Abstract
Background:
MammaPrint was developed as a diagnostic tool to predict risk of breast cancer metastasis using the expression of 70 genes. To better understand the tumor biology assessed by MammaPrint, we interpreted the biological functions of the 70-genes and showed how the genes reflect the six hallmarks of cancer as defined by Hanahan and Weinberg.
Results:
We used a bottom-up system biology approach to elucidate how the cellular processes reflected by the 70-genes work together to regulate tumor activities and progression. The biological functions of the genes were analyzed using literature research and several bioinformatics tools. Protein-protein interaction network analyses indicated that the 70-genes form highly interconnected networks and that their expression levels are regulated by key tumorigenesis related genes such as TP53, RB1, MYC, JUN and CDKN2A. The biological functions of the genes could be associated with the essential steps necessary for tumor progression and metastasis, and cover the six well-defined hallmarks of cancer, reflecting the acquired malignant characteristics of a cancer cell along with tumor progression and metastasis-related biological activities.
Conclusion:
Genes in the MammaPrint gene signature comprehensively measure the six hallmarks of cancer-related biology. This finding establishes a link between a molecular signature and the underlying molecular mechanisms of tumor cell progression and metastasis.
Keywords: bioinformatics, breast cancer, gene signature, functional annotation, tumor biology
Background
The MammaPrint assay was developed as a prognostic tool to predict the recurrence risk of breast cancer.1–3 It has been validated in almost 1,600 patients (reviewed in4,5). The 70 genes that make up the MammaPrint signature were selected from genome-wide expression data using a data-driven approach. This means that the genes were selected in an unbiased fashion; there were no predefined assumptions as to whether certain genes were more likely to be involved in the risk of development of distant metastases in patients with early stage breast cancer. This resulted in a set of 70 genes that was able to predict the risk of recurrence with a high sensitivity.
Functional annotation of the human genome has been greatly facilitated by the availability of new functional genetic approaches to study genes.6 To date, most of the 70 genes in the MammaPrint profile have well-described biological functions. For the remaining genes, we can identify tentative functions and can only speculate about their respective roles in tumor progression and metastasis. It is reasonable to assume that MammaPrint genes whose function is not currently known will, nevertheless, be shown to have important roles in breast cancer biology. One such example is the gene MTDH, which was recently shown to be a key gene in the development of metastasis and to contribute to chemotherapy resistance. The function of this gene in breast cancer was unknown at the time of MammaPrint development.7 As such we anticipate that the tumorigenic role of additional MammaPrint genes of unknown function will be resolved in the coming years. Indeed, we have recently found that one such gene of unknown function in the 70 gene profile, TSPYL5, which is part of a small breast cancer specific amplicon,7 is a key negative regulator of the p53 tumor suppressor protein (R.B. unpublished data).
The fact that gene expression of the primary tumor can predict whether the tumor may metastasize indicates that the metastatic potential is likely hardwired into the tumor cells at a relatively early stage during tumorigenesis and is preserved throughout tumor formation and metastasis development.8–10 This process can be captured in the six steps that are referred to as the “hallmarks of cancer”, which reflect the acquired characteristics of a cancer cell. How and in what order tumor cells acquire these characteristics can differ, but ultimately all cancer cells need these characteristics to successfully metastasize and proliferate.11
To address if the MammaPrint genes indeed capture these six steps, we examined the biological functions of each of the 70 genes. Traditionally, annotation methods focus only on statistically enriched functional categories in gene signatures. Although these categories can reveal major biological processes, they do not always result in a systematic understanding on how all genes in a molecular signature work together. In this study, we used a bottom-up system biology approach that aims to reveal cellular processes reflected by each single gene in MammaPrint and discover how the 70-genes work together to regulate tumor activities. With this knowledge, we investigated the underlying molecular mechanisms linking their biological function with the molecular biology of breast cancer metastasis.
Results and Discussion
The 70-gene profile covers the six hallmarks of cancer
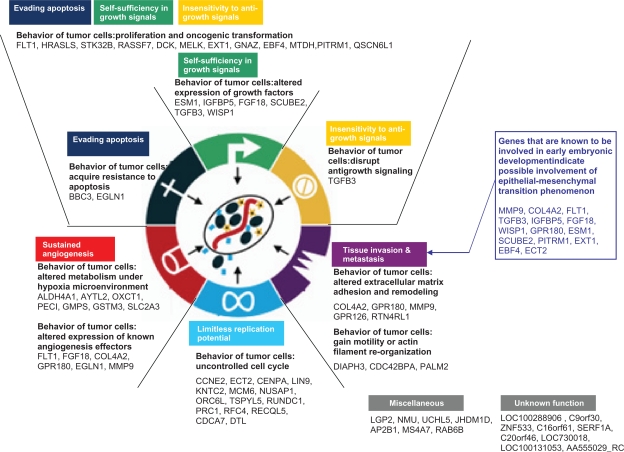
The six hallmarks of cancer describe the acquired characteristics of a cancer cell that collectively dictate malignant growth and thus reflect the potential of a tumor to metastasize.11 These six capabilities are shared by most (and perhaps all) types of human cancer. In breast cancer, the 70 MammaPrint genes are predictive of metastasis development, and we set out to ask whether the biological properties of these genes are correlated with the six hallmarks of cancer (as illustrated in Fig. 1):
Evading apoptosis
Self-sufficiency in growth signals
Insensitivity to anti-growth signals
Limitless replicative potential
Tissue invasion and metastasis
Sustained angiogenesis
Figure 1.
Depicted is how the genes in 70-gene tumor expression profile are involved in the six well-defined hallmarks of cancer, in tumor progression and metastasis related biological processes, as well as epithelial-mesenchymal transition. Adapted from Cell, 100, Hanahan D, Weinberg RA., The Hallmarks of Cancer, 57–70, Copyright (2000) with permission from Elsevier.
The hallmark avoiding apoptosis confers resistance towards programmed cell death. The major converging point of diverse apoptotic signals that a tumor cell may receive are the mitochondria. Mitochondrial death signals are governed by the BCL2 family of proteins that release cytochrome C and, in turn, activates caspases.12 Tumor cells can resist apoptosis amongst others by altering expression level of BCL2 or caspases associated proteins. This mechanism is represented by two of the MammaPrint genes (BBC3 and EGLN1; Table 1).
Table 1.
Biological function of MammaPrint genes and cancer hallmarks. MammaPrint genes are involved in all tumor progression and metastasis-related biological processes, and cover the six well-defined hallmarks of cancer.
| Hallmarks of cancer | Characteristic behavior of tumor cells | Gene name | Gene description |
|---|---|---|---|
| Evading apoptosis | Acquire resistance to apoptosis |
BBC3 EGLN1 |
BCL2 binding component 3 egl nine homolog 1 |
| Insensitivity to anti-growth signals | Disrupt antigrowth signaling | TGFB3 | transforming growth factor, beta 3 |
| Self-sufficiency in growth signals | Altered expression of growth factors | ESM1 | endothelial cell-specific molecule 1 |
| IGFBP5 | insulin-like growth factor binding protein 5 | ||
| FGF18 | fibroblast growth factor 18 | ||
| SCUBE2 | signal peptide, CUB domain, EGF-like 2 | ||
| TGFB3 | transforming growth factor, beta 3 | ||
| WISP1 | WNT1 inducible signaling pathway protein 1, transcript variant 1 | ||
| Evading apoptosis | Proliferation and oncogenic transformation | FLT1 | fms-related tyrosine kinase 1 |
| HRASLS | HRAS-like suppressor | ||
| STK32B | serine/threonine kinase 32B | ||
| Insensitivity to anti-growth signals | RASSF7 | Ras association (RalGDS/AF-6) domain family 7 | |
| DCK | deoxycytidine kinase | ||
| MELK | maternal embryonic leucine zipper kinase | ||
| EXT1 | exostoses 1 | ||
| Self-sufficiency in growth signals | GNAZ | guanine nucleotide binding protein, alpha z polypeptide | |
| EBF4 | early B-cell factor 4 | ||
| MTDH | metadherin | ||
| PITRM1 | pitrilysin metallopeptidase 1 | ||
| QSCN6L1 | quiescin Q6-like 1 | ||
| Limitless replicative potential | Uncontrolled cell cycle | CCNE2 | cyclin E2, transcript variant 1 |
| ECT2 | epithelial cell transforming sequence 2 oncogene | ||
| CENPA | centromere protein A, 17 kDa | ||
| LIN9 | lin-9 homolog | ||
| KNTC2 | kinetochore associated 2 | ||
| MCM6 | MCM6 minichromosome maintenance deficient 6 | ||
| NUSAP1 | nucleolar and spindle associated protein 1 transcript variant 2 | ||
| ORC6L | origin recognition complex, subunit 6 like | ||
| TSPYL5 | TSPY-like 5 | ||
| RUNDC1 | RUN domain containing 1 | ||
| PRC1 | protein regulator of cytokinesis 1, transcript variant 2 | ||
| RFC4 | replication factor C 4, 37 kDa, transcript variant 2 | ||
| RECQL5 | RecQ protein-like 5 | ||
| CDCA7 | cell division cycle associated 7, transcript variant 1 | ||
| DTL | denticleless homolog | ||
| Tissue invasion and metastasis | Altered extracellular matrix adhesion and remodelling | COL4A2 | collagen, type IV, alpha 2 |
| GPR180 | G protein-coupled receptor 180 | ||
| MMP9 | matrix metallopeptidase 9 | ||
| GPR126 | G protein-coupled receptor 126, transcript variant b2 | ||
| RTN4RL1 | reticulon 4 receptor-like 1 | ||
| Gain motility or actin filament re-organization | DIAPH3 | diaphanous homolog 3 | |
| CDC42BPA | CDC42 binding protein kinase alpha, transcript variant B | ||
| PALM2 | paralemmin 2 | ||
| Sustained angiogenesis | Altered metabolism under hypoxia microenvironment | ALDH4A1 | aldehyde dehydrogenase 4 family, member A1 |
| AYTL2 | acyltransferase like 2 | ||
| OXCT1 | 3-oxoacid CoA transferase 1, nuclear gene encoding mitochondrial protein | ||
| PECI | peroxisomal D3,D2-enoyl-CoA isomerase | ||
| GMPS | guanine monphosphate synthetase | ||
| GSTM3 | glutathione S-transferase M3 | ||
| SLC2A3 | solute carrier family 2, member 3 | ||
| Altered expression of known angiogenesis effectors | FLT1 | fms-related tyrosine kinase 1 | |
| FGF18 | fibroblast growth factor 18 | ||
| COL4A2 | collagen, type IV, alpha 2 | ||
| GPR180 | G protein-coupled receptor 180 | ||
| EGLN1 | egl nine homolog 1 | ||
| MMP9 | matrix metallopeptidase 9 | ||
| Unknown function | Unknown function | LOC100288906 | hypothetical protein LOC100288906 |
| C9orf30 | chromosome 9 open reading frame 30 | ||
| ZNF533 | zinc finger protein 533 | ||
| C16orf61 | chromosome 16 open reading frame 61 | ||
| SERF1A | small EDRK-rich factor 1A | ||
| C20orf46 | chromosome 20 open reading frame 46 | ||
| LOC730018 | similar to hCG1980668 | ||
| LOC100131053 | hypothetical LOC100131053 | ||
| AA555029_RC | No significant similarity found | ||
| Miscellaneous (currently no link to hallmarks) | Miscellaneous | LGP2 | likely ortholog of mouse D11lgp2 |
| NMU | neuromedin U | ||
| UCHL5 | ubiquitin carboxyl-terminal hydrolase L5 | ||
| JHDM1D | jumonji C domain containing histone demethylase 1 homolog D | ||
| AP2B1 | adaptor-related protein complex 2, beta 1 subunit, transcript variant 1 | ||
| MS4 A7 | membrane-spanning 4-domains, subfamily A, member 7, transcript variant 3 | ||
| RAB6B | RAB6B, member RAS oncogene family |
The hallmark self-sufficiency in growth signals, refers to tumor cells’ reduced dependence on exogenous growth stimulations by generation of their own growth signals. This can be achieved by manipulating the level of growth factors and their receptors or by mutation/altered expression of signal transduction molecules. This characteristic behavior of tumor cells is captured by six growth factor associated genes in the MammaPrint profile (ESM1, IGFBP5, FGF18, SCUBE2, TGFB3, WISP1; Table 1). They represent the capability of tumor cells to manipulate different signaling pathways, such as the IGF-1 signaling pathway, FGF signaling pathway, cell cycle G1/S checkpoint regulation and Wnt/β-catenin signaling pathway. However, it should be emphasized that when various growth factors produced by tumor cells co-exist, the effect which results from their interplay within the microenvironment13 remains to be elucidated.
Equally important, is the hallmark labeled insensitivity to anti-growth signals. This defines the capability of tumor cells to disrupt responses to antiproliferative signaling. A well-studied example is the disruption of growth inhibiting effect of TGFß during tumorigenesis.13 This pathway is represented by the TGFB3 gene in the MammaPrint profile.
The three hallmarks, evading apoptosis, self-sufficiency in growth signals and insensitivity to anti-growth signals, all lead to growth and proliferation of tumor cells, regardless of the types of exogenous signals received from the tumor microenvironment.11 Although the biological processes by which normal cells acquire these three capabilities can be quite diverse, the biological features of proliferation and oncogenic transformation are shared among malignant tumor cells (see Fig. 1). These shared characteristic behaviors are captured by 12 proliferation or oncogenic transformation-related genes (FLT1, HRASLS, STK32B, RASSF7, DCK, MELK, EXT1, GNAZ, EBF4, MTDH, PITRM1, QSCN6L1; Table 1). Because future metastatic dissemination of a primary tumor is directly associated with the aggressiveness of the primary tumor, it is perhaps not surprising that genes associated with these three hallmarks make up the largest part (21 genes) of the MammaPrint profile (BBC3, EGLN1, TGFB3, ESM1, IGFBP5, FGF18, SCUBE2, TGFB3, WISP1, FLT1, HRASLS, STK32B, RASSF7, DCK, MELK, EXT1, GNAZ, EBF4, MTDH, PITRM1, QSCN6L1; Table 1).
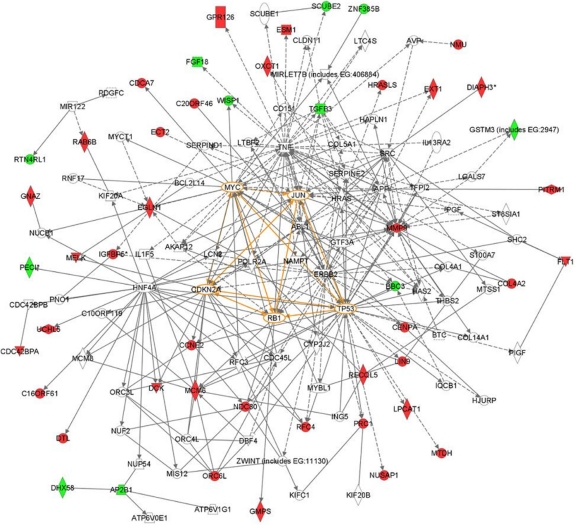
A fourth hallmark of the tumor cell is its limitless replicative potential. Tumor cells bypass two built-in checkpoints that limit the replicative potential of normal cells, the p53 and RB-dependent M1 senescence checkpoint and the telomerase-dependent M2 checkpoint.14 Fifteen MammaPrint genes are cell cycle genes, and can be assigned to this important feature of tumor cells (CCNE2, ECT2, CENPA, LIN9, KNTC2, MCM6, NUSAP1, ORC6L, TSPYL5, RUNDC1, PRC1, RFC4, RECQL5, CDCA7, DTL; Table 1). Interestingly, the protein-protein interaction network analysis also identified TP53 and RB1 to be in the center of this network and confirms that the 70 genes are controlled by key tumorigenesis regulators (vide infra and Fig. 2).
Figure 2.
Protein-protein interaction network analyses indicate that the 70 genes form highly interconnected networks centered on known cancer-related transcription regulators such as TP53, RB1, MYC, JUN and CDKN2A (highlighted in orange). This network indicated that the expression levels of 70 genes are likely regulated by these key tumorigenesis related transcription regulators. The data was analyzed using Ingenuity Pathways Analysis (www.ingenuity.com).
The hallmark of tissue invasion and metastasis is a fifth critical step that involves local invasion of the tumor cells into surrounding tissue, escape from the primary tumor site, entry of metastatic tumor cells into the vasculature (intravasation), transportation and survival into the circulation, and arrest and exit of metastatic tumor cells from the vasculature into distant organs (extravasation).15 During the process of local invasion, tumor cells lose adhesion proteins, remodel extracellular matrix, gain motility by changes in their cytoskeleton and invade adjacent tissue.11 Five of the MammaPrint genes encode adhesion molecules, extracellular matrix constituents and proteins involved in the breakdown of extracellular matrix (COL4A2, GPR180, MMP9, GPR126 and RTN4RL1; Table 1). In addition to changes in cell adhesion and the extracellular matrix, tumor cells also have to acquire enhanced motility to successfully invade the surrounding tissue. A primary mechanism that regulates cell motility is the reorganization of the actin cytoskeleton.16 Actin-binding proteins regulate the dynamic assembly and disassembly of actin filaments that generate a protrusive force at the leading edge of the cell and a contractive force at the trailing edge of cell. These actin dynamics drive cellular motility. This specific malignant characteristic, enhanced motility, is addressed by three genes of the MammaPrint prognostic profile that relate to motility or actin filament organization (DIAPH3, CDC42BPA, PALM2; Table 1). In addition, four transformationrelated genes and growth factors are known to be critical for inducing local invasion,15 and are also represented in the MammaPrint 70 gene expression profile (TGFB3, IGFBP5, FGF18 and WISP1; Table 1).
The last hallmark of cancer is sustained angiogenesis.17 The survival and growth of tumor cells depends on an adequate supply of oxygen and nutrients through blood vessels and by diffusion through the surrounding tissue. Existing vasculature and passive diffusion are sufficient for oxygen supply to tumors of a limited size. However, aggressive solid tumors often grow to a size that can no longer be sustained by the existing tissue vasculature. Tumor cells enhance their glycolytic capability to ensure an energetically efficient metabolism and proliferation rate under hypoxic conditions. These altered metabolic pathways are preserved by the tumor even when the oxygen concentration is sufficient.18 Seven genes implicated in this altered metabolism are represented in the MammaPrint profile (ALDH4 A1, AYTL2, OXCT1, PECI, GMPS, GSTM3 and SLC2 A3; Table 1). Tumor cells induce angiogenesis and vascular remodeling by regulating adhesion proteins and extracellular proteins through binding of hypoxia-inducible factor (HIF) to hypoxia-response elements (HRE).17 At least six genes in the MammaPrint profile are currently known to be direct effectors of angiogenesis and the regulation of vascular remodeling (FLT1, FGF18, COL4 A2, GPR180, EGLN1 and MMP9; Table 1). Together, these genes assess the capability of tumor cells to stimulate the growth of new blood vessels and are likely to contain valuable information about the aggressiveness and malignant potential of a primary tumor.
It should be emphasized that the biological processes associated with the six hallmarks such as proliferation, cell-cell adhesion, angiogenesis and invasion are intrinsically linked. That is, a gene known to play a dominant and critical role in one hallmark might also indirectly be involved in other hallmarks. To better understand interactions between the 70 MammaPrint genes and their transcription regulation, we performed protein-protein interaction network analyses. The networks showed that the 70 genes are highly interconnected and center around known cancer-related transcription regulators such as TP53, RB1, MYC, JUN and CDKN2 A (Fig. 2). This result indicates that the activities of the 70 genes are regulated by these key tumorigenesis-related transcription regulators.
To summarize, MammaPrint has been developed using a data-driven approach and results in a gene profile that has comprehensive coverage of the six hallmarks of cancer, as well as tumor progression and metastasis related biological processes (Table 1, Fig. 1). In addition, protein-protein interaction network analyses presented here, indicate that the 70-genes form highly interconnected networks and that their expression levels are regulated by key tumorigenesis related genes such as TP53, RB1, MYC, JUN and CDKN2 A.
The biological model of acquisition of metastatic competence through epithelial-mesenchymal transition and the 70-Gene Profile
In the previous section, we have shown that malignancy and metastatic competence of tumor cells at the primary tumor site are measured by the expression level of genes in the 70-gene MammaPrint profile. However, this has not provided an answer as to how tumor cells at the primary tumor site initially acquire their metastatic capability. A biological model that is increasingly gaining acceptance is that tumor cells at the primary site might acquire their metastatic capacity through a process similar to epithelelial-mesenchymal transition (EMT): a key epigenetic program that cells undergo during early embryonic development.8 During EMT, epithelial cells lose cell adhesion molecules, reorganize their cytoskeleton, gain increased motility and migrate from an epithelial sheet-like structure to an irregular structure of mesenchyme.19 This change in cellular phenotype is similar to the process that tumor cells undergo to initiate metastasis. Evidence suggests that tumor cells might initiate EMT by turning on or off some of the same transcription factors that are used in early embryonic development.20 These transcription factors regulate the expression of genes that allow tumor cells to lose adhesion, remodel the surrounding extracellular matrix, acquire enhanced motility to enable cellular migration, resist apoptotic signals, and adapt to an unfamiliar microenvironment at the distant site. The biological model based on the assumption that EMT processes are involved in breast cancer metastasis is consistent with the biological functions of the genes in the MammaPrint 70-gene profile identified here (Table 1). A substantial number (ie, 14 genes) of the 70 gene profile encode for proteins that are known to play an role in early embryonic development and are likely involved in EMT (MMP9, COL4 A2, FLT1, TGFB3, IGFBP5, FGF18, WISP1, GPR180, ESM1, SCUBE2, PITRM1, EXT1, EBF4, ECT2; Fig. 1). Within the EMT-associated MammaPrint genes, one gene (EBF4) encodes development-related transcription factors and three genes (TGFB3, FGF18, WISP1) represent the well characterized EMT-mediating TGF-β, FGF and Wnt family proteins.9 It should be noted that in addition to the described 14 genes, other genes within the 70-gene profile might also be involved in early embryonic development. However, their role in early embryonic development has not yet been studied extensively. As outlined above, these 14 EMT-associated MammaPrint genes show a significant overlap with genes that confer the capability of tissue invasion, extracellular matrix remodeling and enhanced motility of tumor cells. These are among the defining characteristics of the EMT phenomenon.9
Conclusions
The MammaPrint 70-gene tumor expression profile was developed using an unbiased data-driven approach without making assumptions that certain genes are more likely to be involved in tumor metastasis. In this study, we have demonstrated how this approach results in a tumor expression profile that contains many genes actually involved in the six well-defined hallmarks of cancer, in tumor progression and metastasis-related biological processes, as well as in epithelial-mesenchymal transition.
It is equally interesting to investigate which genes are not among the 70 genes in the prognostic signature. Most notably, neither ESR1 (encoding estrogen receptor alpha) nor HER2, which is often amplified in breast cancer, are present in the 70 gene profile, whereas both genes are well-established reporters of poor prognosis and are part of other prognostic breast cancer gene signatures. The absence of HER2 is probably best explained by the fact that this gene is not over-expressed in some 80% of breast tumors, and hence is not informative for prognosis in the majority of breast tumors. ESR1 is expressed in the majority of breast tumors, but its presence at the mRNA level is not necessarily informative for functionality of the receptor. The fact that 12 out of the 70 genes in the MammaPrint profile are also part of a 550 gene classifier that reports ER status,2,21 indicates that the 70 genes indirectly report ER status by measuring downstream targets of ER rather than the levels of ER itself.
This study shows how the 70 genes in the MammaPrint gene signature comprehensively measure the six hallmarks of cancer-related biology. This finding establishes a link between a molecular signature and the underlying molecular mechanism of tumor cell progression and metastasis.
Methods
To correlate the probes of the 70 MammaPrint genes2 with the latest NCBI Human genomic plus transcript database (database updated through 18 March 2009), a BLAST search was performed using the corresponding probes at the NCBI BLAST website (http://blast.ncbi.nlm.nih.gov/Blast.cgi22).
After retrieving the genes, they were translated into protein sequences. The transcribed proteins perform functions through conserved functional domains, related small functional site motifs and preserved 3D-structural features. These features were used to investigate the biological function of each of the 70 genes that make up the MammaPrint breast cancer gene expression profile:
- For the annotation of functional domain architecture of individual genes, genes were translated into protein sequences, and the workflow for functional annotation of proteins implemented on the SMART web server25 was followed:
- To identify subcellular localization of a protein, the presence of sorting signals and/or cleavage sites was predicted by the bioinformatics tool SignalP.24
- To identify transmembrane regions of a protein, the TMHMM25 algorithm was used. Regions of the protein separated by transmembrane regions were analyzed separately.
- To analyze the segments that are not covered by highly conserved functional domains and low complexity regions, homologies to other proteins were retrieved with BLAST search.28
For the interpretation of the biological functions of the individual genes within the cellular context (signal transduction pathway, metabolism pathway and protein-protein interaction), Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) was used and a detailed literature review was performed.
Acknowledgments
The study was supported by a grant from TIPharma project T3–108 The authors would like to thank Dr R Bender and Dr B Chan for critically reading the manuscript.
Footnotes
Authors’ contributions
ST carried the functional analysis and drafted the manuscript. PR participated in its design and helped to draft the manuscript. RB and LvV participated in the design of the study and helped to draft the manuscript. FdS conceived of the study, participated in its design and helped to draft the manuscript. AMG conceived of the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors are employed by Agendia BV. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 2.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Glas AM, Floore A, Delahaye LJ, Witteveen AT, Pover RC, Bakx N, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Snoo F, Bender R, Glas A, Rutgers E. Gene expression profiling: decoding breast cancer. Surg Oncol. 2009;18:366–78. doi: 10.1016/j.suronc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Glas AM, Delahaye LJ, Krijgsman O. MammaPrint® Translating Research into a Diagnostic Test. In: Jorgensen JT, Winther H, editors. Molecular Diagnostics: The Key Driver in Personalized Cancer Medicine. Pan Stanford Publishing; 2010. [Google Scholar]
- 6.Brummelkamp TR, Bernards R. New tools for functional mammalian cancer genetics. Nat Rev Cancer. 2003;3:781–89. doi: 10.1038/nrc1191. [DOI] [PubMed] [Google Scholar]
- 7.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 9.Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291–7. doi: 10.1101/sqb.2005.70.033. [DOI] [PubMed] [Google Scholar]
- 10.van’t Veer LJ, Weigelt B. Road map to metastasis. Nat Med. 2003;9:999–1000. doi: 10.1038/nm0803-999b. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 12.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–12. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 13.Fynan TM, Reiss M. Resistance to inhibition of cell growth by transforming growth factor-beta and its role in oncogenesis. Crit Rev Oncog. 1993;4:493–540. [PubMed] [Google Scholar]
- 14.Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock. Cell. 2000;102:407–10. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 15.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 16.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–87. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 17.Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281–90. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 18.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–81. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinberg RA. Mechanisms of malignant progression. Carcinogenesis. 2008;29:1092–5. doi: 10.1093/carcin/bgn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van’t Veer LJ, Dai H, van d V, He YD, Hart AA, Bernards R, et al. Expression profiling predicts outcome in breast cancer. Breast Cancer Res. 2003;5:57–58. doi: 10.1186/bcr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, et al. SMART 4.0:towards genomic data integration. Nucleic Acids Res. 2004;32:D142–4. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen H, Engelbrecht J, Brunak S, von HG. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Krogh A, Larsson B, von HG, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 26.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press; 1998. [Google Scholar]
- 27.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]