotechnology, Birmingham, AL) was used to detect bound 1D05-IgG2. 1D05-IgG2 in unknown samples was compared with a calibrator curve generated with purified 1D05-IgG2 proteins.
Noncompartmental modeling software (WinNonlin, Enterprise version 5.01; Pharsight Corp., Mountain View, CA) was used for pharmacokinetics analysis. The elimination phase terminal half-life (t1/2) was determined using 8–9 data points between day 2 and day 28 postdose. These data points fit well to a monoexponential decay function.
RESULTS
1D05 binds PCSK9 and inhibits the PCSK9/LDLr interaction
Fab 1D05 was isolated by three rounds of panning the Morphosys HuCAL Gold phage display libraries against recombinant murine mouse PCSK9 and identified by ELISA against human and murine mouse PCSK9s. After conversion to an a variant of IgG2-IgG2m4 (20), its binding affinities to PCSK9 proteins were measured using surface plasmon resonance. 1D05-IgG2 binds to recombinant human, mouse, and rhesus PCSK9 proteins with nM affinities. The kinetic and equilibrium constants are reported in Table 1.
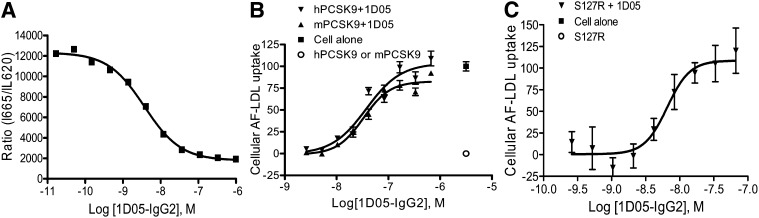
To determine whether the binding of 1D05-IgG2 affects PCSK9 function, we studied its effect on the PCSK9/LDLr interaction. As shown in Fig. 1A, 1D05-IgG2 dose-dependently inhibited PCSK9/LDLr binding in an in vitro TR-FRET assay with an IC50 of 3.7 ± 0.1 nM (n = 3).
Fig. 1.
1D05-IgG2 inhibits PCSK9 function. A: Effect of 1D05-IgG2 on PCSK9-LDLR interaction. TR-FRET experiments were performed using 4 nM Eu3+-LDLr ectodomain and 10 nM AF 647-labeled PCSK9. Increasing concentrations of 1D05-IgG2 was titrated into reaction mixtures containing a preformed complex of AF 647-PCSK9 (10 nM) and Eu3+-LDLr (4 nM) at pH 7.4. Data are means ± SD of triplicates. B: Effect of 1D05-IgG2 on PCSK9-mediated inhibition of LDLc uptake in HEK293 cells. AF 546-LDL uptake was measured in cells incubated with human or mouse PCSK9 (5 µg/ml). C: Effect of 1D05-IgG2 on gain-of-function human PCSK9 S127R mutation-mediated inhibition of LDL uptake. AF 546-LDL uptake was measured in HepG2 cells incubated with 1 µg/ml S127R.
1D05-IgG2 neutralizes the inhibitory effect of wild-type and gain-of-function PCSK9 mutants
We assessed whether binding of 1D05-IgG2 to PCSK9 alters cellular LDLc uptake. As reported earlier (8, 26), addition of purified recombinant human or mouse PCSK9 to cell culture media significantly decreased cellular uptake of AF 546-LDLc in HEK293 cells. 1D05-IgG2 completely blocked the effects of human and mouse PCSK9 with an IC50 of 38 and 33 nM, respectively (Fig. 1B). These values are likely to be an underestimation of the potency of 1D05-IgG2 because they are close to the inhibitor potency-detecting limit of the assay in HEK293 cells (∼30 nM). Similar effects of 1D05-IgG2 on human PCSK9-mediated effect were also observed in HepG2 cells, with an IC50 of approximately 19 nM (data not shown), which is also close to the potency-detecting limit of the assay in HepG2 cells (∼13 nM). Importantly, 1D05-IgG2 can also completely block the effect of proteins carrying the gain-of-function mutations S127R (Fig. 1C) and D374Y (data not shown).
PCSK9/1D05-Fab complex
In order to characterize the molecular details of the PCSK9/1D05 interaction, we determined the crystal structure of the complex between 1D05-Fab and PCSK9ΔC (a truncated form of PCSK9 lacking the C-terminal domain) (12). Crystals were grown at pH 6.5 and belong to space group P212121, with one complex per asymmetric unit and 58% solvent. Data collection and refinement statistics to 2.7-Å resolution are summarized in supplementary Table SI.
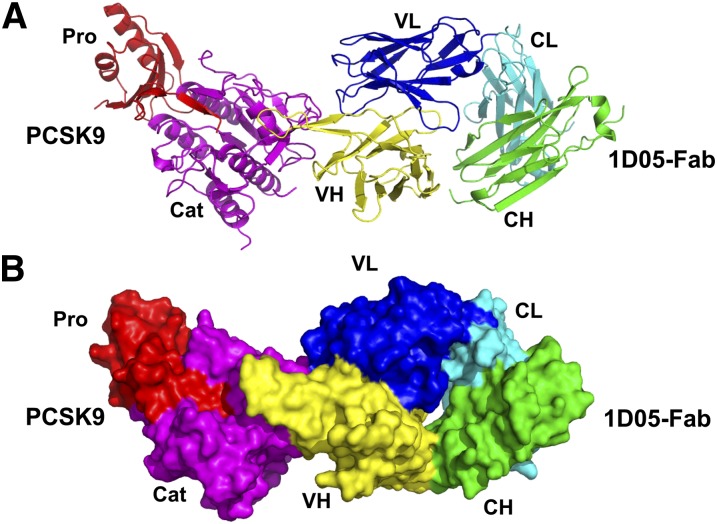
Overall, the complex has an elongated shape with dimensions of ∼130 Å × 50 Å × 40 Å (Fig. 2). 1D05 binds the PCSK9 catalytic domain via both the heavy and light chain variable domains (VH and VL) (Fig. 2 and Fig. 3A–C), although most of the contacts are established by the VH CDR regions. The interface is characterized by the presence of several polar interactions. Ten residues from CDR-H3 (Ile104, Arg106, Tyr107, Met109, Asn110, Val111, Tyr112, Tyr113, Leu114, and Tyr116) and ten residues from CDR-H2 (Trp47, Gln65, Leu55, Gly56, Ile57, Ala58, Asn59, Tyr60, Ala61, and Gln62) contact 18 residues of the PCSK9 catalytic domain: Pro155 (in the P′-helix), Arg194, Glu197, Ala220, Ser221, Arg237, Asp238, Ala239, Ile369, Ser372, Asp374, Cys375, Thr377, Cys378, Phe379, Val380, Ser381, and Gln382. Among these PCSK9 amino acids, Arg194, Glu197, Cys375, and Thr377 also interact with Gln27 and Arg30 from the CDR-L1 region, and with Phe91, Asp92, Gly93, and Asp94 from the CDR-L3 region. In turn, these CDR-L3 residues also interact with PCSK9 amino acids Asp192 and Ser376.
Fig. 2.
Crystal structure of the PCSK9ΔC/1D05 Fab complex. A: Ribbon representation. The PCSK9 catalytic domain (Cat) and prodomain (Pro) are color-coded magenta and red, respectively. 1D05 VH and VL domains are color-coded yellow and blue, respectively; CH and CL domains are color-coded green and cyan, respectively. B: Surface representation (same color codes as in panel A). (Figs. 2 and 3 were prepared using Pymol software;DeLano Scientific).
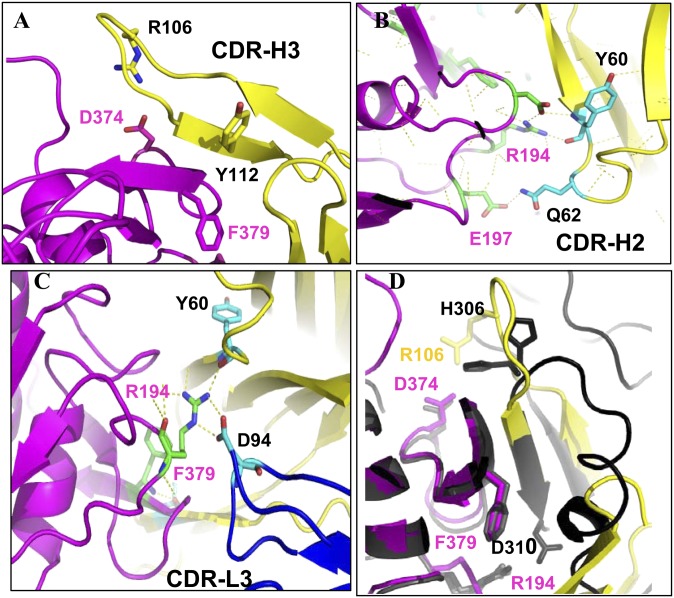
Fig. 3.
PCSK9/1D05-Fab interaction. A–C: PCSK9/1D05 interaction details are shown. Key residues are shown in stick representation. Carbon, oxygen, and nitrogen for the CDR-H3 are colored cyan, red, and blue, respectively. Carbon, oxygen, and nitrogen for PCSK9 are colored green, red, and blue, respectively. Polar interactions are shown as dashed yellow lines. D: Superposition of the 1D05/PCSK9ΔC and EGF(A)/PCSK9ΔC complexes. The PCSK9 catalytic domain (magenta) and VH region of the 1D05 Fab (yellow) are represented by ribbon diagrams. The PCSK9ΔC/EGF(A) complex is represented as ribbon diagram in black.
1D05 Fab CDR-H3 and CDR-H2 structurally mimic the LDLr EGF(A) domain
A prominent feature of the PCSK9ΔC/1D05 complex is the extensive contacts formed between 1D05 CDR-H3 (22 amino acids) and the PCSK9 catalytic domain. Strikingly, superposition of the PCSK9/1D05 and PCSK9/EGF(A) structures (12, 27) reveals that the 1D05 CDR-H3 adopts a hairpin structure that structurally mimics two β-strands of EGF(A) (Val307–Asp310 and Tyr315–Leu318) (Fig. 3D and supplementary Fig. SI). Thus, together with two β-strands of PCSK9 (residues 378–382 and 368–371), CDR-H3 forms an equivalent antiparallel four-stranded β-sheet (Fig. 3D and supplementary Fig. SI). In addition, the tip of the CDR-H2 loop, composed of residues Gly50–Gly56, structurally mimics a helical turn in EGF(A) (residues Gly293–Asp299).
Two residues in PCSK9, Arg194 and Phe379, were shown to be critical for EGF(A) binding, in which Arg194 forms a salt bridge with EGF(A) Asp310, and Phe379 makes several contacts with EGF(A) (27). These PCSK9 amino acids also play an important role in the interaction with 1D05 (Fig. 3D). Arg194 forms a salt bridge with Asp94 in the CDR-L3 region and an H bond with Tyr60 from the CDR-H2 region. Phe379 carbonyl oxygen, on the other hand, forms an H bond with Tyr112 from the CDR-H3 region. Similarly, a salt bridge between Asp374 from PCSK9 and Arg106 from 1D05 CDR-H3 replaces the salt bridge formed at low pH between PCSK9-Asp374 and EGF(A)-His306 (27) (Fig. 3D). Despite these similarities, the interface between PCSK9 and the EGF(A) domain is much smaller than the interface between PCSK9 and 1D05-Fab (total buried surface area is 974 Å2 in the complex with EGF[A] and 1,790 Å2 in the 1D05 complex). Thus, with the exception of Ser153 and Ile154, which are disordered in the PCSK9ΔC/1D05 complex, all the amino acids of PCSK9 that contact EGF(A) also interact with 1D05. In summary, the structural analysis reveals that 1D05 structurally mimics the LDLr EGF(A) domain in its interaction with PCSK9 and, thus, inhibits PCSK9 by sterically preventing it from binding to the receptor.
Generation of a transgenic mouse model with a human-like liproprotein and PCSK9 profile
Most strains of mice have very low levels of LDLc that cannot easily be measured. Therefore, in order to assess in vivo affects of 1D05-IgG2 on LDLc, we generated CETP/LDLr-hemi, a transgenic mouse model which is hemizygous for both human CETP and LDLr alleles. Importantly, the lipid profile of these mice closely resembles that of humans, with relatively high levels of LDLc (80–90 mg/dl) and triglycerides (110–120 mg/dl) and low levels of HDLc (40–50 mg/dl) (supplementary Fig. SII). In comparison, levels of circulating LDLc and HDLc in wild-type C57BL/6J mice are 15–35 mg/dl and 70–120 mg/dl, respectively (data not shown). Moreover, the average plasma PCSK9 level in these mice is 16 nM (∼1,000 ng/ml), which is higher than that in wild-type C57BL/6J mice (supplementary Table SII) and is within the reported range of PCSK9 in human plasma (33–2,988 ng/ml) (28).
1D05-IgG2 reduces circulating LDLc and increases hepatic LDLr levels in rodents
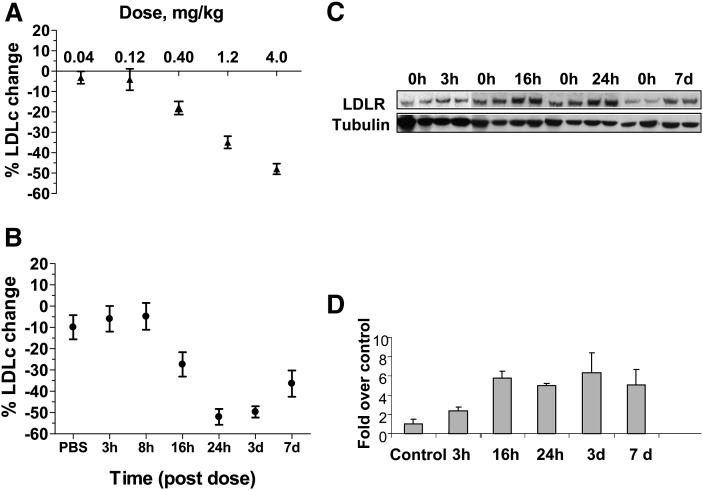
The effect of 1D05-IgG2 on circulating LDLc was studied in the CETP/LDLr-hemi mice. As shown in Fig. 4A, 1D05-IgG2 dose-dependently decreased the level of circulating LDLc. A single 1D05-IgG2 injection (iv, 4 mg/kg body weight) reduced the level of LDLc ∼50% at 48 h postdose. The vehicle PBS by itself, or an irrelevant IgG2, anti-IL13R nonbinding variant 10G5 PL14, had no effect on the level of LDLc (data not shown).
Fig. 4.
1D05-IgG2 reduces LDLc in CETP/LDLr-hemi mice. A: Dose-response study of the effect of 1D05-IgG2. Mice were injected with a single iv dose of 1D05-IgG2 at the indicated doses. Plasma LDLc levels were measured at 48 h postdose. B: The time course of the LDL-lowering effect of 1D05-IgG2 is shown. Mice were injected with a single iv dose of 30 mg/kg 1D05-IgG2. Plasma LDLc was measured at indicated times postinjection. C, D: Time courses of changes in LDLR protein abundance in mice treated with 30 mg/kg 1D05-IgG2 are shown. Liver tissues were collected from mice treated with 1D05-IgG2 and used to prepare tissue lysate. Tubulin was used as loading control. The percentages of changes were calculated relative to those of the PBS vehicle control group. Results are means ± SEM from n = 8–10 replicates.
The time course of the LDLc-lowering effect by 1D05-IgG2 (iv, 30 mg/kg) was also assessed in CETP/LDLr-hemi mice. Decreases in LDLc levels were observed as early as 16 h following the injection, and the maximal effect (∼35%–50%) took place between days 1 and 7 postdose (Fig. 4B). The effect of 1D05 on LDLr protein was evaluated by Western blot analysis of the LDLr protein abundance in the livers from these mice. Consistent with the proposed mechanism of action, 1D05-IgG2 treatment markedly increased the steady-state levels of hepatic LDLr protein (∼fivefold compared with the PBS control group) (Fig. 4C, D). Remarkably, the increase in LDLr protein abundance began as early as 3 h after 1D05-IgG2 injection, which preceded the observed decrease in LDLc levels (16 h postdose) (Fig. 4C, D).
1D05-IgG2 is also a potent LDLc-reducing agent at lower doses: a single 3 mg/kg 1D05-IgG2 iv injection significantly reduced circulating LDLc and total cholesterol by 20%–30% for over 7 days. LDLc and total cholesterol returned to baseline levels approximately 14 days after treatment (supplementary Fig. SIII). It is worth noting that there was a transient decrease in the level of HDLc between day 1 and day 3, which also returned to baseline after 7 days (supplementary Fig. SIIIC). No change was observed in the level of TGs (data not shown).
1D05-IgG2 reduces free PCSK9 in mice
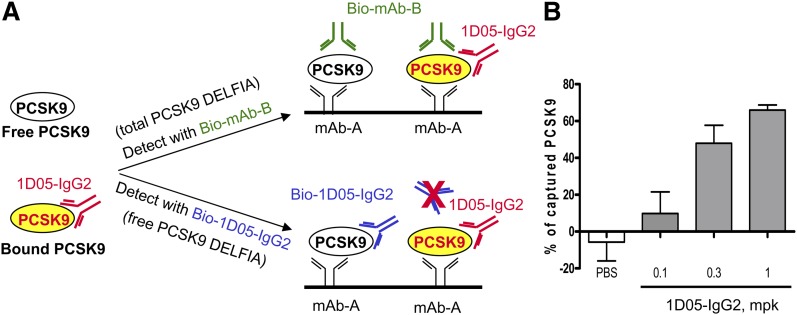
The level of plasma PCSK9 captured by 1D05-IgG2 was monitored by using two PCSK9 DELFIAs that selectively measured 1D05-free PCSK9 and total PCSK9 (which includes free 1D05 and bound PCSK9) in mouse plasma (Fig. 5A, B). Increases in the percentage of captured PCSK9 were observed with increasing doses of 1D05-IgG2 (Fig. 5C). A reduction in LDLc levels of greater than 30% was observed with 0.3 and 1 mg/kg 1D05-IgG2 treatments, which was associated with a fraction of approximately 50% or greater of PCSK9 captured by 1D05-IgG2.
Fig. 5.
Measurement of total and free plasma PCSK9 in CETP/LDLr-hemi mice. A: Schematic illustrates total and free PCSK9 DELFIAs. Plasma PCSK9 was captured by anti-PCSK9 MAb A. Biotinylated anti-PCSK9 MAb B, which does not compete with 1D05-IgG2 for binding to PCSK9, was used for the detection of total PCSK9. Biotinylated 1D05 IgG2 was used to detect free PCSK9. B: Capture of PCSK9 by 1D05-IgG2. Plasma samples were collected at 48 h after 1D05-IgG2 injection. Total and free PCSK9 were measured using two mouse PCSK9 DELFIAs. The percentage of antibody-captured PCSK9 was calculated as described in Experimental Procedures. Results are means ± SEM from n = 8–10 replicates.
1D05-IgG2 reduces circulating LDLc and free PCSK9 in nonhuman primates
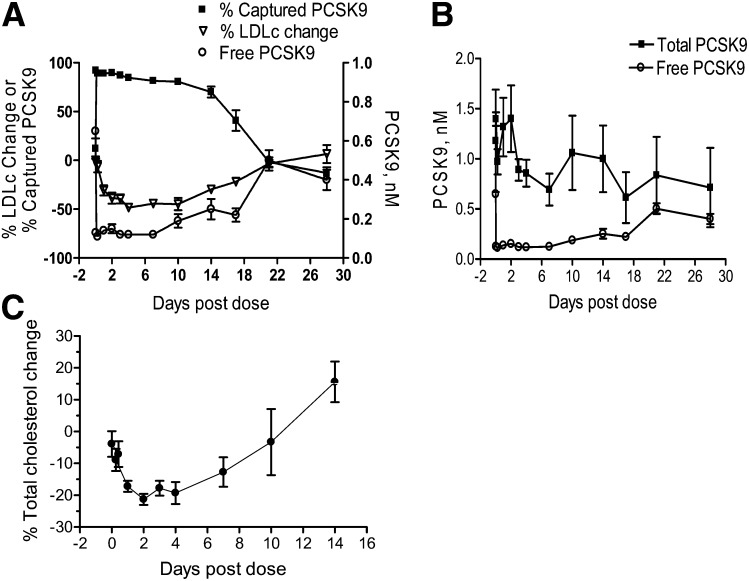
The in vivo efficacy of 1D05 was also evaluated in rhesus monkeys. A single iv injection of 1D05-IgG2 at 3 mg/kg led to a decrease of up to 50% in circulating LDLc. The effect of 1D05-IgG2 was observed within 24 h of dosing and was maintained for over 2 weeks (Fig. 6A). Similar to results from studies with CETP/LDLr-hemi mice, the effects of 1D05-IgG2 on free 1D05 and total plasma PCSK9 levels in rhesus monkeys were determined using two PCSK9 DELFIAs. Administration of 1D05-IgG2 reduced the level of free plasma PCSK9; the level of total plasma PCSK9 did not change significantly (Fig. 6A, B). Maximal reduction in LDLc levels (30%–50%) was associated with greater than 70% capture of plasma PCSK9 by 1D05-IgG2 (or greater than 70% reduction in the level of free PCSK9 in the plasma). Total cholesterol levels were also decreased in these monkeys (Fig. 6C). On day 14, there was an increase in the level of total cholesterol due to an increase in the level of plasma HDLc (data not shown). However, this change was not statistically significant. Circulating serum 1D05-IgG2 concentrations were monitored throughout the study, and terminal t1/2 was determined to be 3.2 days (77 h).
Fig. 6.
1D05-IgG2 reduces LDLc in rhesus monkeys. A: Effects of1D05-IgG2 treatment on levels of plasma LDLc, total cholesterol, and total and free PCSK9. Plasma samples were collected at the indicated times following 1D05-IgG2 iv injection (3 mg/kg). Total and 1D05-IgG2-free PCSK9 levels were measured using two rhesus PCSK9 DELFIAs. The percentage of antibody-captured PCSK9 was calculated as described in Experimental Procedures. B: Change in total and free circulating PCSK9 following 1D05-IgG2 treatment. C: Effect of 1D05-IgG2 on total cholesterol. All results are means ± SEM from n = 3 replicates.
DISCUSSION
Several anti-PCSK9 antibodies have been reported recently. These include monoclonal or polyclonal antibodies directed against the catalytic domain of PCSK9. These antibodies disrupt the PCSK9/LDLr interaction, restore LDLc uptake in cells (15, 16), and reduce circulating LDLc (15). In addition, we have previously reported a human MAb Fab which binds to the C-terminal domain of PCSK9 and partially inhibits its effect on LDLc uptake in hepatocytes through inhibition of PCSK9 internalization (18). We report here that 1D05-IgG2, a high-affinity anti-PCSK9 antibody, neutralizes the PCSK9-mediated inhibitory effect on LDLc uptake by directly interfering with the formation of the PCSK9/LDLr complex and significantly reduces levels of circulating LDLc and total cholesterol in both mice and rhesus monkeys. Functionally, 1D05-IgG2 resembles the MAb reported by Chan et al. (15), as they both bind the catalytic domain of PCSK9, restore LDLc uptake in cells, and reduce circulating LDLc. In addition, these two antibodies have similar terminal t1/2 periods in nonhuman primates: 77 h for 1D05-IgG2 and 61 h for MAb1, as reported by Chan et al (15). While both MAb1 and 1D05-IgG2 sterically inhibit PCSK9 binding to EGF(A), their binding sites on PCSK9 are distinct. Specifically, MAb1 binds to a region on PCSK9 that partially occludes its binding to EGF(A). In comparison, 1D05-IgG2 structurally mimics the LDLr EGF(A) domain in its interaction with PCSK9 and directly inhibits PCSK9 binding to EGF(A) by competitively binding to the same site. Furthermore, while the study by Chan et al. (15) demonstrated the correlation between unbound PCSK9 and circulating LDLc in nonhuman primates, our studies demonstrated that in both rodents and nonhuman primates, the potential therapeutic benefit of the anti-PCSK9 antibody is mediated by the reduction of unbound PCSK9 with a corresponding increase in the percentage of PCSK9 captured by the antibody.
The crystal structure of the PCSK9ΔC/1D05 Fab complex revealed a large interface formed between the PCSK9 catalytic domain and the CDR-H2 and CDR-H3 regions of 1D05. Importantly, the 1D05 binding epitope on PCSK9 completely overlaps that of the LDLr EGF(A) domain and is restricted to the PCSK9 catalytic domain. In contrast, the antibody isolated in the study by Chan et al. (15) interacts with residues from both the PCSK9 prodomain and the catalytic domain; its binding epitope only partially occludes the EGF(A) binding site on the PCSK9 catalytic domain.
Some of the 1D05-binding residues in PCSK9 have been shown to play functional roles in LDLc regulation or to be involved in LDLr EGF(A) binding (27). For instance, Arg237Trp and Asp374Tyr have been reported as loss-of-function and gain-of-function mutations, respectively (29, 30). Consistent with this, 1D05-IgG2 also blocks the interactions between Asp374Tyr and LDLr in hepatocytes.
Changes in both total and free PCSK9 levels following 1D05-IgG2 treatments were monitored in mice and in rhesus monkeys during this study, allowing us to delineate the relationship between changes in circulating LDLc and those in the level of plasma-free PCSK9, and with the percentage of PCSK9 captured by the antibody. As expected, 1D05-IgG2 significantly decreases the level of free plasma PCSK9. This, in turn, results in increases in hepatic LDLr protein abundance and decreases in circulating LDLc. Noticeably, in rhesus monkeys, the onset of decreases in free PCSK9 abundance precede that in LDLc; similar observations were reported by Chan et al. (15) for the effects of MAb1 in cynomolgus monkeys. Overall, the total plasma PCSK9 level (1D05-bound and free PCSK9) did not change significantly, although there was a transient increase (for ∼2 days) following 1D05 treatment in rhesus monkeys. The mechanism of this increase is currently under investigation. It may be related to a decrease in the clearance rate of 1D05-bound PCSK9 or to a compensatory change of PCSK9 synthesis following 1D05-IgG2 exposure.
In this study, the in vivo efficacy of 1D05-IgG2 was assessed in CETP/LDLr-hemi transgenic mice and in rhesus monkeys. In general, wild-type C57BL/6J mice are not ideal for studying cholesterol-reducing agents due to their low circulating LDLc levels. By crossing human CETP transgenic mice with LDLr null mice, we generated a mouse model, CETP/LDLr-hemi, with an overall lipid profile (LDL, HDL, total cholesterol, and TG) closely mimicking that of young healthy humans. Additionally, plasma PCSK9 levels in these mice are also similar to those in humans. Similar mouse models have been used previously in studies of hepatic cholesterol metabolism (31, 32). We demonstrate here that in these mice and in rhesus monkeys, a single low-dose injection of 1D05-IgG2 led to significant reduction in plasma-free PCSK9 levels in parallel with a long-lasting reduction in circulating LDLc.
An anti-PCSK9 MAb is expected to provide therapeutic benefit for patients with hypercholesterolemia. Among patients treated with statins, approximately 2% do not respond. In addition, many statin responders require additional therapy as statin treatment alone does not achieve sufficient LDL reduction. A recent study has also shown that statin treatment promotes a significant increase (∼35%) in plasma PCSK9 levels, potentially attenuating its ability to reduce LDLc (35). Therefore, a PCSK9 inhibitor, either used alone or in combination with a statin, may represent the next major breakthrough for hypercholesterolemia therapy.
Supplementary Material
Acknowledgments
We thank Vijayalakshmi Agnani for production of full-length human PCSK9 and Neil Geoghagen for providing technical support.
Footnotes
Abbreviations:
- AF
- Alexa Fluor
- CETP
- cholesteryl ester transfer protein
- CHD
- coronary heart disease
- DELFIA
- dissociation-enhanced lanthanide fluorescence immunoassays
- HDLc
- high-density lipoprotein cholesterol
- LDLc
- low-density lipoprotein cholesterol
- LDLr
- low-density lipoprotein receptor
- MAb
- monoclonal antibody
- PEG
- polyethylene glycol 6000
- TCEP
- Tris(2-carboxyethyl)phosphine
- TG
- triglycerides
- TR-FRET
- time-resolved fluorescence resonance energy transfer
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and three figures.
REFERENCES
- 1.Horton J. D., Cohen J. C., Hobbs H. H. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J. C., Boerwinkle E., Mosley T. H., Jr., Hobbs H. H. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165. [DOI] [PubMed] [Google Scholar]
- 5.Kotowski I. K., Pertsemlidis A., Luke A., Cooper R. S., Vega G. L., Cohen J. C., Hobbs H. H. 2006. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 78: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., et al. 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. 2008. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. U S A. 105: 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher T. S., Lo Surdo P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., et al. 2007. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 282: 20502–20512. [DOI] [PubMed] [Google Scholar]
- 9.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. 2006. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 116: 2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grefhorst A., McNutt M. C., Lagace T. A., Horton J. D. 2008. Plasma PCSK9 preferentially reduces liver LDL receptors in mice. J. Lipid Res. 49: 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt R. J., Beyer T. P., Bensch W. R., Qian Y. W., Lin A., Kowala M., Alborn W. E., Konrad R. J., Cao G. 2008. Secreted proprotein convertase subtilisin/kexin type 9 reduces both hepatic and extrahepatic low-density lipoprotein receptors in vivo. Biochem. Biophys. Res. Commun. 370: 634–640. [DOI] [PubMed] [Google Scholar]
- 12.Bottomley M. J., Cirillo A., Orsatti L., Ruggeri L., Fisher T. S., Santoro J. C., Cummings R. T., Cubbon R. M., Lo Surdo P., Calzetta A., et al. 2009. Structural and biochemical characterization of the wild type PCSK9-EGF(AB) complex and natural familial hypercholesterolemia mutants. J. Biol. Chem. 284: 1313–1323. [DOI] [PubMed] [Google Scholar]
- 13.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. 2009. Antagonism of secreted PCSK9 increases low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 284: 10561–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan L., Pang L., Zhang R., Murgolo N. J., Lan H., Hedrick J. A. 2008. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem. Biophys. Res. Commun. 375: 69–73. [DOI] [PubMed] [Google Scholar]
- 15.Chan J. C., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., et al. 2009. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc. Natl. Acad. Sci. U S A. 106: 9820–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duff C. J., Scott M. J., Kirby I. T., Hutchinson S. E., Martin S. L., Hooper N. M. 2009. Antibody-mediated disruption of the interaction between PCSK9 and the low-density lipoprotein receptor. Biochem. J. 419: 577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank-Kamenetsky M., Grefhorst A., Anderson N. N., Racie T. S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y., et al. 2008. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. U S A. 105: 11915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni Y. G., Condra J., Orsatti L., Shen X., Di Marco S., Pandit S., Bottomley M. J., Ruggeri L., Cummings R. T., Cubbon R. M., et al. 2010. A proprotein convertase subtilisin-like/kexin type 9 (PCSK9) C-terminal domain antibody antigen-binding fragment inhibits PCSK9 internalization and restores low density lipoprotein uptake. J. Biol. Chem. 285: 12882–12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothe C., Urlinger S., Lohning C., Prassler J., Stark Y., Jager U., Hubner B., Bardroff M., Pradel I., Boss M., et al. 2008. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J. Mol. Biol. 376: 1182–1200. [DOI] [PubMed] [Google Scholar]
- 20.An Z., Forrest G., Moore R., Cukan M., Haytko P., Huang L., Vitelli S., Zhao J. Z., Lu P., Hua J., et al. 2009. IgG2m4, an engineered antibody isotype with reduced Fc function. MAbs. 1: 572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery D. L., Wang Y. J., Hrin R., Luftig M., Su B., Miller M. D., Wang F., Haytko P., Huang L., Vitelli S., et al. 2009. Affinity maturation and characterization of a human monoclonal antibody against HIV-1 gp41. MAbs. 1: 462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collaborative Computational project, N. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- 23.Arnold K., Bordoli L., Kopp J., Schwede T. 2006. The SWISSMODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 57: 1445–1450. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291. [Google Scholar]
- 26.Pandit S., Wisniewski D., Santoro J. C., Ha S., Ramakrishnan V., Cubbon R. M., Cummings R. T., Wright S. D., Sparrow C. P., Sitlani A., et al. 2008. Functional analysis of sites within PCSK9 responsible for hypercholesterolemia. J. Lipid Res. 49: 1333–1343. [DOI] [PubMed] [Google Scholar]
- 27.Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., Deisenhofer J. 2008. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. U S A. 105: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakoski S. G., Lagace T. A., Cohen J. C., Horton J. D., Hobbs H. H. 2009. Genetic and metabolic determinants of plasma PCSK9 levels. J. Clin. Endocrinol. Metab. 94: 2537–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timms K. M., Wagner S., Samuels M. E., Forbey K., Goldfine H., Jammulapati S., Skolnick M. H., Hopkins P. N., Hunt S. C., Shattuck D. M. 2004. A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum. Genet. 114: 349–353. [DOI] [PubMed] [Google Scholar]
- 30.Berge K. E., Ose L., Leren T. P. 2006. Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arterioscler. Thromb. Vasc. Biol. 26: 1094–1100. [DOI] [PubMed] [Google Scholar]
- 31.Harada L. M., Carrilho A. J., Oliveira H. C., Nakandakare E. R., Quintao E. C. 2006. Regulation of hepatic cholesterol metabolism in CETP/LDLr mice by cholesterol feeding and by drugs (cholestyramine and lovastatin) that lower plasma cholesterol. Clin. Exp. Pharmacol. Physiol. 33: 1209–1215. [DOI] [PubMed] [Google Scholar]
- 32.Lie J., de Crom R., van Gent T., van Haperen R., Scheek L., Sadeghi-Niaraki F., van Tol A. 2004. Elevation of plasma phospholipid transfer protein increases the risk of atherosclerosis despite lower apolipoprotein B-containing lipoproteins. J. Lipid Res. 45: 805–811. [DOI] [PubMed] [Google Scholar]
- 33.Welder G., Zineh I., Pacanowski M. A., Troutt J. S., Cao G., Konrad R. J. 2010. High-dose atorvastatin causes a rapid sustained increase in human serum PCSK9 and disrupts its correlation with LDL cholesterol. J. Lipid Res. 51: 2714–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. U S A. 102: 5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubuc G., Tremblay M., Pare G., Jacques H., Hamelin J., Benjannet S., Boulet L., Genest J., Bernier L., Seidah N. G., et al. 2010. A new method for measurement of total plasma PCSK9: clinical applications. J. Lipid Res. 51: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.